ABSTRACT
Cutaneous squamous cell carcinoma (CSCC) is one of the most malignant tumors worldwide. It has been validated that matrix metallopeptidase 1 (MMP1) expression was obviously up-regulated in CSCC tissues. However, its specific role in CSCC is still unclear. RT-qPCR analysis and western blot assays were used to measure the mRNA and protein expressions, respectively. MTT and colony formation assays were conducted to assess proliferative ability. Transwell assays were adopted to evaluate migratory and invasive abilities. Flow cytometry and caspase-3/8/9 activity assays were carried out to evaluate cell apoptosis. Relevant mechanism experiments were finally performed to delineate molecular relationship among genes. We found that the expression of MMP1 was up-regulated in CSCC cells, and knockdown of MMP1 suppressed cell proliferation and invasion in CSCC. Subsequently, miR-361-5p was validated to target MMP1. Moreover, miR-361-5p was proved to be sponged by nuclear paraspeckle assembly transcript 1 (NEAT1) in CSCC. We further demonstrated that NEAT1 could activate Wnt pathway to affect cell proliferation and invasion. Finally, miR-361-5p inhibition rescued the suppressing effects of NEAT1 depletion on cell proliferation, invasion as well as Wnt pathway in CSCC. In summary, MMP1 regulated by NEAT1/miR-361-5p axis facilitated CSCC malignant behaviors via Wnt pathway activation.
Introduction
Cutaneous squamous cell carcinoma (CSCC) is one of the most common skin malignant tumors and may lead to approximately 20% of total skin tumor-associated death globally.Citation1,Citation2 The morbidity of CSCC is rising in the last decade.Citation3 Also, one of the features of CSCC is its metastasis to other organs. The long-term prognosis for CSCC patients in highly metastatic cases is very poor, with a disease-specific survival at 1 year of 44–56%.Citation4 Hence, to find out novel therapeutic targets for improving the poor clinical outcome of CSCC is critical.
Messenger RNAs (mRNAs) are capable of encoding proteins which play a vital role in biological activities. In recent years, it has been well established that the mRNA stability and expression level can be regulated by long non-protein coding RNAs (lncRNAs).Citation5 MRNAs contain partial binding sites, named micro response elements (MRE) that could be recognized and targeted by microRNAs (miRNAs). This target relationship contributes to the significant degradation of mRNA.Citation6 Matrix metalloproteinases (MMPs) are a type of extracellular matrix remodeling proteinase. MMPs have been revealed to play essential roles in the pathology of cancers.Citation7 Matrix metalloproteinase-1 (MMP1) was found to promote early perineural invasion of pancreatic cancer cells.Citation8 Recently, it has been reported that MMP1 has been up-regulated in tissues of patients with CSCC.Citation9 However, whether the differentially expressed MMP1-induced impacts on the development of CSCC have never been elucidated.
With more than 200 nucleotides (nt) in length, lncRNAs represent a group of RNAs with very limited protein-coding ability or without the potential to encode proteins. Nevertheless, lncRNAs can exert regulatory effects on gene expression, thus affecting carcinogenesis process. Similar to mRNA, lncRNAs harbor partial MRE and can competitively bind to specific miRNA. This competitive endogenous RNA (ceRNA) regulatory mechanism creates connections among lncRNA, miRNA and mRNA and further affects disease pathogenesis.Citation10 MiR-361-5p is a relatively well-studied tumor-suppressing gene which is usually lowly expressed in malignancies, such as in prostate cancerCitation11 and lung cancer.Citation12 Furthermore, the overexpression of miR-361-5p will lead to better prognosis for patients suffering from breast cancer.Citation13 Previous researches have uncovered that miR-361-5p suppresses glycolytic metabolism, proliferation and invasion of breast cancer via targeting its downstream FGFR1 and MMP1.Citation14 NEAT1 is an identified carcinogenic gene in various cancers, such as in esophageal cancer and hepatocellular carcinoma.Citation15 Importantly, the link between NEAT1 and miR-361-5p has been revealed in hemangioma. NEAT1 has been elucidated to serve as a ceRNA to increase VEGFA expression via sequestering miR-361-5p in hemangioma.Citation16
Here, we testified whether MMP1 elicited biological function in CSCC and further probed into the possible underlying mechanism of MMP1 in the regulation of CSCC.
Materials and methods
Cell culture
Four human CSCC cells (A431, HSC-5, SCC13, SCL-1) and one human normal cutaneous cell (HaCaT), available from ATCC (Manassas, VA), were incubated under the standard condition of 5% CO2 and 37°C. DMEM medium was commercially acquired from Gibco (Grand Island, NY) for cell culture, with 10% FBS and Pen/Strep antibiotics solution (1%) as supplements.
Quantitative real-time polymerase chain reaction (RT-qPCR)
Total cell RNA was acquired using TRIzol method (Invitrogen, Carlsbad, CA), for cDNA synthesis with Reverse Transcription Kit (TaKaRa Bio, Shiga, Japan). The resulting products were treated with SYBR Green PCR Master Mix (Invitrogen) and then calculated with 2−ΔΔCT method. GAPDH or U6 was used as the internal control.
Cell transfection
The two specific shRNAs and control-shRNAs were produced by Genepharma Company (Shanghai, China) to silence MMP1 or NEAT1 in A431 and HSC-5 cells applying the transfection kit Lipofectamine 2000 (Invitrogen). The miR-361-5p mimics/inhibitor and NC mimics/inhibitor were procured from Genechem (Shanghai, China) for 48 h of cell transfection. In addition, the mutant sequences of MMP 3ʹUTR and NEAT1 were introduced by the authors.
MTT assay
A431 and HSC-5 cells were seeded in 96-well plates at 4 × 103 cells/well after transfection. Cell viability was detected after adding 5 mg/ml MTT for 4 h and then treated with 150 μl of DMSO. The absorbance at 490 nm was examined at indicated times using microplate reader.
Colony formation
Clonogenic cells of A431 and HSC-5 were cultured at 500 cells/well in the 6-well plates for the 14-day culture process. After treatment with 0.5% crystal violet, colonies were counted manually.
Transwell assay
Cells (1 × 105) were transfected into the top of Transwell chamber containing polycarbonate filters (8 μm pore; Corning Incorporated, Corning, NY), which was coated with Matrigel (BD Biosciences, Franklin Lakes, NJ) or not for analyzing cell invasion or migration. The lower chamber was supplied with complete medium. After 24 h, cells on the bottom were fixed in 4% paraformaldehyde for crystal violet staining and then counted under microscope.
Flow cytometry apoptosis analysis
Cells were cultured in PBS washing buffer and suspended in the 100 mL of binding buffer with 5 mL of Annexin V-FITC and 5 mL of propidium iodide (PI). After 15 min, stained cells were detected with flow cytometry (BD Biosciences, CA, USA).
Caspase-3/8/9 activity detection
Based on the standard methods, the activity of caspase-3/8/9 was examined using the caspase-3/8/9 activity kits (Beyotime, Shanghai, China) in A431 and HSC-5 cells. The activities were detected by microplate reader at 405 nm.
RNA immunoprecipitation (RIP) assay
After being washed in PBS, cell lysates were acquired by incubation in RIP lysis buffer and centrifugation and then cultured with the antibody-beads mixture overnight at 4°C. Antibodies specific to human AGO2 and normal control IgG were procured from Abcam (Cambridge, MA). QRT-PCR was conducted for analyzing the precipitated RNAs.
Luciferase reporter assay
The fragments of MMP1 3ʹUTR or NEAT1 covering the wild-type or mutant miR-361-5p binding sites were used to construct the luciferase reporter plasmids using luciferase reporter pmirGLO vector (Promega, Madison, WI). Following the co-transfection with miR-361-5p mimics or NC mimics in A431 and HSC-5 cells for 48 h, luciferase assay was carried out by Luciferase Reporter Assay System (Promega).
Subcellular fractionation
The nuclear or cytoplasmic RNA fraction was isolated from A431 and HSC-5 cells using PARIS™ Kit method (Invitrogen). After RNA purification, qRT-PCR was conducted to analyze the NEAT1 expression in both fractions.
Fluorescence in situ hybridization (FISH) assay
Based on the established method, RNA FISH assay was conducted in hybridization buffer using the specifically designed NEAT1-FISH probe (Ribobio). Then, cells were washed for Hoechst staining, finally observed with microscope.
Statistical analyses
Graphpad Prism 6 software was employed for statistical analysis in all bio-triplicates. The measurement data were represented as the mean ± standard deviation (SD). Comparisons of groups were examined by Student’s t test or one-way ANOVA, with p < .05 seen as significant.
Results
MMP1 promotes cell growth, migration and invasion of CSCC
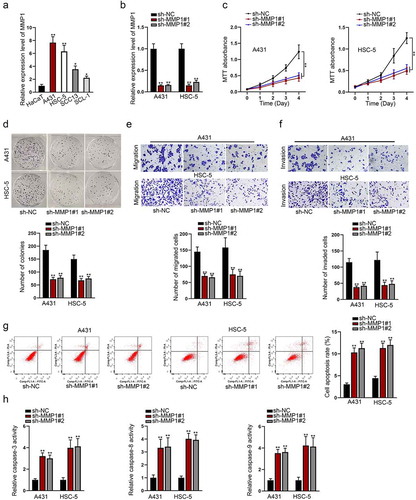
For exploring the potential biological role of MMP1 in CSCC, the expression level of MMP1 was evaluated in CSCC cells (A431, HSC-5, SCC13, SCL-1) and human normal cutaneous cell (HaCaT). Compared with HaCaT cell, MMP1 mRNA level was found distinctively up-regulated in CSCC cells, particularly in A431 and HSC-5 cells ()). Hence, they were selected for following assays. Then, MMP1 was stably silenced in A431 and HSC-5 cells for the purpose of loss-of-function assays ()). After that, MTT assay was conducted to assess CSCC cell vitality. It showed that MMP1 silencing evidently weakened the viability of CSCC cells ()). Consistently, colony formation assay further proved the inhibitory effect of MMP1 silencing on the proliferation of selected CSCC cells ()). Besides, Transwell migration and invasion assays indicated that MMP1 silencing dampened the migratory and invasive capacities of A431 and HSC-5 cells ()). Additionally, flow cytometry assay manifested that MMP1 silencing markedly promoted cell apoptosis of A431 and HSC-5 ()). Caspase-3/8/9 activity test revealed that MMP1 silencing remarkably enhanced activities of caspase-3, caspase-8, and caspase-9 ()). What’s more, pcDNA3.1 vectors were constructed to detect the overexpression efficiency of MMP1 (Fig. S1A). After that, MTT, colony formation and Transwell assays were conducted to verify the effect of MMP1 overexpression on the malignant behaviors of CSCC cells (Fig. S1B-S1E). As shown by the results, after the transfection of pcDNA3.1/MMP1, a totally different effect was achieved on the malignant behaviors of CSCC cells compared with those transfected with sh-MMP1#1/2. Aside from that, to exclude the possibility that the changes in the malignant behaviors of CSCC cells were caused by transfection toxicity, relevant rescue assays were conducted by us. First of all, we compared the expression of MMP1 in CSCC cells transfected with sh-MMP1#1 and sh-MMP1#1 + pcDNA3.1/MMP1 (Figure. S2A). As shown by the figure, the expression of the former one was close to that of cells transfected with sh-NC. Next, MTT and colony formation assays were adopted, the result of which showed that the proliferative ability of CSCC cells transfected with sh-MMP1#1 could be partially reversed by the transfection of pcDNA3.1/MMP1 (Figure. S2B-S2C). What’s more, the same results could be achieved in Transwell, flow cytometry assays and caspase-3/8/9 activity test (Figure S2D-S2G). Therefore, the malignant behavior changes were caused by the silencing and overexpression of MMP1 rather than transfection toxicity. Overall, these results demonstrated that the silencing of MMP1 could suppress cell growth and migration in CSCC.
Figure 1. MMP1 promotes cell growth, migration, and invasion of CSCC
MiR-361-5p is the molecular upstream gene of MMP1
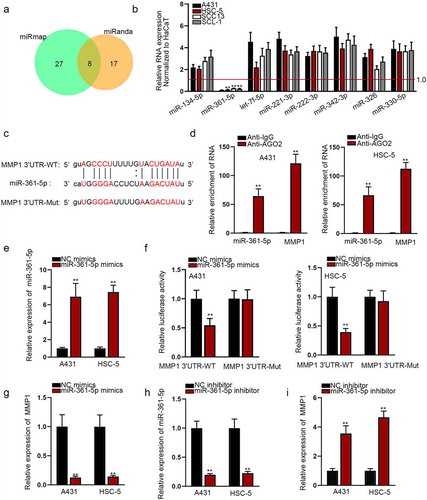
Increasing protein-coding RNAs have been discovered to be mediated by miRNA post-transcriptionally. To probe into the mechanism of MMP1, we firstly found the potential upstream target miRNA of MMP1. Utilizing miRmap and miRanda databases, we obtained eight potential target miRNAs of MMP1 ()). We examined the expression pattern of these miRNAs in CSCC cells and miR-361-5p was found in CSCC cells with markedly low expression ()). The potential wild and mutant sequences of miR-361-5p in the 3ʹUTR sequence of MMP1 were shown in ). RIP assay manifested a significant enrichment of miR-361-5p and MMP1 in the AGO2-bound complexes ()). We ensured that miR-361-5p mimics could effectively increase the level of miR-361-5p in selected CSCC cells ()). Luciferase reporter assay data showed that miR-361-5p mimics obviously suppressed the luciferase activity of MMP1-WT ()). Furthermore, MMP1 mRNA level was down-regulated in miR-361-5p-overepxressed A431 and HSC-5 cells ()). We then confirmed the interfering effect of miR-361-5p inhibitor by qRT-PCR ()). MMP1 mRNA level was up-regulated after the transfection of miR-361-5p inhibitor ()). To conclude, MMP1 could bind to miR-361-5p and was regulated by miR-361-5p in CSCC negatively.
Figure 2. MiR-361-5p is the molecular upstream gene of MMP1
MiR-361-5p inhibits the malignant behaviors of CSCC
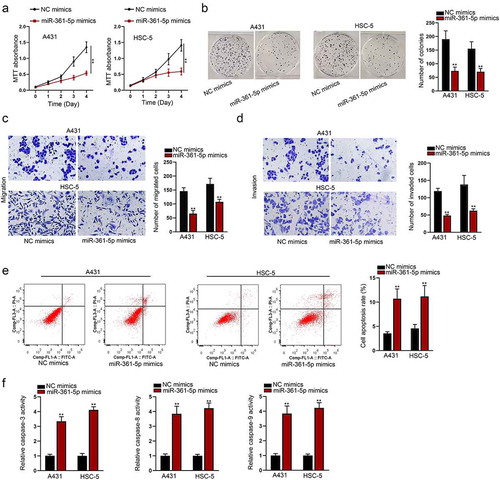
MiR-361-5p has been found to exert an inhibitory role in the development of an array of malignancies. However, its functional role in CSCC remains unclear. First of all, we detected the protein level and activity of MMP1 in CSCC cells after the transfection of miR-361-5p mimics (Fig. S3A) after the reference of two papers.Citation17,Citation18 Functional experiments indicated that miR-361-5p inhibited CSCC cells proliferative ()), migratory ()), and invasive ()) abilities. Meanwhile, miR-361-5p strengthened the apoptotic capacity of CSCC cells ()). To further verify our thought, the protein level and activity of MMP1 in CSCC cells after the transfection of miR-361-5p inhibitor were detected (Figure S3B). After that, functional experiments were carried out in cells transfected with miR-361-5p inhibitor. As shown by the results, it was indicated that the transfection of miR-361-5p inhibitor enhanced the proliferative (Fig. S3C-S3D), migratory (Fig. S3E), and invasive (Fig. S3F) abilities of CSCC cells. Therefore, we could conclude that miR-361-5p could inhibit the malignant behaviors of CSCC.
Figure 3. The function of miR-361-5p in CSCC was explored
To exclude the possibility that all other readouts probably stem from the cell apoptosis, we designed a series of experiments. First we detected the efficiency of anti-miR-361-5p (Fig. S4A-B). Then the cell viability, proliferation, migration and invasion abilities were tested by RT-qPCR respectively when miR-361-5p mimics were transfected into the cells (Fig. S4C-F). We found that all these abilities weakened. However, after co-transfected with anti-miR-361-5p, the above abilities recovered. Next, cell apoptosis was measured by flow cytometry and caspase-3/8/9 activity (Fig. S4G-H). Also, the effects caused by miR-361-5p overexpression were rescued by co-transfection with anti-miR-361-5p. In conclusion, all the phenotypes were caused by miR-361-5p rather than cell apoptosis.
NEAT1 interacts with miR-361-5p
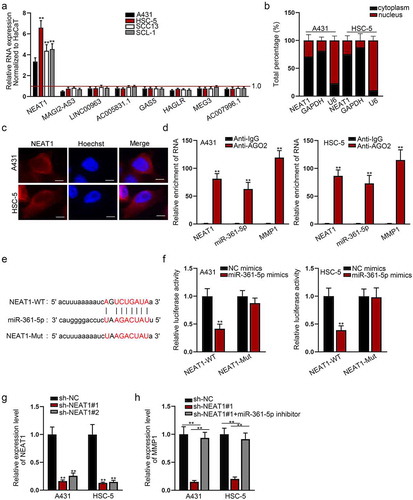
Since miRNAs have been increasingly reported to be regulated by lncRNAs, we detected the potential lncRNAs that might bind to miR-361-5p. With the help of starBase (http://starbase.sysu.edu.cn/), eight lncRNAs that might share binding sites with miR-361-5p were sifted out. The expression of NEAT1 was found to be obviously high in CSCC cells ()). To determine the potential regulatory role of NEAT1, we firstly studied its distribution in A431 and HSC-5 cells. Subcellular fractionation and FISH assays manifested that NEAT1 was located in both cytoplasm and nucleus of A431 and HSC-5 cells, but predominantly in cytoplasm (), Fig. S5A). RIP assay revealed a marked enrichment of NEAT1, miR-361-5p and MMP1 by antibody directly targeting AGO2 ()). We found that NEAT1 had binding sites with miR-361-5p ()). Besides, miR-361-5p mimics significantly decreased the luciferase activity of NEAT1-WT in A431 and HSC-5 cells ()). To determine the effects of NEAT1 on miR-361-5p-regulated MMP1, we silenced NEAT1 by constructing sh-NEAT1 ()). The protein level and activity of MMP1 in CSCC cells after the transfection with sh-NEAT1#1 or the co-transfection with sh-NEAT1#1 and miR-361-5p inhibitor were detected (Figure S5B). MMP1 mRNA was down-regulated in NEAT1-depleted A431 and HSC-5 cells, but restored by co-transfection of miR-361-5p inhibitor ()).
Figure 4. NEAT1 interacts with miR-361-5p
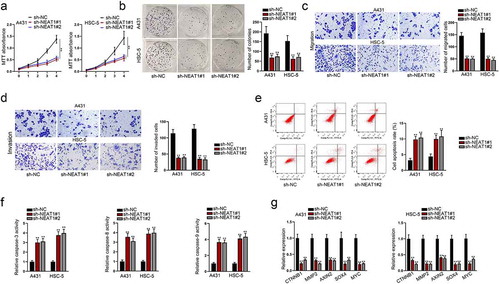
NEAT1 knockdown suppresses cell growth and migration via activation of Wnt pathway in CSCC
To determine whether NEAT1 influences the development of CSCC, we performed loss-of-function assays in selected CSCC cells. As expected, we found that NEAT1 knockdown suppressed cell proliferation ()), migration ()), and invasion ()). Also, NEAT1 silencing would lead to an increase in the number of apoptotic cells in selected CSCC cells ()). Since NEAT1 has been reported to activate Wnt pathway via increasing β-catenin nuclear transport and down-regulated ICAT, GSK3B, and Axin2 expression in the progression of glioblastoma,Citation19 we aimed at verifying whether NEAT1 exerted carcinogenic role in CSCC via the activation of Wnt pathway. QRT-PCR was conducted to evaluate the mRNA expression level of genes associated with Wnt pathway, including CTNNB1, MMP2, AXIN2, SOX4, and MYC. It showed that NEAT1 silencing greatly down-regulated these mRNA expressions in CSCC cells ()). In conclusion, NEAT1 silencing suppressed cell growth and migration via activating Wnt pathway in CSCC.
Figure 5. NEAT1 knockdown suppresses cell growth and migration via activation of Wnt pathway in CSCC
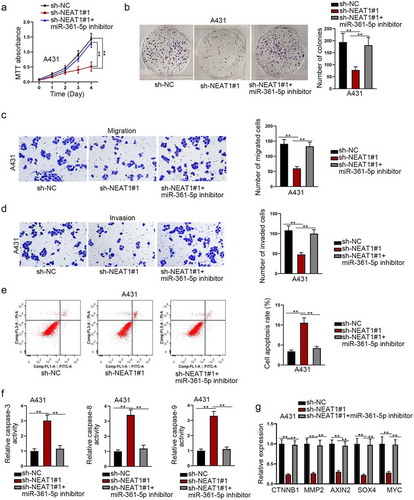
MiR-361-5p inhibitor rescues the oncogenic function of NEAT1 silencing on CSCC
To further elucidate whether the biological function of MMP1 could be indirectly regulated by NEAT1 through miR-361-5p, rescue experiments were then conducted. MTT analysis and colony formation assay showed that the cell viability or proliferation was impaired by NEAT1 knockdown, but partially restored by down-regulating miR-361-5p ()). Additionally, the migration and invasion abilities were weakened by NEAT1 knockdown, while partially restored by down-regulating miR-361-5p ()). Flow cytometry analysis and caspase 3/8/9 activity test indicated that cells apoptosis was promoted by NEAT1 knockdown, yet hampered by down-regulating miR-361-5p ()). Moreover, we measured the expression level alternation of Wnt pathway-relevant genes. We found that the expression levels of CTNNB1, MMP2, AXIN2, SOX4, and MYC were down-regulated by NEAT1 knockdown; however, this interfering outcome was reversed by co-transfecting miR-361-5p inhibitor ()). According to the result, it was suggested that NEAT1 regulated Wnt pathway via sponging miR-361-5p. Overall, above data manifested that miR-361-5p inhibitor rescued the oncogenic function of NEAT1 in CSCC, which was closely related with the restoration of Wnt pathway activity.
Figure 6. MiR-361-5p inhibitor rescues the oncogenic function of NEAT1 in CSCC
Discussion
Extensive reports have revealed the critical role of MMP1 in the metastasis of cancers. MMP1 expression level was reported to be significantly elevated in penile carcinoma tissues and served as a predictive biomarker for lymph node metastasis in patients with penile carcinoma.Citation20 Besides, previous investigation demonstrated that MMP1 overexpression advanced non-small cell lung cancer (NSCLC) progression and may serve as a useful biomarker for NSCLC patients.Citation21 According to our study, we detected an up-regulation of MMP1 in CSCC cell lines and found out that the knockdown of MMP1 hampered cell proliferative, migratory and invasive abilities, while facilitated the apoptotic capacity of CSCC cells.
MiRNAs could restrain the production of proteins via impacting the stability of their target mRNA or translational repression. The expression of MMP1 could be regulated by miRNAs in a variety of diseases. MiR-623 directly repressed MMP1 and relieved MMP1-induced migration, invasion, and metastasis in pancreatic cancer.Citation22 In this study, we explored the potential regulatory mechanism for the aberrant overexpression of MMP1. Also, miR-361-5p was verified to be an upstream molecular gene of MMP1 through bioinformatics analysis. MiR-361-5p expression was uncovered to be inversely correlated with vascular endothelial growth factor A (VEGFA) in CSCC.Citation23 Here, we found the expression of miR-361-5p in CSCC cell lines was low, which was contrary to the expression pattern of MMP1. MiR-361-5p was proved to inhibit the malignant behaviors of CSCC in vitro.
Since miRNAs exert negative impacts on the expression of target mRNA, we deduced that there might be another mechanism of molecular mediation on miR-361-5p. MiR-361-5p was discovered to be epigenetically sponged by many lncRNAs to affect target gene function in the carcinogenesis process. For instance, its repression on target mRNA FOXM1 was offset by the sponge of SBF2-AS1 in cervical cancer.Citation24 Herein, mechanism exploration revealed that the up-regulated NEAT1 was a molecular sponge for miR-361-5p in CSCC and it exhibited oncogenic properties in CSCC. This finding accorded with its carcinogenic role in diverse cancers, including myeloma,Citation25 hepatocellular carcinomCitation26 and thyroid carcinomaCitation27. Moreover, the pro-tumor role of NEAT1 in CSCC was identical to its promoting impacts on other type of skin cancer, such as melanoma cancer.Citation28 Furthermore, inhibition of miR-361-5p countervailed the biological effects induced by silencing NEAT1. NEAT1 was validated to activate Wnt pathway in nasopharyngeal carcinomaCitation29 and non-small cell lung cancer.Citation30 Here, silencing NEAT1 inhibited the expression level of Wnt pathway related genes, such as CTNNB1, MMP2, AXIN2, SOX4 and MYC in CSCC, but this effect was reversed by down-regulating miR-361-5p. This phenomenon implied that the NEAT1/miR-361-5p/MMP1 axis contributed to CSCC development via the activation of Wnt pathway.
In a word, our study firstly uncovered that MMP1 regulated by NEAT1/miR-361-5p axis facilitates cutaneous squamous cell carcinoma cell proliferation and migration via the activation of Wnt pathway. This observation might shed new light on novel therapeutic target for CSCC clinical application. However, our study lacked the specific illustration of how lncRNA NEAT1 affect the activity of Wnt pathway, which needs in-depth study in the future.
Supplemental Material
Download Zip (10.3 MB)Acknowledgments
We sincerely thank all participators for their support to our research.
Disclosure statement
The authors declared no relevant conflicts of interests.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- Languino LR, Singh A, Prisco M, Inman GJ, Luginbuhl A, Curry JM, South AP. Exosome-mediated transfer from the tumor microenvironment increases TGFbeta signaling in squamous cell carcinoma. Am J Transl Res. 2016;8:2432–2437.
- Zhang L, Xiang P, Han X, Wu L, Li X, Xiong Z. Decreased expression of microRNA-20a promotes tumor progression and predicts poor prognosis of cutaneous squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:11446–11451.
- Zhou L, Wang Y, Ou C, Lin Z, Wang J, Liu H, Zhou M, Ding Z. microRNA-365-targeted nuclear factor I/B transcriptionally represses cyclin-dependent kinase 6 and 4 to inhibit the progression of cutaneous squamous cell carcinoma. Int J Biochem Cell Biol. 2015;65:182–191. doi:10.1016/j.biocel.2015.06.009.
- Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol. 2018;78:237–247. doi:10.1016/j.jaad.2017.08.059.
- Hombach S, Kretz M. Non-coding RNAs: classification, Biology and Functioning. Adv Exp Med Biol. 2016;937:3–17.
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi:10.1016/j.cell.2005.07.031.
- Yadav L, Puri N, Rastogi V, Satpute P, Ahmad R, Kaur G. Matrix metalloproteinases and cancer - roles in threat and therapy. APJCP. 2014;15:1085–1091. doi:10.7314/APJCP.2014.15.3.1085.
- Huang C, Li Y, Guo Y, Zhang Z, Lian G, Chen Y, Li J, Su Y, Li J, Yang K, et al. MMP1/PAR1/SP/NK1R paracrine loop modulates early perineural invasion of pancreatic cancer cells. Theranostics. 2018;8:3074–3086. doi:10.7150/thno.24281.
- Wei W, Chen Y, Xu J, Zhou Y, Bai X, Yang M, Zhu J. Identification of Biomarker for Cutaneous Squamous Cell Carcinoma Using Microarray Data Analysis. J Cancer. 2018;9:400–406. doi:10.7150/jca.21381.
- Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi:10.1158/2159-8290.CD-13-0202.
- Ling Z, Liu D, Zhang G, Liang Q, Xiang P, Xu Y, Han C, Tao T. miR-361-5p modulates metabolism and autophagy via the Sp1-mediated regulation of PKM2 in prostate cancer. Oncol Rep. 2017;38:1621–1628. doi:10.3892/or.2017.5852.
- Hou XW, Sun X, Yu Y, Zhao HM, Yang ZJ, Wang X, Cao XC. miR-361-5p suppresses lung cancer cell lines progression by targeting FOXM1. Neoplasma. 2017;64:526–534. doi:10.4149/neo_2017_406.
- Cao ZG, Huang YN, Yao L, Liu YR, Hu X, Hou YF, Shao ZM. Positive expression of miR-361-5p indicates better prognosis for breast cancer patients. J Thorac Dis. 2016;8:1772–1779. doi:10.21037/jtd.2016.06.29.
- Ma F, Zhang L, Ma L, Zhang Y, Zhang J, Guo B. MiR-361-5p inhibits glycolytic metabolism, proliferation and invasion of breast cancer by targeting FGFR1 and MMP-1. J Exper Clin Cancer Res. 2017;36:158. doi:10.1186/s13046-017-0630-1.
- Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT1: a novel cancer-related long non-coding RNA. Cell Prolif. 2017;50(2):e12329.
- Yu X, Liu X, Wang R, Wang L. Long non-coding RNA NEAT1 promotes the progression of hemangioma via the miR-361-5p/VEGFA pathway. Biochem Biophys Res Commun. 2019;512:825–831. doi:10.1016/j.bbrc.2019.03.084.
- Holmberg C, Ghesquière B, Impens F, Gevaert K, Kumar JD, Cash N, Kandola S, Hegyi P, Wang TC, Dockray GJ, et al. Mapping proteolytic processing in the secretome of gastric cancer-associated myofibroblasts reveals activation of MMP-1, MMP-2, and MMP-3. J Proteome Res. 2013;12:3413–3422. doi:10.1021/pr400270q.
- Tjomsland V, Pomianowska E, Aasrum M, Sandnes D, Verbeke CS, Gladhaug IP. Profile of MMP and TIMP Expression in Human Pancreatic Stellate Cells: regulation by IL-1α and TGFβ and Implications for Migration of Pancreatic Cancer Cells. Neoplasia (New York, NY). 2016;18:447–456. doi:10.1016/j.neo.2016.06.003.
- Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, Zhou J, Kang C, Li M, Jiang C. Long Noncoding RNA NEAT1, Regulated by the EGFR Pathway, Contributes to Glioblastoma Progression Through the WNT/beta-Catenin Pathway by Scaffolding EZH2. Clin Cancer Res. 2018;24:684–695. doi:10.1158/1078-0432.CCR-17-0605.
- Kuasne H, Barros-Filho MC, Busso-Lopes A, Marchi FA, Pinheiro M, Munoz JJ, Scapulatempo-Neto C, Faria EF, Guimaraes GC, Lopes A, et al. Integrative miRNA and mRNA analysis in penile carcinomas reveals markers and pathways with potential clinical impact. Oncotarget. 2017;8:15294–15306. doi:10.18632/oncotarget.14783.
- Wang Y, Ding X, Liu B, Li M, Chang Y, Shen H, Xie SM, Xing L, Li Y. ETV4 overexpression promotes progression of non-small cell lung cancer by upregulating PXN and MMP1 transcriptionally. Mol Carcinog. 2020 Jan;59(1):73-86.
- Chen Y, Peng S, Cen H, Lin Y, Huang C, Chen Y, Shan H, Su Y, Zeng L. MicroRNA hsa-miR-623 directly suppresses MMP1 and attenuates IL-8-induced metastasis in pancreatic cancer. Int J Oncol. 2019;55:142–156. doi:10.3892/ijo.2019.4803.
- Kanitz A, Imig J, Dziunycz PJ, Primorac A, Galgano A, Hofbauer GF, Gerber AP, Detmar M. The expression levels of microRNA-361-5p and its target VEGFA are inversely correlated in human cutaneous squamous cell carcinoma. PloS One. 2012;7:e49568. doi:10.1371/journal.pone.0049568.
- Gao F, Feng J, Yao H, Li Y, Xi J, Yang J. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells, Nanomed Biotechnol. 2019;47:776–782. doi:10.1080/21691401.2019.1577883.
- Taiana E, Favasuli V, Ronchetti D, Todoerti K, Pelizzoni F, Manzoni M, Barbieri M, Fabris S, Silvestris I, Gallo Cantafio ME, et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia. 2019;34:234–244. doi:10.1038/s41375-019-0542-5.
- Li X, Zhou Y, Yang L, Ma Y, Peng X, Yang S, Li H, Liu J. LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J Cell Physiol. 2020 Apr;235(4):3402-3413.
- Zeng W, Lin Y, Lin H, Wu X. Silencing NEAT1 suppresses thyroid carcinoma via miR-126/NEAT1/VEGFA axis. Front Biosci (Landmark Edition). 2020;25:564–576. doi:10.2741/4821.
- Ding F, Lai J, Gao Y, Wang G, Shang J, Zhang D, Zheng S. NEAT1/miR-23a-3p/KLF3: a novel regulatory axis in melanoma cancer progression. Cancer Cell Int. 2019;19:217. doi:10.1186/s12935-019-0927-6.
- Ji Y, Wang M, Li X, Cui F. The Long Noncoding RNA NEAT1 Targets miR-34a-5p and Drives Nasopharyngeal Carcinoma Progression via Wnt/beta-Catenin Signaling. Yonsei Med J. 2019;60:336–345. doi:10.3349/ymj.2019.60.4.336.
- Sun SJ, Lin Q, Ma JX, Shi WW, Yang B, Li F. Long non-coding RNA NEAT1 acts as oncogene in NSCLC by regulating the Wnt signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:504–510.
