ABSTRACT
Up until now, cancer refractoriness and distal organ metastatic disease remain as major obstacles for oncologists to achieve satisfactory therapeutic effects for lung adenocarcinoma patients. Previous studies indicated that TRIM55, which participates in the natural development of muscle and cardiovascular system, plays a protective role in hepatocellular carcinoma (HCC) pathogenesis. Therefore, in this study, we aimed to unveil the detailed molecular mechanism of TRIM55 and identify the potential target for lung adenocarcinoma patients. Surgical samples and lung cancer cell lines were collected to detect the TRIM55 expression for patients with or without lymph node/distal organ metastasis. Cellular functional assays including transwell assay, wound healing assay, cellular survivability assay, etc. as well as ubiquitination assay were performed to evaluate the impact of TRIM55/Snail1 regulatory network via the UPP pathway on lung cancer tumor cell migration and chemo-resistance. Lung cancer tissues and tumor cell lines exhibited significantly lower levels of TRIM55 expression. Functional study further indicated that TRIM55 inhibited chemo-resistance, migration, and cancer stem-cell like phenotype of tumor cells. Further detailed molecular experiments indicated that TRIM55 promoted degradation of Snail1 via the UPP pathway, which played an interesting role in the regulation of cancer cell malignancy. This study provided novel theory that TRIM55 acted as a potential tumor suppressor by inhibition of tumor cell malignancy through enhancement of Snail1 degradation via the UPP pathway. Our research will inspire further exploration on TRIM55 to promote therapeutic effects for lung adenocarcinoma patients.
Introduction
Up until now, lung cancer remains as one of the leading cancer types around the globe.Citation1 As a major component of non-small cell lung cancer (NSCLC), lung adenocarcinoma patients have shorter survival time compared to other NSCLC subtypes.Citation2 Despite current development in lung cancer diagnosis and treatment being constantly evolving, cancer refractoriness and distal organ metastatic diseases still remain as major clinical obstacles. For example, nearly 20% of the advanced NSCLC patients suffered from disease refractoriness, which significantly limited overall survival.Citation3 Therefore, it is of clinical significance to fully decipher the underlying detailed molecular mechanism in order to develop novel therapeutic interventions.
Tripartite motif containing 55 (TRIM55), which is also called muscle-specific ring finger 2 (Murf2), has been indicated to participate in the natural development of muscle and cardiovascular system.Citation4 A previous study also provided evidences that TRIM55 played an interesting protective role in hepatocellular carcinoma (HCC) patients and TRIM55 overexpression inhibited migration and invasion of HCC tumor cells mediated by the epithelial–mesenchymal transition (EMT) process.Citation5 However, it is still not clear whether TRIM55 plays any roles in lung adenocarcinoma. Therefore, in this study, through experiments on clinical samples of lung adenocarcinoma patients and several lung adenocarcinoma cell lines, we aimed to discover the regulatory role of TRIM55 in lung adenocarcinoma pathogenesis and underlying molecular mechanism. The cancer stem-like cell markers have been believed to be closely related to self-renewal, differentiation potential, resistance to chemotherapy, high tumorigenicity, and EMT process.Citation6 Therefore, two specific stem-like cell markers, ALDH1 and CD133, were measured in this study.
Results
TRIM55 expression was significantly suppressed in lung adenocarcinoma tumor cells
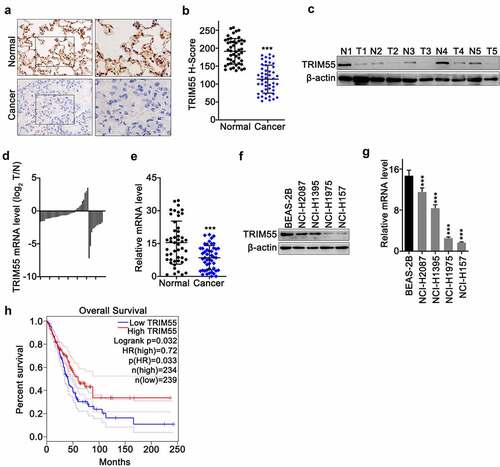
To explore the TRIM55 expression pattern in lung adenocarcinoma tumor cells, immunohistochemical assay on TRIM55 was conducted on lung cancer tissue and matched normal lung tissues. Results indicated that the TRIM55 protein level was significantly lower in cancer tissues (H score comparison p < .0001) []. Western Blot assay on paired samples of lung cancer and normal lung tissue also confirmed the above findings [)]. In addition, qRT-PCR experiments also provided consistent results that the TRIM55 mRNA level was significantly suppressed in lung cancer samples compared to matched normal tissue []. Furthermore, we also demonstrated that the TRIM55 mRNA and protein level was suppressed in several lung cancer cell lines, including NCI-H2087, NCI-H1395, NCI-H1975, and NCI-H157, in comparison with that of normal lung epithelial cell line BEAS-2B []. Subsequently, in order to elucidate the impact of the TRIM55 expression level on lung adenocarcinoma patients’ prognosis, the online platform Gene Expression Profiling Interactive Analysis platform (GEPIA, http://gepia.cancer-pku.cn/index.html) was utilized to compare prognostic differences between lung adenocarcinoma patient groups with TRIM55 high (n = 234) or low expression (n = 239) (LUAD dataset). Results indicated that patients with a high level of TRIM55 exhibited significantly superior overall survival compared with patients with a low TRIM55 expression level (p = .032) ()).
Figure 1. A. IHC experiments to compare protein level of TRIM55 in lung adenocarcinoma tissue and adjacent normal lung tissue. B. Histochemistry score analysis on TRIM55 level between lung adenocarcinoma samples and adjacent normal lung samples. C. Western-blot analysis on TRIM55 protein level between lung adenocarcinoma samples and matched normal tissues. D and E. TRIM55 mRNA expression level comparison between lung adenocarcinoma samples and adjacent normal lung samples. F. Western-blot analysis on TRIM55 protein expression in a normal human lung epithelial cell line (BEAS-2B) and lung adenocarcinoma cell lines (NCI-H2087, NCI-H1395, NCI-H1975, NCI-H157). G. qRT-PCR analysis on TRIM55 mRNA expression in a normal human lung epithelial cell line (BEAS-2B) and lung adenocarcinoma cell lines (NCI-H2087, NCI-H1395, NCI-H1975, NCI-H157). *** p < .001 compared with group BEAS-2B. H. Kaplan–Meier study on the overall survival comparison between the lung adenocarcinoma patient group with high/low TRIM55 expression levels.
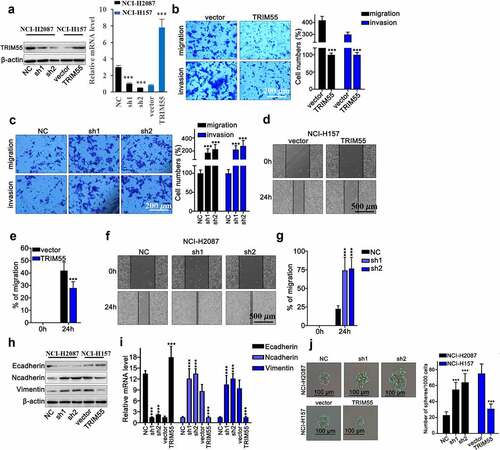
TRIM55 overexpression suppressed the migration and invasion of lung cancer cells
To further evaluate the impact of TRIM55 expression on malignant behavior of lung cancer cells, TRIM55 specific shRNAs (sh1 and sh2) and overexpression vectors were built and their modulative effects on TRIM55 mRNA expression were validated via qRT-PCR [)]. Subsequently, transwell assay was applied, and results demonstrated that sh-TRIM55 significantly enhanced the migrated and invasive capabilities of lung cancer cells and overexpression of TRIM55 markedly inhibited the migration and invasion of lung cancer cells []. Subsequent wound healing assay also provided consistent results that TRIM55 silencing by shRNAs transfection significantly boosted the migration percentage of lung cancer cells []. To understand the underlying mechanism of TRIM55 regulatory effects on tumor cell motility, we then detected mRNA and protein level of several key markers in the cellular epithelial–mesenchymal transition (EMT) process. Results indicated that TRIM55 silencing by shRNAs significantly elevated N-cadherin and vimentin levels, while the E-cadherin level was significantly suppressed (), which indicated that TRIM55 silencing was associated with enhanced mesenchymal phenotype transition of lung cancer cells, which facilitated tumor cell invasion. The cell proliferation was measured through spheroid formation assay. Treatment with sh-TRIM55 significantly increased the proliferation of lung cancer cells, and overexpression of TRIM55 significantly suppressed the proliferation of lung cancer cells [)].
Figure 2. A. TRIM55 mRNA and protein level detection on NCI-H2087 and NCI-H157 cell groups, respectively, transfected with TRIM55-specific shRNA, over-expression vector or control vectors. *** p < .001 compared with group NC or vector. B. Transwell assay detection of tumor cell migration and invasion on NCI-H2087 and NCI-H157 cell groups transfected with TRIM55-specific over-expression vectors or control vectors. *** p < .001 compared with group vector. C. Transwell assay detection of tumor cell migration and invasion on NCI-H2087 and NCI-H157 cell groups transfected with TRIM55-specific shRNAs. *** p < .001 compared with group NC. D–G. Wound healing assay on NCI-H157 and NCI-H2087 cell groups transfected with TRIM55-specific overexpssion vectors or shRNAs, respectively. *** p < .001 compared with group NC. H-I. mRNA and protein level detection of biomarkers of EMT (Ecad, Ncad, Vim) in different NCI-H157 cell groups transfected with TRIM55 specific shRNAs, overexpression vectors or control vector, respectively. *** p < .001 compared with group NC or vector. J. G. Spheroid formation assay was performed to investigate the role of TRIM55 in suppressing tumor cells. *** p < .001 compared with group vector.
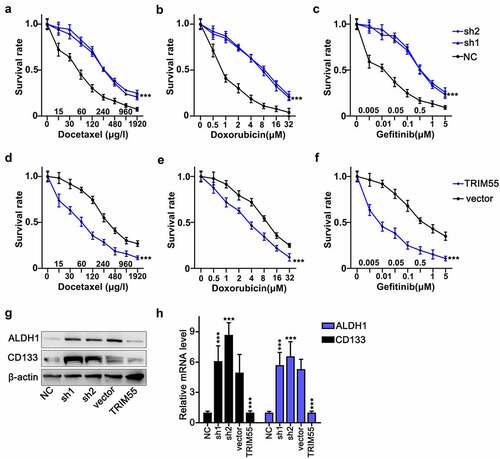
TRIM55 overexpression suppressed tumor cell survivability challenged by chemo-agents and reduced cellular stem-like cell (CSC) biomarkers
Subsequently, we performed cellular survivability assay on NCI-H2087 and NCI-H157 cell lines. These cell groups were challenged by three different kinds of chemo-agents including docetaxel, doxorubicin and gefitinib, each with a different range of concentration. These cancer cells were transfected by TRIM55-specific shRNAs and overexpression vectors, respectively. Results indicated that TRIM55 silencing by shRNAs significantly enhanced tumor cell survivability under the treatment of chemo-agents []. TRIM55 overexpression vector transfection deprived tumor cellular survival superiority compared with tumor cells transfected with control vectors []. In addition, we utilized western blot and qRT-PCR assay to detect tumor stem-like cell (CSC) biomarkers expression, including ALDH1 and CD133, in NCI-H2087 and NCI-H157 cell lines, respectively, transfected with TRIM55 specific shRNAs and overexpression vectors. Results demonstrated that ALDH1 and CD133 were significantly repressed in cell groups transfected with TRIM55 overexpression vectors, while TRIM55 silencing significantly enhanced ALDH1 and CD133 expression compared with the negative control group [].
Figure 3. A–F. Cellular survivability assay on NCI-H157 and NCI-H2087 cell groups challenged by docetaxel, doxorubicin, and gefitinib with different ranges of concentrations. Each cell group was transfected with TRIM55-specific shRNAs or overexpression vectors, respectively. *** p < .001 compared with group NC or vector. G and H. Western Blot and qRT-PCR analysis on ALDH1 and CD133 protein and mRNA expression level in NCI-H157 and NCI-H2087 cell groups, and each cell group was transfected with TRIM55 specific shRNAs or overexpression vectors, respectively. *** p < .001 compared with group NC or vector.
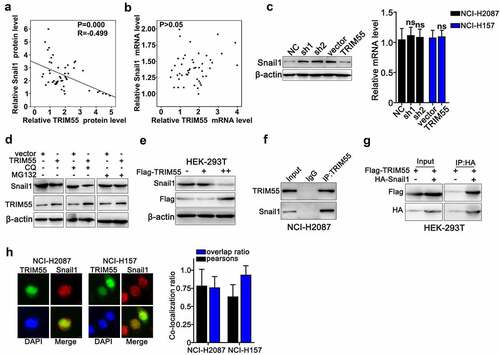
TRIM55 interacted with Snail1 and modulated its protein level via UPP pathway
Moreover, in this study, we made further exploration on TRIM55 molecular regulative pathways. Through western blot and qRT-PCR analysis on lung cancer clinical samples, we discovered that TRIM55 protein expression was negatively correlated with Snail1 protein [)], while we did not find linear correlation between Snail1 and TRIM55 mRNA levels [)]. These results were further confirmed by the following cell line studies, and results indicated that TRIM55 silencing by shRNAs significantly elevated the Snail1 protein level, while TRIM55 overexpression reduced the Snail1 protein level. Notably, neither TRIM55 silencing nor overexpression had significant influences on the Snail1 mRNA expression level [)]. To further clarify the above findings, we utilized autophagy inhibitor (CQ) and proteasome pathway inhibitors (MG132) to explore the role of autophagy and ubiquitin-proteasome pathway (UPP) in Snail1 protein regulation. As a result, TRIM55 overexpression resulted in Snail1 protein level suppression in cell groups treated with CQ, while such suppressive effects were abrogated by MG132 treatment [)], which indicated that TRIM55 overexpression modulated the Snail1 protein level via the UPP pathway. Additionally, after transfecting FLAG-TRIM55 vector into the HEK-293 T cell line, Snail1 protein was suppressed in dose-dependent manner, which depended on the level of FLAG-tagged TRIM55 protein level [)]. A slight higher expression of Snail1 was observed after transfecting the FLAG-TRIM55 vector in the group “+” compared with group “-”. Furthermore, Co-immunoprecipitation assay and immunofluorescence co-localization assay were subsequently performed and results also confirmed direct interaction between two proteins [)].
Figure 4. A and B. Linear regression analysis on the association of Snail1 protein and mRNA expression with TRIM55.C. qRT-PCR and western blot analysis on mRNA and protein expression level of Snail1 on NCI-H2087 and NCI-H157 cell groups, respectively, transfected with TRIM55 specific shRNAs and overexpression vectors or control vectors/negative control. D. Western blot analysis on TRIM55 and Snail1 protein expression on NCI-H2087 cell groups, respectively, transfected with TRIM55 specific overexpression vectors/control vectors, in combinatory treatment of two different UPP pathway inhibitors (CQ and MG132). E. Western blot analysis on the association between Flag-tagged TRIM55 and Snail1 protein expression level in HEK-293 T cell line. ++ indicated higher concentration of Flag-tagged TRIM55 compared to +. F. Co-immunoprecipitation analysis on the interaction of TRIM55 with Snail1 protein in NCI-H2087 cell line. G. Co-immunoprecipitation analysis on the interaction of TRIM55 (Flag-tagged) with Snail1 (HA-tagged) in HEK-293 T cell line. H. Immunofluorescent colocalization analysis on TRIM55 and Snail1 protein in NCI-H2087 and NCI-H157 cell lines after MG132 tretament.
TRIM55 silencing significantly inhibited ubiquitin-mediated protein degradation of Snail1
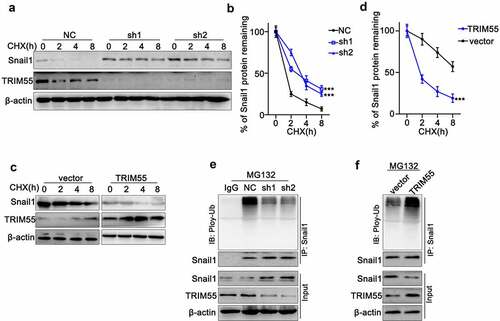
To further explore whether TRIM55 modulated the Snail1 protein level via the ubiquitination pathway, we utilized cycloheximide (CHX) chase assay to detect protein degradation of TRIM55 and Snail1 in NCI-H2087 cell line. As shown in , both TRIM55 and Snail1 protein levels decreased eventually under CHX treatment. Notably, when transfected with TRIM55 specific shRNAs, protein degradation of Snail1 was significantly suppressed. Consistently, when transfected with TRIM55 overexpression vector, the degradation of Snail1 was significantly accelerated []. Moreover, by co-immunoprecipitation assay, we discovered that under the treatment of MG132, NCI-H2087 cell groups transfected with TRIM55 shRNAs exhibited significantly suppressed ubiquitination of Snail1, resulting in increased Snail1 protein level [)]. Meanwhile, when NCI-H157 cell groups were transfected with TRIM55 overexpression vectors, ubiquitination of Snail1 was significantly enhanced and Snail1 protein level exhibited significant decline [)].
Figure 5. A and B. Cycloheximide (CHX) chase assay to detect TRIM55 and Snail1 protein expression level in NCI-H2087 cell groups treated with CHX for different timespan (0 h–8 h). Each cell group was transfected with negative control/TRIM55 specific shRNAs (sh1 and sh2). *** p < .001 compared with group NC. C and D. Cycloheximide (CHX) chase assay to detect TRIM55 and Snail1 protein expression levels in NCI-H157 cell groups treated with CHX for different timespans (0 h–8 h). Each cell group was transfected with control vectors or TRIM55 specific overexpression vectors. *** p < .001 compared with group vector. E. Co-Immunoprecipitation assay to detect ubiquitylation of Snail1 influenced by TRIM55 on NCI-H2087 cell groups treated with MG132. Each cell group was treated with control/TRIM55 specific shRNAs. F. Co-Immunoprecipitation assay to detect ubiquitylation of Snail1 influenced by TRIM55 on NCI-H157 cell groups treated MG132. Each cell group was treated with control vectors/TRIM55 overexpression vectors.
TRIM55 exerted its regulatory effects on tumor malignancy by suppressing the expression of Snail1
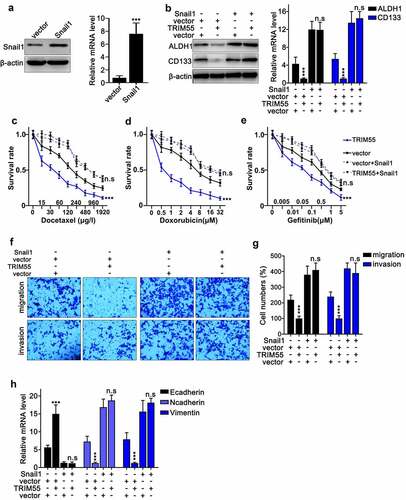
To validate our hypothesis that TRIM55 suppressed the malignant behavior of lung cancer cells by inhibiting the expression of Snail1, we designed the Snail1 specific overexpression vector and validated its promotive effects on Snail1 mRNA and protein expression level [)]. Then, western blot assay was conducted and results indicated that the suppressive effects of TRIM55 overexpression vector transfection on mRNA and protein expression of cancer stem-like cell markers (ALDH1 and CD133) can be abrogated through combinatory transfection of Snail1 overexpression vectors [)]. Additionally, cellular survivability assay also demonstrated that combinatory transfection of Snail1 overexpression vectors reversed the suppressive effects of TRIM55 overexpression vectors on tumor cell survivability under chemo-agents challenges []. Consistently, our subsequent transwell assay also provided evidences that combinatory transfection of Snail1 overexpression vectors abolished the suppressive effects of TRIM55 overexpression vector transfection on tumor cell migration and invasion []. Finally, the qRT-PCR assay on EMT related markers also demonstrated that combinatory transfection of Snail1 overexpression vectors abrogated the significantly regulatory effects of TRIM55 overexpression vectors on E-cadherin, N-cadherin and vimentin mRNA expression [)].
Figure 6. A. Design and modulatory effect evaluation of Snail1 specific overexpression vector on Snail1 mRNA and protein expression level. B. Western blot and qRT-PCR analysis to detect CD133 and ALDH1 protein and mRNA expression in NCI-H2087 cell groups. Each cell group was transfected with Snail1 or TRIM55 specific overexpression vectors. C–E. Cellular survivability assay on the NCI-H2087 cell line challenged by different concentrations of chemo-agents (docetaxel, doxorubicin and gefitinib). Each cell group was transfected with TRIM55 overexpression vector with or without combinatory transfection of Snail1 overexpression vectors. *** p < .001 compared with group vector. F–G. Cellular migration and invasion assay on NCI-H2087 cell lines. Each cell group was transfected TRIM55 overexpression vector, with or without combinatory transfection of Snail1 overexpression vector. *** p < .001 compared with group vector. H. qRT-PCR assay to detect the mRNA expression level of E-cadherin, N-cadherin and vimentin in NCI-H2087 cell line; each cell group was transfected with TRIM55 overexpression vector, with or without combinatory transfection of Snail1 overexpression vectors. *** p < .001 compared with group vector.
Discussion
As previously demonstrated, in this study, we provided consistent evidences that TRIM55 suppression was characteristic in lung adenocarcinoma tumor cells. Both clinical samples and cell line experiments indicated that the TRIM55 expression level was positively correlated with inhibited malignant biological behavior of tumor cells and better overall prognosis of lung adenocarcinoma patients. More detailed analysis indicated that the expression of TRIM55 also inhibited several markers of cancer stem-like cells (CSC), including ALDH1 and CD133. Recent studies have demonstrated that cancer stem cells played a vital role in tumor growth. CD133 is a glycoprotein of five-transmembrane domains, which was originally used for hematopoietic stem cell (HSC) identification,Citation7 and CD133 has been utilized to identify CSC in several malignancies including renal, cerebral, and lung cancers.Citation8–11 CD133 has been shown as a potential prognostic marker for NSCLC.Citation12 As for aldehyde dehydrogenase 1 (ALDH1), it functions as a catalyzer for intracellular aldehyde oxidation, which mediates tumor cell drug-resistance,Citation13 and ALDH1 has also been considered as an important marker for CSC in breast, colorectal and lung cancer.Citation14–16 Our study provided novel evidence that TRIM55 suppressed ALDH1 and CD133, which could suggest new clues in targeted intervention against CSCs in lung adenocarcinoma patients. More detailed experiments are warranted for further thorough investigation.
Notably, in this study, we discovered that TRIM55 exerted its tumor suppressive function via the UPP pathway through increasing the degradation of Snail1 protein. As is well known, ubiquitination plays a vital role in multiple biological processes including cell cycle,Citation17,Citation18 cellular apoptosis,Citation19,Citation20 and DNA damage repairing.Citation21 Ubiquitination dysregulation of several signaling pathways including PI3K-AKT-mTOR, RAS-RAF-MEK-ERK and EGFR has been proved to contribute in NSCLC pathogenesis.Citation22 Moreover, as a member of Snail superfamily, Snail1 is a zinc-finger transcription factor that played a central role in promoting the EMT process.Citation23 Snail1 has also been shown to regulate CSC phenotype in lung cancer cells.Citation24 Our study provided consistent results that as the downstream molecular target of TRIM55, Snail1 protein degradation via the UPP pathway might serve as a potential intervention approach for lung adenocarcinoma patient treatment.
However, it is worth mentioning the limitations existed in this study. First, our study was based on single-centered clinical cohort and in-vitro cell line experiments, more expanded multi-centered validation clinical studies as well as in-vivo studies utilizing TRIM55 knock-out animal models are warranted for the confirmation of our findings. Moreover, due to the complexity of the signal regulation network, whether TRIM55 modulated other crucial downstream molecules in lung adenocarcinoma cells via other pathways remains unanswered. More thorough experiments based on knock-out cell line and animal models combined with high-throughput bioinformatic analysis are required to fully unveil the mystery.
In this study, through integrative analysis of clinical samples as well as lung cancer cell models, we demonstrated for the first time that TRIM55 served as an interesting tumor suppressor in lung adenocarcinoma pathogenesis. Subsequent experiments provided evidences that TRIM55 exerted its anti-tumor effects by enhancing the protein degradation of Snail1 via promoting the ubiquitination pathway. Our findings contributed novel evidence of lung adenocarcinoma molecular pathogenesis, which would enlighten future exploration and potential therapeutic intervention development.
Material and methods
Patient recruitment and sample collection
Lung adenocarcinoma patients newly diagnosed in our clinical center between June 2019 and June 2020 were retrospectively enrolled in this study. No prior surgical treatment, radiotherapy or chemo-therapy was conducted before sample collection. Clinical samples of lung cancer tissues and adjacent normal tissues were gathered post-surgical treatment. Samples were stored in liquid nitrogen for the following processing procedures. All patients’ informed consents were obtained for this research, and this research was approved by the ethical reviewing board of Fujian Provincial Hospital.
RNA extraction and qRT-PCR experiments
Cell line samples and clinical tissue RNAs were extracted by RNAiso Plus agent (TaKaRa, Dalian, China) according to the standardized protocol provided by the manufacturer. After RNA level quantification, reverse transcription polymerase chain reaction was performed to generate cDNAs. The levels of mRNA expression were quantified by real-time PCR qRT-PCR reaction with condition setting as 95°C for 25s, 56°C for 30s, and 72°C for 90s with total cycles of 45. Primers used in this study were listed in detail (supplementary table 1).
Cell line culturing
Lung cancer cell lines (NCI-H2087, NCI-H1395, NCI-H1975, NCI-H157) and human normal lung epithelial cell (BEAS-2B) were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). RPMI-1640 medium plus 10% fetal bovine serum (FBS; Hyclone, South Logan, UT, USA). 100 IU/mL penicillin with 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA, USA) were used for culture, with environmental setting under 37°C, with 5% CO2.
Lentiviral transfection
The pLVX lentiviral vector was purchased from Clontech (USA). Polymerase chain reaction (PCR) was performed utilizing templates of TRIM55 specific shRNA and full-length cDNA sequences of TRIM55 and Snail1. First, PCR products were purified using 1% agarose gel and double digested by BamHI and EcoRI as well as empty pLVX vectors. Then, ligation reaction between vector and the purified PCR products was performed by T4DNA ligase. The ligation product was subsequently utilized for the E.coli DH5α competent cells transformation. Third, cell clones were subsequently placed into the LB plate with ampicillinum and incubated at 37°C overnight. Positive clones were then collected, and the plasmid was extracted and sequenced (Shanghai Invitrogen Biotech Co., Ltd). Subsequently, the lentiviral vectors with TRIM55/Snail1 shRNA/cDNA and control vectors were packaged and added into lung cancer tumor cell groups with multiplicity of infection (MOI) of 15 (control group with empty vectors, overexpression group with TRIM55/Snail1 cDNA vectors, suppressed group with TRIM55 shRNA vectors), and the vectors titer was set as 2 × 109/ml. Then, the fluorescent protein expression level was detected and transfection efficiency was evaluated 24–48 h post-transfection.
Western Blot
The 2 x 106 cells per cell group was first washed twice with cold PBS, resuspended and then treated with ice-cold cell lysis buffer RIPA agent (Beyotime, Shanghai, Chian) to extract total protein from the samples. Then, the BCA protein assay kit was used for sample protein level quantification. Then, protein samples were first separated by SDS-PAGE and 10% separating gels and samples were subsequently electroblotted onto PVDF membranes (Immobilon-P; Millipore, Billerica, MA). After blocking in Tris buffer (50 mM Tris, pH 7.5) containing 5% skim milk, the membranes were then incubated overnight at 4°C with primary antibodies. After rinsing with the Tris-buffered salin and Tween buffer solution (TBST; Sigma-Aldrich, St. Louis, MO, USA), they were then incubated with the secondary antibody for 2 h. Chemiluminescence was used to expose the protein bands on the membrane.
Cellular migration assay
For transwell study, each group of cell samples was first placed with a density of 6 × 104 in the upper chamber (8-μm) (Corning, Lowell, MA, USA). The bottom chamber was added with complete medium. Then, after incubation for 48 h under 37°C, 5% CO2 condition, cells in the lower chamber were fixed and stained in crystal violet solution for observation using a microscope (Olympus). For calculation of the number of penetrating cells, twelve randomly chosen fields per sample were selected.
For wound healing assay, each group of cells was correspondingly seeded into the 6-well plate. Cellular monolayers were scraped with a sterile micropipette tip. The wounded monolayers were then washed with phosphate buffer solution (PBS) to remove debris. The distance between the two edges of the wound was calculated at three different positions. The distance between the two edges should be measured again after 48 h of incubation.
Cellular survivability assay
Each of the cell group was respectively challenged with different concentrations of docetaxel, doxorubicin, and gefitinib for 4 h, and then, cells were further washed by PBS and treated by 75% ethanol at −20°C overnight. Then, at room temperature, these cells were treated with Annexin V-FITC (Thermo Fisher Scientific) and propidium iodide (PI) for 20 min. Afterward, flow cytometric analysis was performed (FACScan™, BD Biosciences, Franklin Lakes, NJ, USA) to detect apoptotic cells.
Co-immunoprecipitation (Co-IP)
HEK-293 T, NCI-H2087, and NCI-H157 cells transfected with Flag-tagged TRIM55 or HA-tagged Snail1 were cultured in 10-cm dishes. Then, cell samples were treated in immunoprecipitation buffer (containing 20 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA, pH 8.0; 1% NP-40; 1× Protease and Phosphatase Inhibitor). Then, 1 μl FLAG/HA antibody or IgG and 10 μl of protein A/G magnetic beads (pre-washed with lysis buffer) were added and incubated for 5 h under 4°C condition. Afterward, the beads were sequentially rinsed with wash buffer. Finally, proteins were eluted from the beads using 2× SDS loading buffer and subsequently treated at 95°C for 10 min prior to SDS-PAGE and immunoblotting with FLAG and HA antibodies.
Immunofluorescence (IF)
Cells samples were first washed with phosphate-buffered saline (PBS), and then, paraformaldehyde was used for cell fixation for 15 min. Afterward, cells were permeabilized with 0.5% NP-40 for 20 min, and then, cells were blocked with 5% bovine serum albumin for 30 min. Subsequently, cells were incubated with primary antibody for 2 h at RT and subsequently incubated with fluorescein conjugated secondary antibody for 1 h. Finally, the DAPI and confocal microscopy were used to stain and image the slides, respectively.
CHX-chase assay
CHX-chase assay was conducted via CHX (Selleck Chemicals). The cells in each group were mixed with 12.5 μg/mL of CHX, and the expression of Snail1 and TRIM55 protein was detected by western blot analysis at 0, 2, 4, 8 h.
Ubiquitination assay
Cells were transfected with ubiquitin (Ubbiotech, Changchun, China) and TRIM55 overexpression vectors or shRNAs by jetPRIME (Polyplus, Strasbourg, France). Then, 20 μM of MG132 (Selleck Chemicals, Houston, TX, USA) was added to the medium after 36 h of transfection. Then, protein from each cell group was extracted for subsequent western blot analysis. Cell lysates were immunoprecipitated (IP) with the labeled antibodies and incubated at 4°C overnight. Then, the eluted proteins were determined by western blotting.
Spheroid Forming Assays
The cells (300 cells/well) were cultured with spheroid media after transfection treatments in the 96 well plates for 5 d. PBS was used to reduce evaporation. The spheroids were defined as rounded aggregates of cells with a smooth surface . Loose cellular aggregates of well-defined cells were not viewed as spheroids.
Statistical analysis
Statistical analysis was conducted by software package (SPSS 21.0 for Windows; IBM-SPSS Inc., Chicago, IL). Data were presented as the mean SD of three independent experiments. Statistical test of differences between numerical data was performed by a standard t-test. Pearson test was conducted to compare gene correlation. P < .05 was considered to indicate statistical significance.
Ethical approval
All patients’ informed consents were obtained for this research, and this research was approved by the ethical reviewing board of Fujian Provincial Hospital.
Supplemental Material
Download MS Word (13.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Siegel RL, Miller KD, Fuchs HE, Jemal A. 2021 Jan. Cancer Statistics. CA Cancer J Clin. 71(1):7–33. 10.3322/caac.21654.
- Williams DE, Pairolero PC, Davis CS, Bernatz PE, Spencer Payne W, Taylor WF, Uhlenhopp MA, Fontana RS. Survival of patients surgically treated for stage I lung cancer. J Thorac Cardiovasc Surg. 1981 Jul;82(1):70–76. doi:10.1016/S0022-5223(19)39389-4.
- Wang X, Adjei AA. 2015 Jun. Lung cancer and metastasis: new opportunities and challenges. Cancer Metastasis Rev. 34(2):169–171. 10.1007/s10555-015-9562-4.
- Perera S, Holt MR, Mankoo BS, Gautel M. 2011 Mar 1. Developmental regulation of MURF ubiquitin ligases and autophagy proteins nbr1, p62/SQSTM1 and LC3 during cardiac myofibril assembly and turnover. Dev Biol. 351(1):46–61. 10.1016/j.ydbio.2010.12.024.
- Li X, Huang L, Gao W. Overexpression of Tripartite Motif Conaining 55 (TRIM55) Inhibits Migration and Invasion of Hepatocellular Carcinoma (HCC) Cells via Epithelial-Mesenchymal Transition and Matrix Metalloproteinase-2 (MMP2). Medical Sci Monitor: Int Medical J Exp Clinical Res. 2019 Jan 27;25:771–777. doi:10.12659/msm.910984.
- Leng Z, Xia Q, Chen J, Li Y, Xu J, Zhao E, Zheng H, Ai W, Dong J. 2018. Lgr5+CD44+EpCAM+ Strictly Defines Cancer Stem Cells in Human Colorectal Cancer. Cell Physiol Biochem. 46(2):860–872. 10.1159/000488743.
- Irollo E, Pirozzi G. CD133: to be or not to be, is this the real question? Am J Transl Res. 2013 Sep 25;5(6):563–581.
- Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C, De Maria R, et al. 2008 Mar. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 15(3):504–514. DOI:10.1038/sj.cdd.4402283.
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. 2004 Nov 18. Identification of human brain tumour initiating cells. Nature. 432(7015):396–401. 10.1038/nature03128.
- Bussolati B, Bruno S, Grange C, Buttiglieri S, Deregibus MC, Cantino D, Camussi G. 2005 Feb. Isolation of renal progenitor cells from adult human kidney. Am J Pathol. 166(2):545–555. 10.1016/s0002-9440(10)62276-6.
- Florek M, Haase M, Marzesco AM, Freund D, Ehninger G, Huttner WB, Corbeil D. 2005 Jan. Prominin-1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res. 319(1):15–26. 10.1007/s00441-004-1018-z.
- Qu H, Li R, Liu Z, Zhang J, Luo R. Prognostic value of cancer stem cell marker CD133 expression in non-small cell lung cancer: a systematic review. Int J Clin Exp Pathol. 2013;6(11):2644–2650.
- Yoshida A, Hsu LC, Davé V. 1992. Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme. 46(4–5):239–244. 10.1159/000468794.
- Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. 2009 Apr 15. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 69(8):3382–3389. 10.1158/0008-5472.Can-08-4418.
- Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA, et al. 2009 Mar. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Molecular Cancer Res: MCR. 7(3):330–338. DOI:10.1158/1541-7786.Mcr-08-0393.
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. 2007 Nov. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 1(5):555–567. DOI:10.1016/j.stem.2007.08.014.
- Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, Battaglia V, Vivat V, Sippl W, et al. 2012 Apr 20. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 19(4):467–477. 10.1016/j.chembiol.2012.02.007.
- Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez-Martin N, Matsumoto ML, Dixit VM, et al. 2017 Nov 2. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell. 171(4):918–933.e20. 10.1016/j.cell.2017.09.040.
- Heger K, Wickliffe KE, Ndoja A, Zhang J, Murthy A, Dugger DL, Maltzman A, de Sousa E Melo F, Hung J, Zeng Y, et al. 2018 Jul. OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature. 559(7712):120–124. DOI:10.1038/s41586-018-0256-2.
- Zou Q, Jin J, Hu H, Li HS, Romano S, Xiao Y, Nakaya M, Zhou X, Cheng X, Yang P, et al. 2014 Jun. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol. 15(6):562–570. DOI:10.1038/ni.2885.
- Zeman MK, Lin JR, Freire R, Cimprich KA. 2014 Jul 21. DNA damage-specific deubiquitination regulates Rad18 functions to suppress mutagenesis. J Cell Biol. 206(2):183–197. 10.1083/jcb.201311063.
- Fan Q, Wang Q, Cai R, Yuan H, Xu M. 2020. The ubiquitin system: orchestrating cellular signals in non-small-cell lung cancer. Cell Mol Biol Lett. 25(1):1. 10.1186/s11658-019-0193-6.
- Barrallo-Gimeno A, Nieto MA. 2005 Jul. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development (Cambridge, England). 132(14):3151–3161. 10.1242/dev.01907.
- Wang H, Zhang G, Zhang H, Zhang F, Zhou B, Ning F, Wang H-S, Cai S-H, Du J. Acquisition of epithelial-mesenchymal transition phenotype and cancer stem cell-like properties in cisplatin-resistant lung cancer cells through AKT/β-catenin/Snail signaling pathway. Eur J Pharmacol. 2014 Jan 15;723:156–166. doi:10.1016/j.ejphar.2013.12.004.
