ABSTRACT
Radiotherapy plays important roles in the treatment of breast cancer (BC), which develops from malignant cells in the breast. Long non-coding RNAs (lncRNAs) have been reported to be implicated in radio-resistance or radio-sensitivity of human cancer, which includes breast cancer. Nevertheless, long intergenic non-protein coding RNA 0504 (LINC00504) has not been investigated in BC. In our study, from RT-qPCR analysis, LINC00504 was found to be up-regulated in BC cells. By conducting in vitro assays, it was confirmed that the knockdown of LINC00504 could enhance the radio-sensitivity of BC cells. The regulatory mechanism of LINC00504 in BC was also verified by chromatin immunoprecipitation (ChIP), RNA immunoprecipitation (RIP) and luciferase reporter assays. From the experimental results, we knew that the up-regulation of LINC00504 was mediated by signal transducer and activator of transcription 1 (STAT1). Moreover, LINC00504 stabilized the expression of cytoplasmic polyadenylation element-binding protein 2 (CPEB2) via binding to TATA-box binding protein associated factor 15 (TAF15). Furthermore, rescue assays validated that LINC00504 participated in regulating the radio-sensitivity of BC cells via up-regulating CPEB2. In summary, our study disclosed that STAT1 could mediate LINC00504 and weaken the radio-sensitivity of BC cells via binding to TAF15 and stabilizing CPEB2 expression.
Introduction
As one of the most common malignancies, breast cancer (BC) has been identified as a common cancer among women. Moreover, it can not only occur among women but also among men.Citation1,Citation2 Nowadays, great breakthroughs have been made in the treatment for BC including surgery, radiation therapy (RT) and drug therapy.Citation3 Among them, RT has been recognized as one of the most effective treatment options.Citation4,Citation5 Hence, seeking effective factors in the regulation of radio-sensitivity of cancers is much more crucial to BC treatment.
Long non-coding RNAs (lncRNAs) are defined as a group of transcripts longer than 200 nucleotides. Increasing evidence has suggested that lncRNAs can influence the progression of various cancers.Citation6,Citation7 In recent years, the aberrant expression of lncRNAs has also been demonstrated to be associated with the radio-sensitivity or radio-resistance of cancers. For instance, Gao et al. have proposed that lncRNA GAS5 serves as a sponge for miR-106b and influences cervical cancer cell radio sensitivity.Citation8 Zou et al. have proved that OIP5-AS1 decreases radio-resistance of colorectal cancer via targeting miR-369-3p/DYRK1A axis.Citation9 Hu et al. have elucidated that HOTAIR is involved in breast cancer radio-sensitivity by activating miR-218.Citation10 According to previous studies, LNC00504 has been proved to be involved in the progression of NSCLC,Citation11 but whether it could influence the radio-sensitivity of breast cancer is yet to be verified. In the present study, we aimed to unveil the role of LINC00504 and explore the underlying mechanism of LINC00504 in BC cell radio-sensitivity.
Materials and methods
Cell culture
BC cell lines (MCF7, MDA-MB-453, MDA-MB-468 and MDA-MB-231) and normal epithelial cell line (MCF10A) were all obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). MDA-MB-453, MDA-MB-468 and MDA-MB-231 cell lines were cultured in L-15 Medium. All mediums contained 10% FBS (16140071, Thermo Fisher Scientific) and were maintained under 37°C in 5% CO2.
Cell transfection
The target shRNAs against LINC00504, STAT1, or TAF15, as well as their corresponding control shRNA plasmids, were all synthesized by Genechem (Shanghai, China). Those sequences were provided in for reference. Besides, pcDNA3.1 vectors were subcloned with STAT1 or CPEB2 and empty pcDNA3.1 vectors were regarded as the negative control. Lipofectamine 3000 (L3000075, Invitrogen, Carlsbad, CA, USA) was used for cell transfection.
Table 1. Sequences of shRNAs were provided
Quantitative real-time PCR (RT-qPCR) analysis
Using TRIzol Reagent (15596026, Invitrogen), total RNA was extracted, and cDNA was synthesized by RevertAid First-Strand cDNA Synthesis Kit (K1621, Thermo fisher, IL, USA). To evaluate the expression of genes (LINC00504, STAT1, CPEB2, and TAF15), SYBR Green PCR Master Mix (4309155, Applied Biosystems, Foster City, CA, USA) was applied to conduct PCR analysis. The quantitative analysis was determined by 2−ΔΔCt method and all results were normalized to GAPDH or U6. The sequences of the primers were provided in for reference. The experiments were independently conducted at least three times.
Table 2. Sequences of primers were provided
Colony formation assay
In brief, cells were cultured in complete growth medium after being added into 6-well plates at a density of 200 cells. Fourteen days later, the fixed cells were stained with 0.1% crystal violet. The stained cell colonies were counted manually. The experiments were independently conducted at least three times. The GraphPad Prism version 8 software was used to select the line chart template for drawing. The number of clones detected in all samples of the control group was set as 1 when the radiation amount was 0.
In vitro irradiation
Cells were seeded into 6-well plates (600 cells/well) and exposed to 0, 2, 4, 6, and 8 Gy 6-MV X-rays from linear accelerators (Varian2300EX; Varian, Palo Alto, CA, USA) at a dose rate of 5 Gy/min. After incubation at 37°C for 14 days, the plates were fixed with 100% methanol and then stained with 0.1% crystal violet. Colonies containing >50 cells were counted by microscopic inspection. The surviving fraction (SF) was calculated. A multi-target single-hit model was fitted to the data to generate survival curves using the following formula: SF = 1 − (1 − e−D/D0)N. The experiments were independently conducted at least three times.
Transferase-mediated dUTP nick end labeling (TUNEL) assay
Cells (1 × 104) on culture slides were fixed with 4% paraformaldehyde and washed in PBS, then treated with 0.1% TritonX-100 for re-suspension. Apoptotic cells were examined by applying 50 μl TUNEL Detection Kit (11772457001, Roche, Basel, Switzerland) and incubated at 37°C for 60 min. Cell nuclei were dyed with DAPI. Optical microscopy (Olympus) was used to analyze labeled samples. TUNEL-positive cells were evaluated in a randomly selected field with no significant necrosis. The TUNEL index was calculated based on the total number of nuclei and cells with green nucleus.
Immunofluorescent (IF) staining
IF staining was conducted to examine the formation of gamma histone 2AX (γ-H2AX) foci. Cells were cultured in 24-well plates at a density of 1 × 104 cells/well to receive 0 and 4 Gy of irradiation. Twenty-four hours later, the cells were fixed with 4% paraformaldehyde and permeabilized in 0.1% Triton X-100 (T8787, Sigma). Afterward, the cells were blocked with 1% goat serum, followed by incubation with primary anti-γ-H2AX antibodies (ab124781, 1:100; Abcam). Then, the cells were incubated with a tetramethylrhodamine isothiocyanate–conjugated secondary antibody (diluted 1:200; Jackson ImmunoResearch, West Grove, Pennsylvania) conjugated to fluorescein isothiocyanate after primary antibodies were washed off. Finally, nuclei were counterstained with DAPI. The percentage of γ-H2AX-positive cells was detected under a fluorescent microscope. The experiments were independently conducted at least three times.
Subcellular fractionation
Subcellular fractionation assay was conducted with the employment of Cytoplasmic and Nuclear RNA Purification Kit (Cat. 21000, 37400, NORGEN, Thorold, ON, Canada). GAPDH and U6 were separately treated as the cytoplasmic control or the nuclear control. The experiments were independently conducted at least three times.
RNA immunoprecipitation (RIP) assay
RIP assay was performed with RNA-binding protein immunoprecipitation kit (17–700, Millipore, Burlington, MA, USA). BC cells were collected and then lysed in RIPA lysis buffer (R0010, Solarbio, Beijing, China). Subsequently, lysates were immunoprecipitated with anti-Ago2 (Abcam) antibody or anti-IgG (Abcam) antibody. Similarly, rabbit anti-TAF15 (Abcam) antibody and immunoglobulin G (IgG) antibody (Abcam) were also precipitated. Finally, the RNA precipitates were extracted and then analyzed by RT-qPCR. The experiments were independently conducted at least three times.
Chromatin immunoprecipitation (ChIP) assay
The EZ ChIP™ Chromatin Immunoprecipitation Kit (17–408, Millipore, Burlington, MA, USA) was used to carry out ChIP assay. Briefly, BC cells were fixed with 1% formaldehyde and sonicated into fragments. Rabbit anti-STAT1 (ab234400, Abcam, UK) antibody and IgG Rabbit antibody (ab133470, Abcam, UK) were precipitated with chromatin. The immunoprecipitated DNA was extracted for RT-qPCR analysis. The experiments were independently conducted at least three times. The antibodies used for ChIP assay were listed as follows: anti-Ago2 (SAB4200085, Merck, Shanghai, China), anti-RNA polymerase II (ab193468, Abcam, UK), together with the above STAT1 and IgG antibodies.
Luciferase reporter assay
The LINC00504 promoter region containing the binding sites (wild type or mutant type) was constructed into the pGL3 vector (E1751, Promega, Madison, WI) and co-transfected along with pcDNA3.1/STAT1 or the empty vector into BC cells. After 48 h, the Dual-Luciferase Reporter Gene Assay Kit (E1910, Promega, Madison, WI) was applied to measure luciferase activity. The experiments were independently conducted at least three times.
Western blot analysis
Total protein samples were separated with SDS-PAGE and later transferred to PVDF membranes (88585, Thermo fisher, IL, USA). Then, membranes were incubated with primary antibodies at 4°C overnight after being blocked in milk without fat. After being washed with PBS for three times, the blots were then incubated with secondary antibody for 1 h in a dark room. The quantification analysis of protein was conducted by Chemiluminescence system (GE Healthcare, Chicago, USA). The experiments were performed at least three times. The antibodies were listed as follows: anti-STAT1 (ab234400, 1:1000), anti-CPEB2 (ab222070, 1:1000), anti-TAF15 (ab134916, 1:10000), anti-P-Chk1 (ab79758, 1:1000), anti-P-Chk2 (ab32148, 1:1000), and GAPDH (ab8245, 1:1000) as the control. The above antibodies were bought from Abcam.
Statistical analysis
Experimental data were analyzed by SPSS version 22.0 statistical software. All data were expressed as mean ± standard deviation (SD). The differences between two groups or more groups were analyzed with the employment of Student’s t-test or ANOVA. All the experiments were independently conducted at least three times. Differences were considered statistically significant when P < .05.
Results
LINC00504 is up-regulated and related to radio-sensitivity of BC
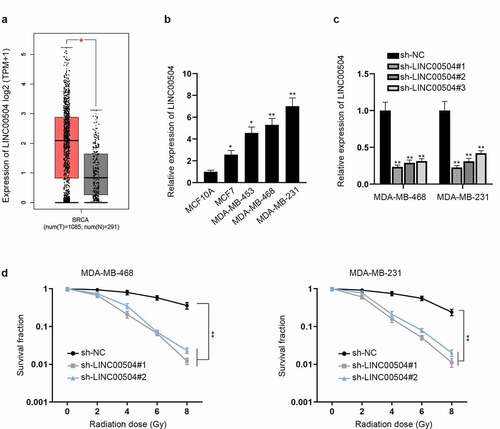
From GEPIA database (http://gepia2.cancer-pku.cn/#index), we found that compared with normal tissues, LINC00504 was notably overexpressed in tumor tissues (). Next, we further analyzed the expression level of LINC00504 in BC cell lines (MCF7, MDA-MB-453, MDA-MB-468 and MDA-MB-231) and normal epithelial cell line (MCF10A). The result demonstrated that the expression level of LINC00504 is obviously higher in BC cell lines than that in normal cell line (). Moreover, MDA-MB-468 and MDA-MB-231 were kept for further assays as the expression of LINC00504 were the highest among BC cell lines. To further investigate the function of LINC00504 in BC, we firstly knocked down LINC00504 and examined the interference efficiency of sh-LINC00504 (). As the interference efficiency of sh-LINC00504#1/2 was better, they were selected for the following assays. By conducting colony formation assay, we surprisingly discovered that after LINC00504 was silenced, cell survival fraction showed a descending tendency with the increase in radiation doses (). Overall, LINC00504 was involved in the radio-sensitivity of BC cells.
Figure 1. LINC00504 is up-regulated and related to radio-sensitivity of BC. (a) The expression level of LINC00504 in tumor tissues and normal tissues was displayed by GEPIA database. (b) The expression levels of LINC00504 in BC cell lines and normal cell line were assessed by RT-qPCR assay. (c) The interference efficiency of LINC00504 was analyzed. (d) Survival fraction of BC cells exposed to different doses of radiation was demonstrated. *P < .05, **P < .01.
Knockdown of LINC00504 enhances cell radio-sensitivity of BC
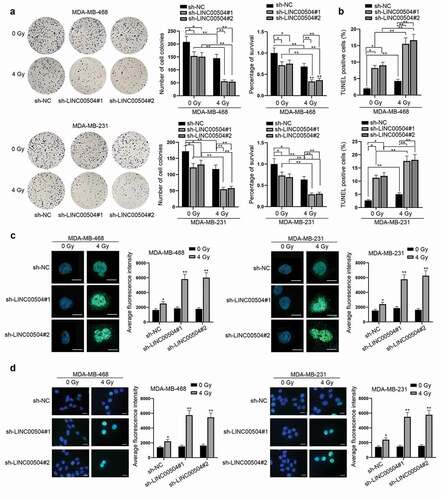
Functional assays were then conducted to assess the influence of LINC00504 on cell radio-sensitivity of BC. Via colony formation assay, we found that the down-regulation of LINC00504 inhibited cell proliferation and decreased the survival rate of BC cells while this effect could be enhanced when cells were exposed to 4 Gy of radiation (). Also, colony formation assay was conducted in MCF10A cell transfected with sh-LINC00504#1/2 for further verification. As shown by the result, no obvious change was found in the number of cell colonies in MCF10A cell line transfected with sh-LINC00504#1/2 compared with that in the sh-NC group. Therefore, we drew the conclusion that sh-LINC00504 could not affect normal BC cells after radiation (Fig. S1A). According to the results from TUNEL assay, it was observed that LINC00504 depletion enhanced the apoptotic capacity of BC cells. Similarly, the number of apoptotic cells was larger under the condition of 4 Gy of radiation (). In addition, γ-H2AX, which is one of the proteins related to DNA damage, has been identified as a crucial indicator in tumor radio-sensitivity.Citation12 From the experimental results of IF staining, we found that the average fluorescence intensity of γ-H2AX positive cells was dramatically augmented due to the down-regulation of LINC00504 (). In addition, after referring to related researches, we found that ATM and ATR have a close relation with the repair of DNA damage. Also, the irradiation of tumor cells may cause the activation of ATM/ATR, thus leading to the phosphorylation of Chk1 and Chk2, which are the downstream proteins of ATM/ATR. Therefore, we conducted western blot assay to detect how the down-regulation of LINC00504 would affect the downstream proteins of ATM/ATR after the BC cells were radiated (Fig. S1B). As shown by the figure, the expression of P-Chk1 and P-Chk2 after the down-regulation of LINC00504 with 4 Gy of radiation was higher than that of the control group. To conclude, LINC00504 could affect ATM/ATR and influence the radio-sensitivity of BC cells by affecting DNA damage.
Figure 2. Knockdown of LINC00504 enhances cell radio-sensitivity of BC. (a,b) Colony formation and TUNEL assays were performed to observe the proliferation, apoptosis and survival rate of BC cells under 0 and 4 Gy of radiation. (c) IF assay was used to quantify the fluorescence intensity of γ-H2AX positive cells in BC cells (magnification: ×200; scale bar: 10 μm). (d) IF images containing more cells per field were presented to show the representative number of γ-H2AX positive cells under different treatment (magnification: ×200; scale bar: 10 μm). *P < .05, **P < .01.
LINC00504 up-regulation is induced by STAT1
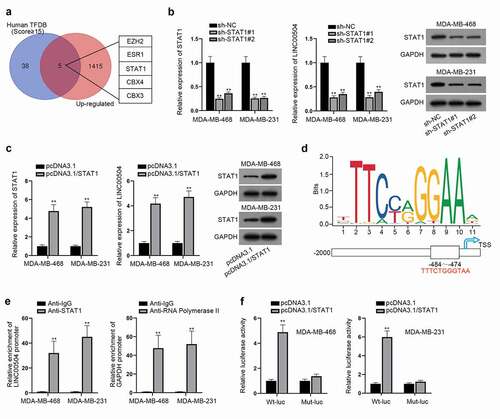
Transcription factors have been reported to specifically binding to the promoter of lncRNAs and are concerned with aberrant gene expression.Citation13,Citation14 Hence, we supposed that the up-regulation of LINC00504 might be activated by certain transcription factors. Using Human TFDB (http://bioinfo.life.hust.edu.cn/HumanTFDB#!/) and GEPIA databases, we sifted out five potential transcription factors of LINC00504 (EZH2, ESR1, STAT1, CBX4, and CBX3) which were also dramatically overexpressed in BC cells (). Among them, STAT1 has been widely reported to act as a transcription factor in multiple cancers.Citation15,Citation16 Subsequently, we chose STAT1 for further study. Firstly, the interference efficiency of sh-STAT1#1/2 was measured. After that, RT-qPCR was then conducted and the result suggested that the expression level of LINC00504 was reduced after STAT1 was silenced (). Besides, the overexpression efficiency of pcDNA3.1/STAT1 was measured and the result of RT-qPCR assay suggested that STAT1 overexpression could induce the up-regulation of LINC00504 (). After that, we adopted RT-qPCR assay to verify how the inhibition of STAT1 (by using pravastatin) would affect the expression of LINC00504 in BC cells (Fig. S1C). According to the result, the inhibition of STAT1 induced by the adding of pravastatin would lead to a decrease in the expression of LINC00504 in BC cells. As shown in , the potential-binding sites between STAT1 and LINC00504 promoter were predicted according to the data from JASPAR database (http://jaspar.genereg.net/). For further validation, ChIP assay was implemented to demonstrate the affinity between LINC00504 promoter and STAT1 with IgG group as the negative control and RNA polymerase II as the positive control (). Moreover, the results of luciferase reporter assay also proved that the luciferase activity of LINC00504 promoter was greatly elevated after the overexpression of STAT1 (). In summary, STAT1 could enhance the expression of LINC00504 by activating the transcription of LINC00504.
Figure 3. LINC00504 up-regulation is induced by STAT1. (a) Venn diagram showed 5 predicted up-regulated transcription factors in GEPIA database. (b,c) RT-qPCR assay was conducted to examine the expression of LINC00504 and STAT1 under different treatment and western blot assay was utilized to detect the STAT1 protein level. (d) The predicted binding regions of STAT1 on LINC00504 promoter were presented. (e,f) ChIP and luciferase reporter assays further certified the binding relationship between STAT1 and LINC00504 promoter. **P < .01.
LINC00504 regulates CPEB2 expression
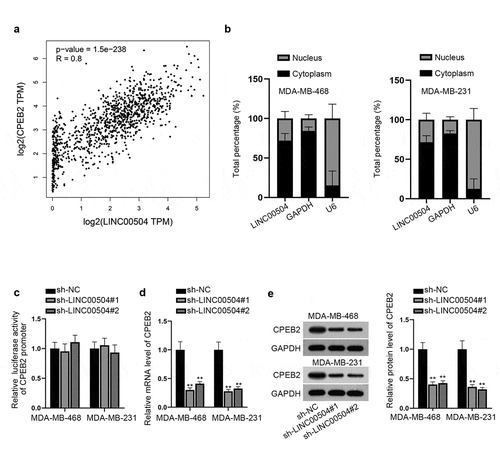
Then we aimed to explore the mechanism of LINC00504 in BC. Through GEPIA database, we found that LINC00504 had an extremely positive correlation with CPEB2 (). In addition, UCSC database (http://genome.ucsc.edu/) also demonstrated that CPEB2 was a nearby gene of LINC00504. According to previous studies, lncRNAs have been reported to regulate nearby genes by cis-acting and trans-acting.Citation17 Thus, we hypothesized that LINC00504 could regulate CPEB2. First of all, subcellular fractionation assay was carried out and the result suggested that LINC00504 was mainly located in the cytoplasm (). Luciferase reporter assay was then carried out to unveil the regulatory pattern of LINC00504. Intriguingly, no obvious change was found in the luciferase activity of CPEB2 promoter before and after LINC00504 was knocked down (). Therefore, we excluded cis-acting of LINC00504. At the same time, RT-qPCR and western blot assays were utilized to analyze the expression level and protein level of CPEB2. As shown by the result, both levels were reduced due to LINC00504 down-regulation (). Therefore, we concluded that LINC00504 could regulate CPEB2 expression through trans-acting.
Figure 4. LINC00504 regulates CPEB2 expression. (a) GEPIA database showed the positive correlation between LINC00504 and CPEB2. (b) Subcellular fractionation assay was used to confirm the location of LINC00504 in BC cells. (c) Luciferase reporter assay was performed to detect the relationship between LINC00504 and CPEB2 promoter. D-E. RT-qPCR and western blot assay were utilized to analyze the expression level and protein level of CPEB2 after LINC00504 depletion. **P < .01.
LINC00504 recruits TAF15 and stabilizes CPEB2 expression
Previous studies have verified that LINC00504 regulated CPEB2 expression via trans-acting. Therefore, out next step was to investigate the detailed mechanism of LINC00504. In trans-acting, lncRNAs primarily act as a competing endogenous RNA (ceRNA) or bind to proteins to achieve the regulation of mRNAs.Citation18 From the results of RIP assay, we found that compared with lncRNA HOTIR, which has been widely reported to bind to Ago2 in diverse cancersCitation19 and AC010733.2 which has been reported to be unable to bind to Ago2,Citation20 LINC00504 showed no binding ability to Ago2 protein in BC cells (), which meant that LINC00504 couldn’t bind to any miRNA in BC cells. Therefore, we conjectured that LINC00504 might bind to certain proteins to stabilize CPEB2 expression. Through Venn diagram, we sifted out two RNA-binding proteins (RBPs) which could both bind to LINC00504 and CPEB2 (). Between the above two RBPs (U2AF2 and TAF15), only TAF15 has been proved to be able to stabilize mRNAsCitation21 while U2AF2 has been reported to influence mRNA splicing.Citation22 Consequently, we chose TAF15 for further study. The strong-binding ability between TAF15 and LINC00504, TAF15, and CPEB2 were, respectively, substantiated by RIP assay (). To detect the effect of TAF15 on CPEB2 expression and protein levels, we firstly knocked down TAF15 by transfecting sh-TAF15#1/2 (). Then, RT-qPCR and western blot assays were used and we found that both the expression level and protein level of CPEB2 were significantly decreased when TAF15 was down-regulated (). In addition, we found that the expression of CPEB2 mRNA was declined after LINC00504 knockdown or TAF15 deletion in BC cells treated with Actinomycin D (Act D), suggesting that the stability of CPEB2 was evidently reduced on account of the depletion of LINC00504 or TAF15 (). Collectively, LINC00504 could stabilize CPEB2 by recruiting TAF15.
Figure 5. LINC00504 recruits TAF15 and stabilizes CPEB2 expression. (a) RIP assay was carried out to verify whether LINC00504 could bind to Ago2 in BC cells. (b) Venn diagram showed potential RBPs that could both bind to LINC00504 and CPEB2. (c) The interaction between LINC00504 and TAF15, TAF15 and CPEB2 was verified by RIP assay. (d,e) The efficiency of TAF15 depletion was examined. (f,g) The expression level and protein level of CPEB2 after TAF15 depletion was detected by RT-qPCR and western blot assays. (h) The stability of CPEB2 mRNA was detected after silencing LINC00504 and TAF15. **P < .01.
LINC00504 decreases cell radio-sensitivity of BC cells via regulating CPEB2 expression
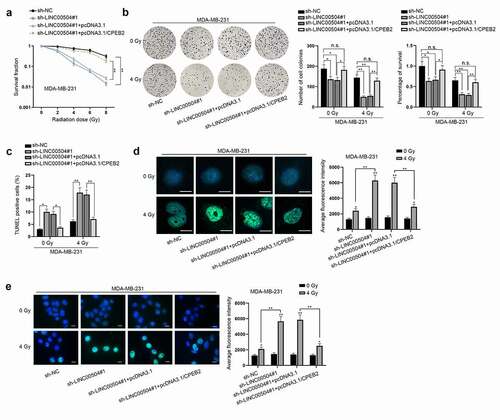
Finally, to figure out that whether LINC00504 could regulate the radio-sensitivity of BC cells via enhancing CPEB2 expression, several rescue assays were conducted. As shown in , with the increasing radiation doses, the survival fraction of BC cells was decreased in a steady manner. Besides, LINC00504 down-regulation could cut down the survival rate of BC cells while CPEB2 up-regulation could partially restore this effect. After that, colony formation and TUNEL assays were implemented to observe cell proliferation and apoptosis under different conditions. The findings indicated that the knockdown of LINC00504 could suppress cell proliferation and decreased the survival rate of BC cells while promoting cell apoptosis. Also, at 4 Gy of radiation, such effect was greatly strengthened. At the same time, it was found that the co-transfection of pcDNA3.1/CPEB2 could counteract those effects induced by LINC00504 depletion (). Immunofluorescence staining was then conducted by us and it was uncovered that γ-H2AX positive cells showed higher fluorescence intensity after LINC00504 was knocked down, while such result could be partially reversed after the co-transfection of CPEB2 (). To sum up, LINC00504 could enhance CPEB2 expression, thereby regulating the radio-sensitivity of BC cells.
Figure 6. LINC00504 decreases cell radio-sensitivity of BC via regulation of CPEB2 expression. (a) Survival fraction of BC cells in sh-NC, sh-LINC00504, sh-LINC00504+pcDNA3.1 and sh-LINC00504+pcDNA3.1/CPEB2 groups was observed. (b,c) The proliferation, apoptosis and survival rate of BC cells were examined under 0 and 4 Gy of radiation in different transfection groups. (d) The fluorescence intensity of γ-H2AX positive cells among different groups (sh-NC, sh-LINC00504, sh-LINC00504+ pcDNA3.1 and sh-LINC00504+ pcDNA3.1/CPEB2) was shown under 0 and 4 Gy of radiation (magnification: ×200; scale bar: 10 μm). (e) IF images containing more cells per field were presented to show the representative number of γ-H2AX positive cells under different treatment (magnification: ×200; scale bar: 10 μm). *P < .05, **P < .01.
Discussion
Radiation therapy has been widely considered to be an effective therapeutic method for BC.Citation23 Plenty of researches have uncovered that lncRNAs are crucial biomarkers indicating the radio-sensitivity of cancers including BC.Citation8,Citation24 For instance, Gao et al. have found that through regulating miR-106b/IER3 axis, lncRNA GAS5 enhances the radio-sensitivity of cervical cells.Citation8 Lai et al. have proved that lncRNA CCAT1 enhances the radio-sensitivity through sequestering miR-148b in breast cancer.Citation24 Hu et al. have validated that lncRNA HOTAIR is involved in breast cancer radio-sensitivity through activating miR-218.Citation10 LINC00504 has been reported to regulate the progression of colon cancer, ovarian cancer, and non-small cell lung cancer.Citation11,Citation25,Citation26 All the researches declared the oncogenic role of LINC00504 in cancers. Unlike the previous studies, the focusing point was on the role of LINC00504 in cell radio-sensitivity of BC. Consistent with those references, LINC00504 was also found to be obviously overexpressed in BC cell lines. Moreover, loss-of-function assays proved that LINC00504 depletion could enhance the radio-sensitivity of BC cells.
As shown by the previous studies, the aberrant expression of LINC00504 is always related to transcription factors.Citation13,Citation14 Moreover, STAT1 has been clarified to be a common transcription factor of lncRNAs in cancers, such as colorectal carcinoma, lung adenocarcinoma and so on.Citation27,Citation28 In our study, we found that LINC00504 could be activated by STAT1. In other words, STAT1 could bind to LINC00504 promoter, thus inducing LINC00504 up-regulation.
RBPs have been identified to act as crucial regulators in cancers via regulating cellular processes.Citation29 It has been reported that TAF15 functions as a RBP to participate in the progression of colorectal cancer.Citation21 Our study disclosed that LINC00504 recruited TAF15 to stabilize CPEB2 mRNA and enhance CPEB2 expression. More importantly, the experimental results of rescue assays validated that CPEB2 overexpression could restore the enhanced radio-sensitivity mediated by LINC00504 silencing in BC cells.
In conclusion, our study elucidated that STAT1 induced LINC00504 to exert influence on BC cell radio-sensitivity through modulating TAF15/CPEB2 axis, which might improve the curative effect of radiation therapy and make contributions to BC treatment in the future.
However, it was a pity that we just verified that after the mediation of STAT1, LINC00504 could influence the radio-sensitivity of BC via binding to TAF15 and stabilizing CPEB2 expression. We didn’t delve into the way by which LINC00504 affected DNA damage. Therefore, further investigations on whether LINC00504 could regulate other pathways would be conducted in our future studies.
Supplemental Material
Download Zip (362.4 KB)Acknowledgments
We are grateful to the help provided by all lab personnel in this research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Elbachiri M, Fatima S, Bouchbika Z, Benchekroun N, Jouhadi H, Tawfiq N, Sahraoui S, Benider A. Breast cancer in men: about 40 cases and literature review. Pan Afr Med J. 2017;28:287.
- Anastasiadi Z, Lianos GD, Ignatiadou E, Harissis HV, Mitsis M. Breast cancer in young women: an overview. Updates Surg. 2017;69:313–317. doi:https://doi.org/10.1007/s13304-017-0424-1.
- Wörmann B. Breast cancer: basics, screening, diagnostics and treatment. Med Monatsschr Pharm. 2017;40:55–64.
- Boyages J. Radiation therapy and early breast cancer: current controversies. Med J Aust. 2017;207:216–222. doi:https://doi.org/10.5694/mja16.01020.
- Castaneda SA, Strasser J. Updates in the treatment of breast cancer with radiotherapy. Surg Oncol Clin N Am. 2017;26:371–382. doi:https://doi.org/10.1016/j.soc.2017.01.013.
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi:https://doi.org/10.1016/j.ccell.2016.03.010.
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi:https://doi.org/10.1038/onc.2017.184.
- Gao J, Liu L, Li G, Cai M, Tan C, Han X, Han L. LncRNA GAS5 confers the radio sensitivity of cervical cancer cells via regulating miR-106b/IER3 axis. Int J Biol Macromol. 2019;126:994–1001. doi:https://doi.org/10.1016/j.ijbiomac.2018.12.176.
- Zou Y, Yao S, Chen X, Liu D, Wang J, Yuan X, Rao J, Xiong H, Yu S, Yuan X, et al. LncRNA OIP5-AS1 regulates radioresistance by targeting DYRK1A through miR-369-3p in colorectal cancer cells. Eur J Cell Biol. 2018;97:369–378.
- Hu X, Ding D, Zhang J, Cui J. 2019. Knockdown of lncRNA HOTAIR sensitizes breast cancer cells to ionizing radiation through activating miR-218. Biosci Rep. 39. DOI:https://doi.org/10.1042/BSR20181038
- Ma HP, Wang LX, Li W, Guo HH, Wu Y, Li XY. Upregulation of LINC00504 is associated with aggressive progression and poor prognosis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2020;24:699–703.
- Beaton LA, Marro L, Malone S, Samiee S, Grimes S, Malone K, Wilkins RC. Investigating γ H2AX as a biomarker of radiosensitivity using flow cytometry methods. ISRN Radiol. 2013;2013:704659. doi:https://doi.org/10.5402/2013/704659.
- Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177–186. doi:https://doi.org/10.1016/j.gpb.2016.12.005.
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi:https://doi.org/10.1038/nrg2957.
- Kostanian IA, Vonarshenko AV, Lipkin VM. [STAT1: a many-sided transcription factor]. Bioorg Khim. 2010;36:15–28.
- Adámková L, Soucková K, Kovarík J. Transcription protein STAT1: biology and relation to cancer. Folia Biol (Praha). 2007;53:1–6.
- Yan P, Luo S, Lu JY, Shen X. Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Curr Opin Genet Dev. 2017;46:170–178. doi:https://doi.org/10.1016/j.gde.2017.07.009.
- Gardiner AS, Twiss JL, Perrone-Bizzozero NI. Competing interactions of RNA-binding proteins, MicroRNAs, and their targets control neuronal development and function. Biomolecules. 2015;5:2903–2918. doi:https://doi.org/10.3390/biom5042903.
- Chiyomaru T, Fukuhara S, Saini S, Majid S, Deng G, Shahryari V, Chang I, Tanaka Y, Enokida H, Nakagawa M, et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. J Biol Chem. 2014;289:12550–12565. doi:https://doi.org/10.1074/jbc.M113.488593.
- Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7:10982. doi:https://doi.org/10.1038/ncomms10982.
- Pan L, Li Y, Jin L, Li J, Xu A. TRPM2-AS promotes cancer cell proliferation through control of TAF15. Int J Biochem Cell Biol. 2020;120:105683. doi:https://doi.org/10.1016/j.biocel.2019.105683.
- Lin CL, Taggart AJ, Lim KH, Cygan KJ, Ferraris L, Creton R, Huang YT, Fairbrother WG. RNA structure replaces the need for U2AF2 in splicing. Genome Res. 2016;26:12–23. doi:https://doi.org/10.1101/gr.181008.114.
- Minafra L, Bravatà V, Cammarata FP, Russo G, Gilardi MC, Forte GI. Radiation gene-expression signatures in primary breast cancer cells. Anticancer Res. 2018;38:2707–2715.
- Lai Y, Chen Y, Lin Y, Ye L. Down-regulation of LncRNA CCAT1 enhances radiosensitivity via regulating miR-148b in breast cancer. Cell Biol Int. 2018;42:227–236. doi:https://doi.org/10.1002/cbin.10890.
- Feng J, Ma J, Liu S, Wang J, Chen Y. A noncoding RNA LINC00504 interacts with c-Myc to regulate tumor metabolism in colon cancer. J Cell Biochem. 2019;120:14725–14734. doi:https://doi.org/10.1002/jcb.28733.
- Liu Y, He X, Chen Y, Cao D. Long non-coding RNA LINC00504 regulates the Warburg effect in ovarian cancer through inhibition of miR-1244. Mol Cell Biochem. 2020;464:39–50. doi:https://doi.org/10.1007/s11010-019-03647-z.
- Shen Y, Gao X, Tan W, Xu T. STAT1-mediated upregulation of lncRNA LINC00174 functions a ceRNA for miR-1910-3p to facilitate colorectal carcinoma progression through regulation of TAZ. Gene. 2018;666:64–71. doi:https://doi.org/10.1016/j.gene.2018.05.001.
- Lin XL, Zhu J, Wang LM, Yan F, Sha WP, Yang HL. MiR-92b-5p inhibitor suppresses IL-18 mediated inflammatory amplification after spinal cord injury via IL-18BP up-regulation. Eur Rev Med Pharmacol Sci. 2019;23:1891–1898.
- Mohibi S, Chen X, Zhang J. Cancer the’RBP’eutics-RNA-binding proteins as therapeutic targets for cancer. Pharmacol Ther. 2019;203:107390.
