Abstract
Cell cycle arrest, senescence and apoptosis are commonly regarded as the major tumor suppression mechanisms of p53. However, accumulating evidence indicates that loss of these canonical functions is not sufficient for tumor formation, highlighting the complexity of p53-mediated tumor suppression. PCDH10 belongs to a proto cadherin protein family and is a potential tumor suppressor protein as the dysregulation of PCDH10 gene frequently existed in multiple human tumors. Here, we found that PCDH10 is a transcriptional target of p53 and that the levels of PCDH10 expression can be induced by wild type p53 but not mutant p53 in a number of human cancer cell lines. Moreover, we identified a p53 consensus binding site located in the PCDH10 promoter region that is responsive to p53 regulation. Although upregulation of PCDH10 has no obvious effect on growth arrest or apoptosis in human cells, PCDH10 exhibits inhibitory roles in cancer cell motility and cell migration. These results suggest an important role of p53 in regulating tumor cell migration through activating PCDH10 expression and support the notion that non-canonical activities of p53 may contribute to its tumor suppressor function in vivo.
Introduction
The tumor suppressive function of p53 has never been contested and the “three major classic” functions of p53, cell-cycle arrest, apoptosis, and senescence, have been ascribed to the primary actions of p53 in tumor prevention.Citation6 However, an unanticipated and possibly intriguing work challenged this dogma by showing tumor free mice which carried p53 mutations leading to the disruption of all 3 fundamental functions of p53.Citation8 Certainly this work implied that the molecular mechanism of p53-mediated tumor suppression is far beyond being understood fully; this work as well generated the motivation to reexamine the p53 non-canonical functions that have been proposed from numerous former studies and to carefully re-analyze the p53 transcriptional program, for identifying novel targets, which may contribute as critical components to the tumor suppressive role of p53. In fact, a growing number of non-canonical p53 target genes have been added to the p53 transcription program, and these genes encode for proteins ranging over a broad spectrum of cellular functions in cell metabolism, stem cell differentiation, tumor cell invasion, and yet unknown functions.Citation1
The aberrantly increased tumor cell mobility contributes to tumor invasiveness and metastasis potential, which account for the most of the resultant mortality of cancer patients.Citation15,17 This has not escaped the attention of p53 and the accumulating evidence recently linked p53 to key stages of metastatic progression, including cell adhesion, cell migration and cell invasion.Citation17 It has been observed for long time that the deletion of p53 allows fibroblast to migrate faster in wound healing assays, potentially due to the altered cell morphology and polarity.Citation14,21 This observation is not only limited to fibroblast, but also expanded to multiple cancer epithelial cell lines.Citation14,21 However, the underlying molecular mechanism of this regulation is, at least partially, kept unknown and requires delineated studies. It was discovered that the loss of p53 activates Rho signaling pathway, a well-established pathway to influence the way in which tumor cells migrate, to promote cell migration.Citation4 In addition, p53 also induces the degradation of slug, a key transcript factor in regulating epithelial-mesenchymal transition (EMT), to suppress tumor cell invasion.Citation17,26 Further, tumor derived mutant p53 enhances integrin and epidermal growth factor receptor (EGFR) trafficking therefore promotes cell migration and invasion.Citation14,16 Numerous proteins have been linked to the p53's ability in controlling cell migration and metastasis, such as Rho GTPases (Cdc42,RhoE,Caldesmon), EMC(extra cellular matrix)protein MMP-2, growth factor EGFR, and PTEN etc.; however, most of these mediators of p53 in regulating cell migration and metastasis are repressed targets of p53, secondary transcriptional targets of p53, or regulated molecules by p53 in a transcriptional-independent manner.Citation14 The direct transcriptional targets of p53 that functions as cell migration and invasion regulators are scarce. In this study, we report that Protocadherin 10(PCDH10) is a direct p53 transcriptional target and regulate tumor cell motility and further cell migration. PCDH10 is a protocadherin protein and protocadherins are a group of transmembrane proteins that belong to the cadherin superfamily.Citation24 PCDH10 contains 6 cadherin domains and one transmembrane region in the carboxyl-terminus ().Citation7 PCDH10 has been proved essential for neuronal development.Citation24 Moreover, PCDH10 is designated as an autism-spectrum disorder gene.Citation23 Recently, the strong link has been built between PCDH10 gene and tumor development. PCDH10 inactivation through genetic deletion or promoter methylation has been presented in multi-kind of human cancers including colon cancer, gastric cancer, prostate cancer and lymphoid malignancies etc.Citation10–12,Citation18–20,Citation30 Moreover, that the methylation level of PCDH10 correlates with clinical parameters manifests the significance of PCDH10 in the tumor progression process.Citation29 For example, methylation status of PCDH10 is associated with poor prognosis in patients with bladder cancer.Citation12 Similarly, downregulation of PCDH10 expression correlates with malignant behavior and poor prognosis in human bladder cancer.Citation13 PCDH10 downregulation is widely involved into many types of cancers, suggesting PCDH10 as a potential novel tumor suppressor. The cellular functions of PCDH10 have been reported highly diversified and probably differential in tissue specific manner. The re-expression of PCDH10 could induce cell cycle arrest, apoptosis, and inhibit cell clonogenicity, though with underlying working mechanism undiscovered; however these proposed functions substantiate the tumor suppressive role of PCDH10.Citation19,20,Citation28,29 The basic transcriptional regulatory mechanism of PCDH10 gene is absent from the previous studies except for the profound epigenetic studies from tumor samples.
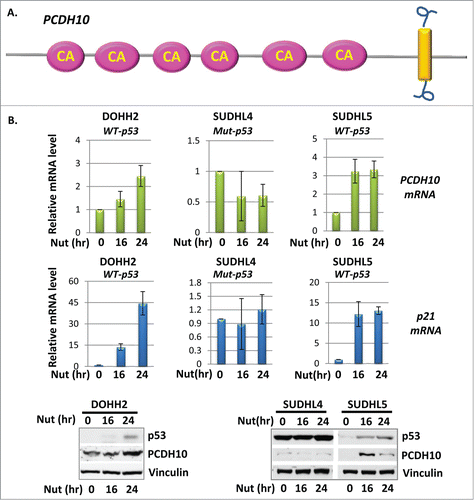
Figure 1. PCDH10 is induced by p53 in B cell lymphoma cells. (A) Diagram of PCDH10 proto cadherin protein. The pink oval represents for cadherin (CA) domain and the yellow rectangular represents for the transmembrane domain. (B) The expression level of PCDH10 and p21 in DOHH2, SUDHL4 and SUDHL5 B cell lymphoma cells. Cells were treated with 10 μM Nutlin-3A for 0, 16 and 24 hours and total RNA were extracted, and cDNA was prepared by reverse transcription. mRNA abundance for PCDH10 and p21 was assessed using quantitative real time PCR. The western blot analysis of PCDH10 protein expression was done at indicated time points.
In this study, we find that p53 is a bona find transcription factor of PCDH10 gene and serves as a generic transcription regulatory factor in tested multiple tumor cell lines. The exogenous overexpression of PCDH10 does not induce significant apoptosis or cell growth arrest. Instead, PCDH10 performed non-canonical function of p53. We report here that PCDH10 restrains cancer cell migration in non-small cell lung cancer cells, potentially contributing to the tumor suppressive function of p53.
Results
PCDH10 is induced by p53 in B-cell lymphoma cells
The linkage between p53 and PCDH10 was firstly built up through a RNA sequencing screening project performed in H1299 non-small cell lung cancer cells with mouse p53 exogenously overexpressed. PCDH10 mRNA level was up-regulated at multiple time points upon p53 overexpression, therefore PCDH10 gene was suspected as a p53 inducible gene. To confirm that PCDH10 is a truly p53-inducible gene, we examine PCDH10 expression level in B-cell lymphoma cell lines, in which p53 gene alterations are relatively rare, in response to p53 activation. In DOHH2 B-cell lymphoma cells containing endogenous wild type p53, the expression level of p21, a classic p53 target gene, was stimulated by multi-tenfold upon p53 activation when cells were incubated with 10 uM Nutlin-3a for 16 and 24 hours (). PCDH10, probably as a weak p53 induced gene target, has been observed with 1.5 and 2.5 times of induction at the similar time points (). Similarly, PCDH10 mRNA level increased in a time-dependent manner, as well as other p53 gene target p21 in SUDHL5 wild type p53 containing B-cell lymphoma cell line with Nutlin-3a treatment (). In contrast, PCDH10 expression level remains at the basal level in mutant p53 containing SUDHL4 lymphoma cells under the similar treatment of p53 activating reagent (). Similar to PCDH10 mRNA, the protein levels of PCDH10 were augmented upon Nutlin-3A treatment in DOHH2 and SUDHL5 cells but not in SUDHL4 cells ().
PCDH10 is induced by p53
The previous data showed that the activation of endogenous p53 could induce PCDH10 gene expression. We next explore if PCDH10 gene induction will recapitulate when p53 is exogenously expressed. The induction fold of PCDH10 gene upon p53 activation in lymphoma cells in nature is modest. To mimic physiological condition and maintain sustained p53 activation for extended time period, we established an inducible cell line expressing tetracycline regulated wild-type mouse p53 in ptripz vector. These cells express relatively low level of p53 upon addition of tetracycline in short time period and p53 activation could continue for days without inducing apoptosis for cell to demise until late time point. Therefore, the activation of p53's “weak," or low DNA-binding targets could be amplified more than that under transient p53 overexpression condition. As expected, enforced wild-type p53 expression activates p21 for hundreds of fold when p53 expression was induced by addition of 0.5 ug/ml tetracycline for 16 hour, 24 hour and 48 hours (data not shown). At same indicated time point, PCDH10 mRNA was induced to 15, 20, and 22 folds, demonstrating a dramatic activation by p53 (). As PCDH10 gene showed quite abundant basal level in H1299 cells, we performed western blot analysis in H1299 cells with p53 induction. Similar to PCDH10 mRNA, PCDH10 protein level increased in a p53 dose-dependent manner along with other classic p53 target genes, including p21, PUMA and MDM2().
Figure 2. p53 induces PCDH10 expression. (A) PCDH10 mRNA expression in tet-on-p53 H1299 cells. Tet-on-p53 H1299 cells were induced by addition of 0.5 ug/ml tetracycline (Tet) for 0, 16, 24, and 48 hours. Total RNA were extracted, and cDNA was prepared by reverse transcription. mRNA abundance for PCDH10 was assessed using quantitative real time PCR. (B) Tet-on-p53 H1299 cells were induced by 0.5 ug/ml tetracycline(tet) at the indicated time point and the total cell extracts were analyzed by Western blotting using antibodies against p53(CM5), PCDH10, Mdm2, Puma, p21, and β-actin. (C) PCDH10 expression level in MEF cells. PCDH10 mRNA levels were analyzed in p53+/+ and p53−/- MEFs either left untreated or treated with 1 μM Dox (doxorubicin) for 12 hours via RT-PCR. (D,E) The expression level of PCDH10 in H460 and wm2664 cells. H460 and wm2664 cells were treated with doxorubicin at 1 uM for 0, 12 , 24, and 36 hours. Total RNA were extracted, and cDNA was prepared by reverse transcription. mRNA abundance for PCDH10 was assessed using quantitative real time PCR.
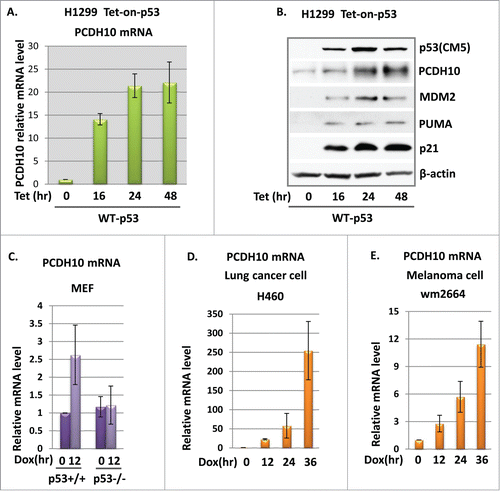
To confirm whether PCDH10 induction by p53 is limited to certain cell type or not, we examined PCDH10 mRNA expression in response to p53 activation in other types of cancer cell lines containing wild type p53. PCDH10 has been reported to be silenced by promoter methylation to differential extent in multiple human cancers. Therefore PCDH10 induction upon p53 activation is not expected entirely universal. For example, HepG2 (Human hepatocellular carcinoma) cells have been tested for PCDH10 transcripts by real time PCR and PCDH10 mRNA was undetectable probably due to hypermethyletion status of its promoter region. In lung cancer cell line H460 and melanoma cell line wm2664, both of which bear wild type p53, PCDH10 mRNA levels were measured by real-time PCR and exhibited measurable quantity. In both cell lines, PCDH10 mRNA level were induced at multiple time points when cells were treated with 1 uM doxorubicin for 12, 24 and 36 hours for p53 activation (). In contrast, in AU48 renal carcinoma cells and A375 melanoma cells, PCDH10 expression failed to show even slight induction in response to p53 activation (data not shown). Collectively, p53 activates PCDH10 gene expression in a cell line or tissue dependent manner. To explore if the regulation of PCDH10 expression by p53 is limited to cancer cell lines, we treated wild type and p53 knockout MEFs with 1 uM doxorubicin for 12 hours and examined PCDH10 mRNA level. Although only 2-fold induction of PCDH10 was observed in p53 wild type MEF upon stress, the silent induction of PCDH10 in p53 null MEF cells demonstrated that PCDH10 is modulated by p53 in normal fibroblast cells (). Collectively, our data indicated that PCDH10 is a novel induced gene target of p53.
PCDH10 is a transcriptional target of p53
In turn, we investigate if p53 directly regulates PCDH10 gene expression as its transcriptional factor. 3000 base pair genomic DNA sequence upstream of transcription start site (TSS) site of PCDH10 gene was carefully examined for p53 binding sites (p53 BS) and 4 potential p53 BSs were identified and their locations were illustrated in . To verify the binding of p53 protein to the PCDH10 promoter, we performed chromatin-immunoprecipitation (CHIP) assay in H1299 cells transfected with wild type or mutant p53 expression vectors, followed by quantitative real-time PCR (qPCR) amplification of the pulled-down DNA fragments. The relative wild type p53 enrichment, but not mutant p53 (R175H) enrichment, was detected at one of the p53 potential BSs, p53 BS-3, 1.2 kb upstream of TSS, while no enrichment was detected at the other 3 potential p53 BS, providing that p53 indeed binds to the PCDH10 promoter ().
Figure 3. PCDH10 is a transcriptional target of p53. (A,B) ChIP analysis of p53 enrichment at the promoter region of PCDH10. (A) Upper panel: the schematic representation of human PCDH10 gene and its promoter. The four potential p53 binding sites (p53 BS) are located at 520 bp, 1200 bp, 1800 bp, 2200 bp upstream of TSS (transcription start site) of PCDH10 gene. Lower panel: the schematic representation of luciferase construct pLucPCDH10 containing the potential p53 binding site p53 BS3. (B) ChIP-qPCR analysis of p53 enrichment at the 4 potential binding sites in the promoter regions of PCDH10 in H1299 cells expressing wild type p53 and R175H mutant p53 protein. (C) Gel shift assay shows p53 binding on oligonucleotide containing p53-binding site p53 BS-3 in the PCDH10 promoter region. The DNA binding activity of purified p53 protein presents in the radiolabeled PCDH10 probe and p53 protein complex. Specificity of the binding was confirmed by competition with non-radiolabeled PCDH10 mRNA levels. (D) p53 activates luciferase activity of reporter construct containing p53 BS-3 in PCDH10 promoter. SAOS2 cells were transfected with 500 ng PCIN4 empty vector (EV), titrated increasing amounts(100 ng, 200 ng, 500 ng) of PCIN4-WTp53 expression vector, or 500 ng PCIN4-R175H p53 mutant vector along with luciferase construct pLucPCDH10 for 24 hours before measuring luciferase activity by luciferase reporter gene assays (Promega, Dual-Luciferase Assays).
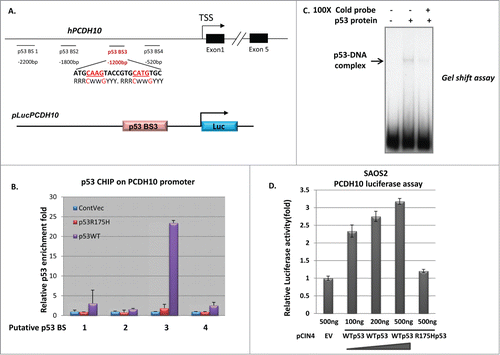
To validate the binding specificity of p53 protein to the identified PCDH10 promoter region, we next performed gel mobility shift assay to determine if p53 is able to bind to the PCDH10 promoter region containing p53 BS-3 in this reconstituted in vitro system, as compared to CHIP assay in whole cell crude mixture. We amplified a 170 bp DNA fragment, covering p53 BS-3 and locating 1200 bp upstream from TSS, from genomic DNA. Using Flag/M2 immunopurified human p53 protein, we observed a super shift of this specific PCDH10 promoter fragment (). In addition, binding of p53 to the radiolabled PCDH10 fragment was outcompeted by the excessive amount (100×) of addition of same cold probe (). Taken together, these results indicate that PCDH10 gene is a p53 target and the consensus p53 binding site, p53 BS-3, is responsible for PCDH10 gene activation.
Since p53 is a transcription factor, we tested whether p53 was able to activate transcription through the PCDH10 promoter using luciferase assay. Luciferase constructs pLuc-PCDH10, containing the verified p53 binding site p53 BS-3 and flanking over 600 base pair nucleotides in PCDH10 promoter region, was generated as illustrated in (lower panel). Co-transfection of pLucPCDH10 with wild type p53 expression plasmid into p53 null SAOS2 cells increased the luciferase activity in a p53 dosage dependent manner (). In contrast, the co-transfection of mutant p53 R175H, which is defective in DNA binding, failed to do so (). Similar stimulated luciferase activity by wild type p53 was also observed in H1299 cells. This result demonstrated that p53 activated gene expression of PCDH10 through the promoter region as well as confirmed that the potential p53 binding site is in the predicted region, p53 BS-3, of PCDH10 promoter region.
PCDH10 does not convey the classic functions of p53
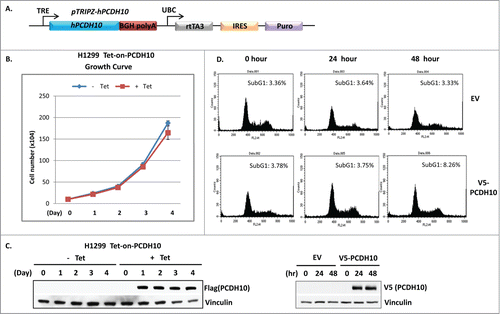
The dysregulation of PCDH10 gene in multiple tumor samples and cell lines implied that it might play a suppressive role in tumorigenesis. As a potential p53 target, we are interested in exploring whether PCDH10 plays any role in the p53 mediated canonical cellular functions, such as cell growth arrest and apoptosis. In support of its prospective tumor suppressive role, PCDH10 has been proposed and demonstrated to induce G1 cell cycle arrest to suppress tumor cell growth in myeloma cells.Citation9 Since p53 is the major cell cycle arrest effector in G1 phase and the linkage has been built up between p53 and PCDH10 from the preceding evidence, we next investigated whether PCDH10 virtually inhibited tumor cell growth as a general phenomenon. To this end, we established a human PCDH10 containing tetracycline-inducible cell line in H1299 cells as illustrated in . After the treatment of 5 ug/ml tetracycline for PCDH10 induction, we extracted cell lysates on every day of 4 successive days and lysates were subjected to western blotting analysis. The sustained and equal protein expression of PCDH10 gene was perceived at each indicated time point (). From the cell growth curve, it is fairly apparent that the cell proliferation of PCDH10 induced H1299 cells showed insignificant difference in comparison with that of control H1299 cells (). In addition, H1299 cells were transiently transfected with V5-tagged PCDH10 expression vector or empty control vector and the transfected cells were subjected to FACS analysis to measure subG1 population at 24 hours or 48 hours post transfection. Compared to empty vector transfection, PCDH10 overexpression does not exhibit apparent impact on the abundance of subG1 peak at any time point, implying that PCDH10 does not modulate p53 induced apoptosis (). Together, PCDH10 expression does not exhibit any impact onto p53 regulated classis cellular functions.
Figure 4. PCDH10 does not convey the classic functions of p53. (A) The schematic representation of the construct design of Ptripz human PCDH10 plasmid used to make tet-on-PCDH10 H1299 cells. The image is modified from pTripz vector map on GE Dharmacon website. (B and C) Up-regulation of PCDH10 does not affect cell growth rate. (B) Cell growth rate analysis of control and PCDH10 induced H1299 cells.Tet-on-PCDH10 H1299 cells were induced or not by addition of 5 ug/ml tetracycline for 4 successive days and cell numbers were counted on each day for 4 d for growth curve. (C) The western blot analysis of PCDH10 protein expression was done at indicated time points after the addition of tetracycline. (D) Upregulation of PCDH10 does not induce apoptosis.H1299 cells were transfected with control empty vector (EV) or V5 tag containing PCDNA3 p53 expression vector( V5-PCDH10). The cells were harvested at 0, 24, and 48 h post transfection, fixed in cold 80% methanol, stained with propidium iodide and subjected to DNA content analysis by flow cytometry. Cells with sub-G1 DNA content were scored as apoptotic cells.
A potential role of p53 in regulating cell migration
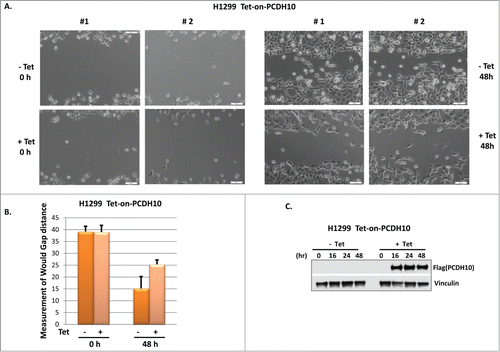
PCDH10 belongs to the protocadherin gene family, a subfamily of the cadherin superfamily, and its role is presumably involved in the establishment of cell-cell connections and in the control of cell migrations.Citation7 Using our established PCDH10 inducible H1299 cells, we are able to test the hypothesis that PCDH10 influences tumor cell migration without the interference of lipofectamine induced cell toxicity. As shown in , PCDH10 induced cells exhibit visibly and measurably slower migration rate than control cells in a wound healing assay with a statistical significance, suggesting that PCDH10 potentially plays an important role in tumor cell mobility and migration. So far, the functional assays we have exploited pointed out that PCDH10 potentiate the cell migratory capability, in accordance with the frequent loss of PCDH10 gene expression in relatively later stage of tumor progression rather than in initiation step.
Figure 5. PCDH10 inhibits cell migration in wound healing assay. (A) Tet-on-PCDH10 H1299 cells were plated into 6 well plates at 30% confluence. After 24 hours, cells were scraped to create a wound in the middle of each well. Tet-on-PCDH10 H1299 cells were induced or not by addition of 5 ug/ml tetracycline for 48 hours and 2 pictures (#1,#2) were taken and presented at both 0 hour and 48 hours. (B) Wound gaps were measured using Photoshop software from pictures taken under 10× microscopic field. Three measurements were obtained for each sample to calculate mean gap distance. Error bars indicate ±1 standard deviation. (C) PCDH10 expression level in Tet-on-PCDH10 H1299 cells, treated or not by tetracycline, was evaluated in Western blot analysis at indicated time points.
Discussion
As one of the most prominent tumor suppressors, p53 protein has non-canonical functions being kept unveiled or not fully understood. These functions might contribute to the tumor suppressor role of p53 at different tumor developmental stages and pathological processes. The inhibitory effect of tumor cell migration, as a novel p53 function, has been demonstrated involved with tumor's invasive and metastatic potential. The control of cell mobility through p53 pathway has been mostly linked to the acquired function of mutant p53 deficient in transcriptional activity.Citation16,27 We found that PCDH10, a likely direct target of wild type p53, suppresses tumor cell migration in non-small cell lung cancer cells. The data in our study provided evidence that PCDH10 might link p53 to its function in tumor cell migration.
PCDH10 belongs to the non-clustered protocaderin protein family, a subtype of cadherin family distinctive from classical cadherins.Citation7 Protocadherins are believed to possess important functions such as intracellular signaling and growth control, in addition to the generic cell-to-cell adhesion activities of cadherin family proteins.Citation7 However, the exact functions of most protocadherins have not been well elucidated.Citation7 PCDH10 appears to have cell-cell adhesion property and plays roles in neuronal circuit formation.Citation24 The deficiency in PCDH10 gene has been linked to neuronal disease such as autism spectrum disorder.Citation23 PCDH10 has 6 cadherin domains, one transmembrane domain, and one cytoplasmic sequence, suggesting that PCDH10 could stimulate intracellular signaling.Citation24 PCDH10 protein is widely expressed but its function in promoting cell death, inhibiting tumor cell proliferation, and suppressing cell migration seems dependent on the cellular context and tissue type. In recent years, the association of PCDH10 with human malignancies has been proposed. Numerous studies have demonstrated that PCDH10 expression is partially or completely silent through promoter methylation or homozygous deletion in several human cancer types, including lymphoid malignancies, colorectal cancer, lung cancer, bladder cancer, and prostate cancer.Citation2,3,Citation10–12, Citation20,25,Citation30 In our study, it appears that the suppressed cell migration is the most prominent consequence when PCDH10 induction is introduced. The reported cell death and cell cycle arrest are insignificant effects of PCDH10 overexpression in H1299 lung cancer cells. The molecular mechanism of how PCDH10 inhibits tumor cell migration has been suggested to associate with Wnt/b-catenin signaling pathway, however, it is beyond fully understood and requires delineated studies.Citation29
p53 mediated activation of PCDH10 is limited to certain cell types, indicating that the transcriptional regulatory machinery of PCDH10 functions in probably multi-module manner. The understanding of how PCDH10 is regulated at transcriptional level stays in the infancy stage and requires future studies. Interestingly, a long-non-coding RNA HOTAIR, whose aberrantly increased expression frequently shown up in breast cancer metastases, repressed PCDH10 gene expression.Citation5 The interaction between PCDH10 and other cancer genes are yet to be discovered and the advanced knowledge will help to understand the role of PCDH10 in tumor pathogenesis. Of note, other proteins in cadherin protein family have also been reported as candidate tumor suppressor in human cancers such as prostate cancer and esophageal carcinoma.Citation2 Among these proposed tumor suppressor cadherin proteins, E-cadherin (CDH1), like PCDH10, is positively regulated by p53 at transcriptional level.Citation22 Hypothetically, p53 may directly and indirectly modulate various cadherin proteins to interfere with cell mobility and further tumor metastasis. Obviously, it is not reasonable to attribute entire function of p53 in controlling cell migration to any one of its targets. Instead, the direct and indirect p53 targets in regulating cancer cell migration and invasion likely form nodes of a multi-component molecular network to control this particular function of p53.
Accumulating evidence suggests that p53 has broader roles in tumor suppression than previously thought. It is well established that p53 responds to oncogenic insult, DNA damage stress, and other challenging conditions such as metabolic stress and epithelial-mesenchymal transition (EMT) process. It is interesting to see how p53 programs to coordinately control diverse genes in various cellular functions and how p53 determines when to elicit individual gene at different tumor developmental stages to prevent tumorigenesis.
Materials and Methods
RNA Isolation and qRT-PCR
Total RNA was isolated from cells using TRIzol (Invitrogen). One μg of total RNA was reverse-transcribed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen) following manufacturer's protocol. PCR was performed in triplicate using SYBR green mix (Applied Biosystems), and a 7500 Fast Real-Time PCR System (Applied Biosystems) under the following conditions: 15 min at 95°C followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Primers used for qRT-PCR were listed as follows, human p21 forward 5′- CCATGTGGACCTGTCACTGTCTT-3′ human p21 reverse 5′- cggcCTCTTGGAGAAGATCAGCcG-3′; human PCDH10 forward 5′- AGGCCCTTCACAGCACTCT-3′ human PCDH10 reverse 5′- AGCATATCCTTTTCCGTG; human PCDH10 (total transcripts) forward 5′-GAAGGAGTCTTTTCCCAGCTTCA-3′ human PCDH10 (total transcripts) reverse 5′- AGACCCAGATCTTCAGCGATA-3′; mouse PCDH10 forward 5′- CAAGAGAAGGCCCTGCAT-3′ mouse PCDH10 reverse 5′-AGCTGCATCCACAGCATTT-3′
Western Blot Analysis
For Western blot analysis, cells were lysed in cold FLAG lysis buffer [50 mM Tris·HCl (pH 7.9), 137 mM NaCl, 10 mM NaF, 1 mM EDTA, 1% Triton X-100, 0.2% Sarkosyl, 10% glycerol, and freshly supplemented protease inhibitor(Roche)]. Antibodies used for Western blot analysis are p21 (C-19 and SX118), and PUMA (H-136) from Santa Cruz, β-actin (AC-15), and Flag(M2) from Sigma, Vinculin (ab18058) from Abcam,p53(CM5) from Novusbio, Mdm2 (Ab-5) from EMD Biosciences, V5 from Invitrogen and PCDH10 (PA5–31042) from Thermo Scientific.
Plasmids
The full-length PCDH10 cDNA was amplified by PCR from Human MGC Verified FL cDNA (Open Biosystems) and subcloned into pCDNA3.1 V5-his vector (Invitrogen) and self-modified pTripZ includible vector (Dharmacon).
Cell culture
SUDHL4, SUDHL5 and DOHH2 cells were maintained in IMDM (Iscove's modified Dulbecco's medium, Gibco). H1299, SAOS2, WM2664, H460 and MEF cells were maintained in DMEM (Cellgro) medium (Cellgro). All media were supplemented with 10% fetal bovine serum (Gibco). Transfections with plasmid DNA were performed by Lipofectamine2000 (Invitrogen) according to the manufacturer's protocol.
Luciferase activity assay Promoter-containing fragments were amplified from human genomic DNA of H1299 cells and cloned into the pGL3 luciferase reporter vector using KpnI and Xhol restriction digest enzyme cutting sites. pLucPCDH10 containing the p53 BS site 3was amplified using forward primer 5′ – CGGGGTACCACTATCACGCCCATGGACAC – 3′ and reverse primer 5′ – CCGCTCGAGTTTCTGCCAATCCTGG GGTC – 3′. Transfection of SAOS2 cells were performed in 24-well plate using 0.2 μg luciferase reporter constructs, 0.05 μg pRL-tk Renilla construct and various amounts of mediated p53 plasmid. Luciferase activities were measured 24 h post-transfection using Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activities were normalized with Renilla luciferase activities to obtain the relative luciferase activity.
Gel shift assay
Flag-p53 protein was purified from the transfected 293 cells. The protein-DNA binding reactions (20 μl) contained 20 mM HEPES (pH 7.6), 80 mM NaCl, 0.1 mM EDTA, 12.5% glycerol, 2 mM MgCl2, 2 mM spermidine, 0.7 mM DTT, 200 ng/μl BSA, 20 ng/μl sheared sperm DNA, 10–20 fmol DNA probe, 20 ng Flag-p53. The 170 bp DNA fragment used as probe was obtained by PCR amplification from the PCDH10 promoter, labeled by T4 kinase (NEB, M0201S) and purified using the Bio-Spin 30 columns (Bio-Rad). Primers for probe synthesis: forward primer 5′-GTTGGGGCTTACACAGAGCTA-3′; reverse primer 5′- ACCACTGATTTCTGCCAATCCT-3′.
Chromatin immunoprecipitation (ChIP)
Cells were incubated in culture media containing 1% formaldehyde with gentle shaking for 10 min at room temperature, and crosslinking was stopped by addition of 2.5 M glycine to a final concentration of 0.125 M glycine. After two washes with cold PBS, cells were harvested in ice cold lysis buffer (10 mM Tris-Cl [pH 8.0], 85 mM KCl, 0.5% NP-40, 5 mM EDTA, and fresh proteinase inhibitor cocktail) and incubated on ice for 10 min. Nuclei were collected, suspended in cold RIPA buffer (10 mM Tris-Cl (pH 8.0), 150 mM NaCl, 0.1% SDS, 0.1% DOC, 1% Triton X-100, 5 mM EDTA), and sonicated to shear the genomic DNA to an average of 100–500 bp. Cleared extracts were precleared with protein A/G beads (Upstate Biotechnology), and the supernatants were used for immunoprecipitation by p53 FL393(Santa Cruz Biotech) antibody. After five times of wash by RIPA buffer with gentle rotation for 5 min each time, the proteins were eluted from the beads by 0.5 ml elution buffer (0.1 M NaHCO3 and 1% SDS). The DNA samples were recovered by phenol extraction and ethanol precipitation after reversal of crosslinking. The purified DNA was then analyzed either by PCR within linear amplification range followed by agarose gel electrophoresis or by quantitative real-time PCR using Applied Biosystems 7300. Primers used for the p53 ChIP assay:
PCDH10 p53 putative binding site1 forward 5′- GAGCTC TCCAAAGCAAAATTCTCT-3′, reverse 5′- TCCGCAAACGT GGGTTACA -3′, PCDH10 p53 putative binding site2 forward 5′- AACCCTCTTCCCCAGTTTCTG-3′, reverse 5′- CGCG CGTGTTTGCATGT-3′, PCDH10 p53 putative binding site3 forward 5′- CAGGGAGGATGGATGCAAGT-3′, reverse 5′- GCACCGCTGCGGGTATC-3′, PCDH10 p53 putative binding site4 forward 5′- TTTTAAAATGCCCTGCCAGTCT-3′, reverse 5′- CCCACTAATTGGCAAGGGTTAC-3′
Cell motility and migration assays
For wound healing assays, equal numbers of cells were plated at 30% confluence in 6-well plates, and 24 hours after plating, gently and slowly scratch the cell monolayer with a new 1 ml pipette tip across the center of the well. The cells were added with tetracycline and incubated for 48 hours. Captured images were used to compare and quantitatively evaluate the gap distance at each time point. Three independent 10× fields were measured for each experiment.
Disclosure of Potential Conflicts of Interest
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding
This work was supported by the National Cancer Institute of the National Institutes of Health under Award 5RO1 CA172023, 5RO1 CA166294, 5RO1CA085533 and 2P01CA080058 to W. G. D. Shi is also supported by NIH cancer biology training grant T32-CA09503.
References
- Aylon Y, Oren M. New plays in the p53 theater. Curr Opin Genet Deve 2011; 21: 86-92; PMID:21317061; http://dx.doi.org/10.1016/j.gde.2010.10.002
- Bertrand KC, Mack SC, Northcott PA, Garzia L, Dubuc A, Pfister SM et al. PCDH10 is a candidate tumour suppressor gene in medulloblastoma. Child's Nervous System 2011; 27: 1243-9; PMID:21597995; http://dx.doi.org/10.1007/s00381-011-1486-x
- Fang S, Huang SF, Cao J, Wen YA, Zhang LP, Ren GS. Silencing of PCDH10 in hepatocellular carcinoma via de novo DNA methylation independent of HBV infection or HBX expression. Clin Exp Med 2013; 13: 127-34; PMID:22543497; http://dx.doi.org/10.1007/s10238-012-0182-9
- Guo F, Zheng Y. Rho family GTPases cooperate with p53 deletion to promote primary mouse embryonic fibroblast cell invasion. Oncogene 2004; 23: 5577-85; PMID:15122327; http://dx.doi.org/10.1038/sj.onc.1207752
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071-6; PMID:20393566; http://dx.doi.org/10.1038/nature08975
- Hock AK, Vousden KH. Tumor suppression by p53: fall of the triumvirate? Cell 2012; 149: 1183-5; PMID:22682240; http://dx.doi.org/10.1016/j.cell.2012.05.024
- Kim SY, Yasuda S, Tanaka H, Yamagata K, Kim H. Non-clustered protocadherin. Cell Adhesion Migration 2011; 5: 97-105; PMID:21173574; http://dx.doi.org/10.4161/cam.5.2.14374
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 2012; 149: 1269-83; PMID:22682249; http://dx.doi.org/10.1016/j.cell.2012.04.026
- Li Y, Yang ZS, Song JJ, Liu Q, Chen JB. Protocadherin-10 is involved in angiogenesis and methylation correlated with multiple myeloma. Int J Mol Med 2012; 29: 704-10; PMID:22245948
- Li Z, Chim JC, Yang M, Ye J, Wong BC, Qiao L. Role of PCDH10 and its hypermethylation in human gastric cancer. Biochim Biophys Acta 2012; 1823: 298-305; PMID:22206871; http://dx.doi.org/10.1016/j.bbamcr.2011.11.011
- Lin YL, Li ZG, He ZK, Guan TY, Ma JG. Clinical and prognostic significance of protocadherin-10 (PCDH10) promoter methylation in bladder cancer. J Int Med Res 2012; 40: 2117-23; PMID:23321168; http://dx.doi.org/10.1177/030006051204000609
- Lin YL, Li ZG, Guan TY. The clinical significance of PCDH10 promoter methylation in patients with bladder transitional cell carcinoma. Urol Int 2013; 90: 219-24; PMID:23171734; http://dx.doi.org/10.1159/000345053
- Ma JG, He ZK, Ma JH, Li WP, Sun G. Downregulation of protocadherin-10 expression correlates with malignant behaviour and poor prognosis in human bladder cancer. J Int Med Res 2013; 41: 38-47; PMID:23569128; http://dx.doi.org/10.1177/0300060513476989
- Mak AS. p53 in cell invasion, podosomes, and invadopodia. Cell Adh Migr 2014; 8: 205-214; PMID:24714032; http://dx.doi.org/10.4161/cam.27841
- Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer 2006; 6: 449-58; PMID:16723991; http://dx.doi.org/10.1038/nrc1886
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 2009; 139: 1327-1341; PMID:20064378; http://dx.doi.org/10.1016/j.cell.2009.11.026
- Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol 2011; 192: 209-218; PMID:21263025; http://dx.doi.org/10.1083/jcb.201009059
- Narayan G, Scotto L, Neelakantan V, Kottoor SH, Wong AH, Loke SL, Mansukhani M, Pothuri B, Wright JD, Kaufmann AM, et al. Protocadherin PCDH10, involved in tumor progression, is a frequent and early target of promoter hypermethylation in cervical cancer. Genes, Chromosomes & Cancer 2009; 48: 983-92; http://dx.doi.org/10.1002/gcc.20703
- Narayan G, Freddy AJ, Xie D, Liyanage H, Clark L, Kisselev S, Un Kang J, Nandula SV, McGuinn C, Subramaniyam S, et al. Promoter methylation-mediated inactivation of PCDH10 in acute lymphoblastic leukemia contributes to chemotherapy resistance. Genes, Chromosomes Cancer 2011; 50: 1043-53; http://dx.doi.org/10.1002/gcc.20922
- Narayan G, Xie D, Freddy AJ, Ishdorj G, Do C, Satwani P, Liyanage H, Clark L, Kisselev S, Nandula SV, et al. PCDH10 promoter hypermethylation is frequent in most histologic subtypes of mature lymphoid malignancies and occurs early in lymphomagenesis. Genes, Chromosomes Cancer 2013; 52: 1030-41; http://dx.doi.org/10.1002/gcc.22098
- Powell E, Piwnica-Worms D, Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov 2014; 4: 405-14; PMID:24658082; http://dx.doi.org/10.1158/2159-8290.CD-13-0136
- Tamura M, Sasaki Y, Koyama R, Takeda K, Idogawa M, Tokino T. Forkhead transcription factor FOXF1 is a novel target gene of the p53 family and regulates cancer cell migration and invasiveness. Oncogene 2014; 33: 4837-46; PMID:24186199; http://dx.doi.org/10.1038/onc.2013.427
- Tsai NP, Wilkerson JR, Guo W, Maksimova MA, DeMartino GN, Cowan CW, Huber KM. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell 2012; 151: 1581-94; PMID:23260144; http://dx.doi.org/10.1016/j.cell.2012.11.040
- Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S. OL-Protocadherin is essential for growth of striatal axons and thalamocortical projections. Nature neuroscience 2007; 10: 1151-9; PMID:17721516; http://dx.doi.org/10.1038/nn1960
- Wang L, Xie PG, Lin YL, Ma JG, Li WP. Aberrant methylation of PCDH10 predicts worse biochemical recurrence-free survival in patients with prostate cancer after radical prostatectomy. Med Sci Monitor 2014; 20: 1363-8; PMID:25086586; http://dx.doi.org/10.12659/MSM.891241
- Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol 2009; 11: 694-704; PMID:19448627; http://dx.doi.org/10.1038/ncb1875
- Xiong S, Tu H, Kollareddy M, Pant V, Li Q, Zhang Y, Jackson JG, Suh YA, Elizondo-Fraire AC, Yang P, et al. Pla2g16 phospholipase mediates gain-of-function activities of mutant p53. Proc Natl Acad Sci U S A 2014; 111: 11145-11150; PMID:25024203; http://dx.doi.org/10.1073/pnas.1404139111
- Ying J, Li H, Seng TJ, Langford C, Srivastava G, Tsao SW, Putti T, Murray P, Chan AT, Tao Q. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene 2006; 25: 1070-80; PMID:16247458; http://dx.doi.org/10.1038/sj.onc.1209154
- Zhao Y, Yang Y, Trovik J, Sun K, Zhou L, Jiang P, Lau TS, Hoivik EA, Salvesen HB, Sun H, et al. A Novel Wnt regulatory axis in endometrioid endometrial cancer. Cancer Res 2014; 74:5103-17
- Zhong X, Zhu Y, Mao J, Zhang J, Zheng S. Frequent epigenetic silencing of PCDH10 by methylation in human colorectal cancer. J Cancer Res Clin Oncol 2013; 139: 485-90; PMID:23180019; http://dx.doi.org/10.1007/s00432-012-1353-5
