Abstract
CEP161 is a novel component of the Dictyostelium discoideum centrosome which was identified as binding partner of the pericentriolar component CP250. Here we show that the amino acids 1-763 of the 1381 amino acids CEP161 are sufficient for CP250 binding, centrosomal targeting and centrosome association. Analysis of AX2 cells over-expressing truncated and full length CEP161 proteins revealed defects in growth and development. By immunoprecipitation experiments we identified the Hippo related kinase SvkA (Hrk-svk) as binding partner for CEP161. Both proteins colocalize at the centrosome. In in vitro kinase assays the N-terminal domain of CEP161 (residues 1-763) inhibited the kinase activity of Hrk-svk. A comparison of D. discoideum Hippo kinase mutants with mutants overexpressing CEP161 polypeptides revealed similar defects. We propose that the centrosomal component CEP161 is a novel player in the Hippo signaling pathway and affects various cellular properties through this interaction.
Introduction
The centrosome is the main microtubule organizing center (MTOC) and nucleates, anchors and organizes microtubules. It also plays an important role in mitotic spindle orientation and genome stability.Citation1 In animal cells it comprises a pair of centrioles and a surrounding pericentriolar matrix (PCM). The PCM is a key structure of the centrosome and is responsible for microtubule nucleation and anchoring. Its major components are large coiled coil proteins like Pericentrin and proteins of the AKAP450 family that provide docking sites for γ-tubulin ring complexes and regulatory molecules. They allow the centrosome to function as MTOC and to carry out its regulatory roles during cell cycle transitions, cellular responses to stress, and organization of signal transduction pathways.Citation2,3
The centrosome morphology varies largely in protozoa, algae, and fungi. In D. discoideum it is a nucleus associated body consisting of a box-shaped core surrounded by the corona, an amorphous matrix functionally equivalent to the PCM.Citation4 More than 70 candidates of the D. discoideum centrosome have been identified.Citation5 In higher vertebrates, sequence database searches resulted in the identification of more than 2,000 peptides representing more than 500 proteins in the peak centrosome fraction.Citation6
Increasing evidence indicates that the centrosome is well designed for the organization of multiprotein scaffolds that can anchor a diversity of activities ranging from protein complexes involved in microtubule nucleation to multicomponent pathways in cellular regulation.Citation7 The centrosome is also an indispensable component of the cell-cycle machinery of eukaryotic cells, and perturbation of core centrosomal or centrosome-associated proteins is linked to cell-cycle misregulation and cancer.Citation8
The Hippo signaling pathway is a tumor-suppressive pathway and is inactive at low cell density.Citation9 It primarily affects the number of cells produced and has only minor effects on tissue patterning.Citation10 It is however known as a key regulator of organ growth and tissue size in Drosophila and mouse.Citation11-14 At the center of the Hippo pathway is a core kinase cassette that consists of a pair of related serine/threonine kinases, mammalian STE20-like protein kinase 1 and 2 (MST1 and MST2), which are homologues of D. melanogaster Hippo (HPO), and large tumor suppressor 1 (LATS1) and LATS2 together with the adaptor proteins Salvador homolog 1 (SAV1) and MOB kinase activator 1A (MOB1A) and MOB1B.Citation10,11,15-18 These proteins limit tissue growth by facilitating LATS1- and LATS2-dependent phosphorylation of the homologous oncoproteins Yes-associated protein (YAP) and Transcriptional co-activator with PDZ-binding motif (TAZ)Citation19 which represses their transcriptional activity. The Hippo pathway is conserved throughout evolution and core pathway components like Hippo related kinases KrsA, KrsB and SvkA and a LATS homolog have also been detected in D. discoideum.Citation20-22 Analysis of D. discoideum Hippo kinase mutants revealed growth independent roles of the Hippo pathway such as an involvement in cytoskeletal activities regulating cell adhesion and migration and in multicellular pattern formation.Citation21 Transcription factors on which these kinases act have not yet been identified.Citation23
In this study we investigated the D. discoideum centrosomal component CEP161. Our results confer roles for CEP161 in growth and development. Furthermore, we identified the kinase SvkA as its interaction partner which is a D. discoideum Hippo related kinase designated here as Hrk-svk and which is a direct homolog of human MST1. We found that CEP161 has an inhibitory effect on the kinase activity of Hrk-svk and may through this activity regulate the Hippo pathway in D. discoideum.
Results
Identification and characterization of the pericentriolar matrix protein CEP161, the D. discoideum ortholog of CDK5RAP2
We identified CEP161 as interaction partner of the pericentriolar matrix component CP250 in immunoprecipitation experiments using GFP-tagged CP250 followed by mass spectrometry analysis.Citation24 The binding site of CEP161 for CP250 was located in its N-terminus. A GST-tagged polypeptide encompassing residues 1-763 could pull down GFP-CP250 from whole cell lysates (data not shown). A direct interaction of the proteins was shown by yeast-2-hybrid experiments in which residues 1-763 of CEP161 interacted with residues 1-1148 of CP250. The gene encoding CEP161 (DDB_G0282851) is located on chromosome 3 and has 2 exons. The open reading frame comprises 4146 base pairs and codes for a protein of 1381 aa with a molecular mass of 161,600. We named the protein CEP161 based on its molecular mass and location (see below). The BLAST prediction program revealed an N-terminal γ-tubulin ring complex (γ-TuRC) domain (residues 99-174); the SMART prediction tool indicated the presence of 4 coiled-coil domains in the protein (). The γ-TuRC domain (pfam07989) of CEP161 is most closely related to the one in centrosomin from insects and its mammalian homolog CDK5RAP2 which are both centrosomal proteins. The highest conservation is in a consensus 10 amino acid motif ().
Figure 1 (See previous page). CEP161 as a novel centrosomal protein in D. discoideum. (A) CEP161 protein and domain structure. (B) γ-TuRC domain sequence alignment. Protein accession numbers: Dictyostelium discoideum (Dd) (DDB_G0282851), Drosophila melanogaster (Dm) centrosomin (CNN) (NP_725298.1) and Human (Hs) (NP_060719). The numbers indicate the amino acid position of the γ-TuRC domain in the respective proteins. Color code: Red background, identical residues; yellow background, similar residues. (C) Localization of CEP161 at the centrosome in AX2 cells. CEP161 was detected with mAb K83-632-4. (D) Colocalization of CEP161 and GFP-CP250 at the centrosome (upper panel). mAb K83-632-4 recognizes GFP-CEP161 at the centrosome (lower panel). (E) Colocalization of GFP-CEP161 with comitin at the Golgi complex. Comitin was detected with mAb 190-340-8. DAPI was used to stain the nucleus. Scale bar, 10 μm. (F) Overview of N-terminally GFP-tagged CEP161 proteins. (G) Expression of GFP-tagged CEP161 proteins in AX2 cells. Proteins from whole cell lysates were separated by SDS-PAGE (10% acrylamide). mAb K3-184-2 was used to detect the GFP-fusion proteins Alpha-tubulin detected with mAb YL1/2 was used as the loading control. (H) Detection of endogenous CEP161 and GFP-tagged proteins with polyclonal antibodies specific for CEP161. The proteins were resolved by SDS-PAGE (10% acrylamide). The arrow indicates the position of endogenous CEP161. (I) Quantification of the protein gel blot results for amounts of endogenous CEP161 and GFP-tagged proteins. (J) Subcellular localization of GFP-tagged proteins. DAPI was used to stain the nucleus, the centrosome in D3 cells was stained with CP250 specific mAb K68-439-8. Scale bar, 5 μm.
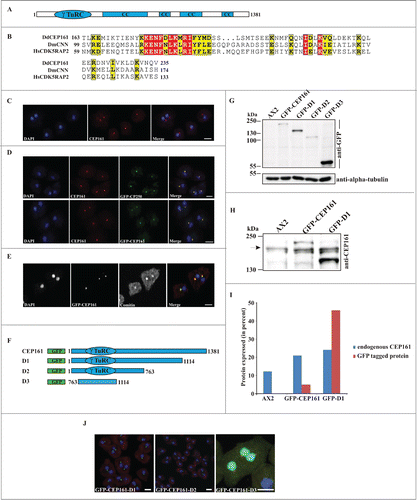
DdCEP161 is a centrosomal protein
To determine the subcellular localization of DdCEP161, we generated monoclonal antibodies against a recombinant polypeptide (CEP161-D2, residues 1-763). mAb K83-632-4 showed in immunofluorescence studies a bright punctate staining near the nucleus suggestive of the centrosome (). This staining persisted throughout the cell cycle (data not shown). The centrosomal localization was confirmed by labeling GFP-CP250 knock-in cells where the antibody staining coincided with the GFP positive centrosome (). GFP-tagged CEP161 also was present in a single dot near the nucleus and was recognized by mAb K83-632-4 (). The centrosome and the Golgi apparatus co-localize in the vicinity of the nucleus. When we stained GFP-CEP161 cells with mAb190-340-8 for comitin, a marker for the Golgi, we found CEP161 in the center of the Golgi apparatus ().Citation25
To identify the region of CEP161 that mediates the centrosome association of CEP161, we generated shortened proteins (GFP-CEP161-D1 designated D1, residues 1-1114; GFP-CEP161-D2 designated D2, residues 1-763; and GFP-CEP161-D3 designated D3, residues 763-1114) (). Expression and molecular weights of the GFP-tagged proteins were analyzed on a SDS-PA gel (). The expression levels varied for the GFP-tagged proteins. GFP-CEP161 was present in lower amounts than endogenous CEP161 whereas GFP-D1 levels were 1.8 fold higher than those of CEP161 (). In immunofluorescence analysis the D1 and D2 proteins behaved like a centrosomal protein and appeared as a single dot near the nucleus. In contrast, the D3 protein was present throughout the nucleus although the prediction programs did not reveal nuclear localization signals or any DNA binding domain (). We conclude that amino acids 1-763 harboring the γ-TuRC domain are sufficient for localization of CEP161 to the centrosome. Based on its overall domain structure, the presence of the conserved γ-TuRC and the localization at the centrosome we presume that CEP161 is the D. discoideum ortholog of CDK5RAP2.Citation26
DdCEP161 mutant cells are viable, but have impaired growth and development
Isolation of a knockout mutant was not successful. Therefore we overexpressed the GFP-tagged versions of CEP161 full length and truncated proteins in AX2 cells in order to analyze the role of DdCEP161 for cell organization, growth and development. First we determined the nuclei number of the cells to understand the role of CEP161 in cell division and cell cycle. The D1, D2 and D3 cells were mostly mononucleated with their centrosome located very close to the nucleus in a distance of <500 nm. By contrast, CEP161 affected the centrosome nucleus distance and approximately 80% of the CEP161 cells had the centrosome far away from the nucleus with a distance >900 nm (). We further observed that the nuclei-centrosome ratio was aberrant in a low percentage of the D1 and D2 expressing cells. AX2 cells had one centrosome per nucleus whereas 0.6% of the D1 cells had 2 centrosomes per nucleus, for D2 cells this number increased to 0.74%, and in 0.24% of the cells we observed 3 and more centrosomes per nucleus (). Ectopic expression of CEP161 led to an increase in cell size with approximately 70% of the cells having a diameter greater than 12 μm. D1 and D2 expressing cells were similar to AX2, D3 expressing cells were smaller (70% with a diameter <10 μm) ( ).
Figure 2 (See previous page). CEP161 regulates nuclear and centrosomal number and affects cell size and growth. (A) Nuclei number per cell. (B) Nucleus-centrosome distance. The percentage of cells with the indicated distances is given. (C) Nucleus-centrosome ratio. The percentage of cells is given. A total of 600 cells were counted for AX2 and mutant strains for each of the experiments in A, B, and C. (D) Confocal images for D1 and D2 cells showing increased centrosome numbers. DAPI was used to stain the nucleus. Scale bar, 5 μm. (E) Cell size of the strains in micrometers. (F) Growth on K. aerogenes. (G) Growth in axenic medium in shaking culture. (H) Phagocytosis was assayed using the strains from (G) and TRITC-labeled yeast. Approximately 200 cells from each strain were counted. The percentage of cells which had engulfed yeast particles after 15 min is shown in the graph (***P < 0.001).
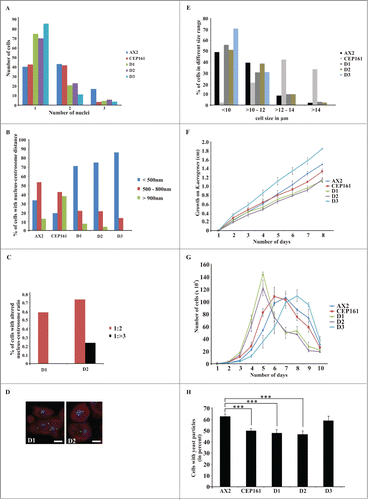
We examined growth on a bacterial lawn and in axenic culture. On a lawn of Klebsiella aerogenes growth was significantly reduced for all mutants except for D3 which showed increased growth (). In suspension culture, the growth curves of AX2, CEP161 and D3 resembled each other and the doubling times were comparable (9 hrs to 12 hrs) whereas the D1 (5 hrs) and D2 (7 hrs) mutants showed a significantly shorter doubling time. Furthermore, after the highest cell densities were reached the cell numbers dropped dramatically for D1 and D2 mutants (). We also monitored uptake of yeast particles and found that fewer CEP161 (50%), D1 (48%) and D2 cells (47%) had ingested one or more yeast particles after 15 min than AX2 cells (63%). D3 (59%) was similar to AX2 ().
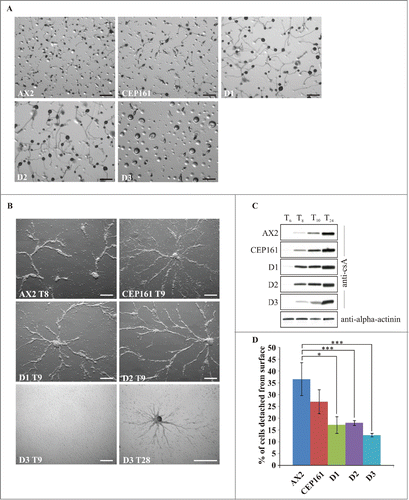
When D. discoideum cells starve, they enter a developmental cycle that ends with the formation of fruiting bodies. The CEP161, D1 and D2 cells plated on phosphate agar aggregated and formed fruiting bodies timely, the D3 cells formed loose aggregates that did not develop further. For D1 and D2 we noted a marked increase in the size of the fruiting bodies both with regard to the length of the stalks and the size of the spore heads, whereas the CEP161 fruiting bodies were only slightly bigger than those of AX2 (). Development was also studied on a plastic surface in Soerensen phosphate buffer which only allows stream and aggregate formation. Stream formation occurred slightly later for CEP161, D1 and D2 (9 h) as compared to AX2 (8 h). For the D3 strain we noted a severe delay in the streaming and aggregation behavior as they formed streams only after 28 hours (). We then analyzed the expression levels of the contact site A (csA), an aggregation stage specific cell surface glycoprotein, in cells that were starved in shaken suspension.Citation27 The pattern of csA expression was comparable for all strains with the exception of D3 which showed lower amounts of the protein up to 10 h and reached high levels only at the 24 h time point (). Streaming and aggregation of the cells is also affected by the adhesion of the cells to the substratum. In adhesion assays we observed that the mutants exhibited an altered adhesion and bound more strongly to the plastic surface than AX2 with D3 exhibiting the strongest adhesion ().
Figure 3. CEP161 plays an important role in development. (A) Development of wild type and mutant cell strains on phosphate agar plates. Pictures were taken after 24 hours. Scale bar, 500 μm. (B) Stream formation on a plastic surface under phosphate buffer. The time points (in hours) indicate the occurrence of stream formation. Scale bar, 250 μm. (C) Expression of csA during starvation in shaken suspension. Cells were collected at the indicated time points and cell lysates analyzed by SDS-PAGE and western blot. csA was detected by mAb 33-294, mAb 47-16-1 detected α-actinin which was used as loading control. Since α-actinin blot was similar for all strains, only the α-actinin blot for AX2 is shown. (D) Cell-substrate adhesion. Percentage of cells detached from a plastic surface after shaking at 200 rpm for 1 hour. The symbol * indicates the significance of the p-Value. *<0.5, ***<0.001.
Cell motility is an important feature during growth and development of D. discoideum. To assess the role of CEP161 and mutant proteins for motility we analyzed the motility of single cells during the aggregation phase. AX2 and the CEP161 strain had a similar speed, whereas the speed of D1, D2 and D3 was significantly decreased. Persistence, which gives an estimation of the movement along the total path, was decreased for all mutant strains, whereas directionality which indicates migration straightness was only affected for D3 (). We further noted an altered cell polarization for CEP161 and D3 which appeared more rounded, whereas AX2 cells were elongated and highly polarized ().
Figure 4. CEP161 affects unicellular motility and multicellular migration. (A) Analysis of chemotactic cell motility of AX2 and mutants. Time-lapse image series were captured and stored on a computer hard drive at 30-second intervals. Images were taken at magnifications of 10X every 30 s. The DIAS software was used to trace individual cells along image series and calculate motility parameters. Speed refers to the speed of the cell's centroid movement along the total path; directionality indicates migration straightness; direction change refers to the number and frequency of turns; persistence is an estimation of movement in the direction of the path. Values are mean ± standard deviation of >30 cells from 3 or more independent experiments. The symbol * indicates the significance of the p-Value. *<0.5, **<0.01, ***<0.001. (B) Images showing the cell shape of aggregation competent cells of AX2, CEP161 and D3 cells. Scale bar, 50 μm. (C) Phototaxis assay. Slugs and slug trails were transferred to nitrocellulose filters and stained with amido black. The red arrow indicates the source of light. (D.) Angle of deviation of the slugs in response to light source. Slugs from four individual experiments were analyzed.
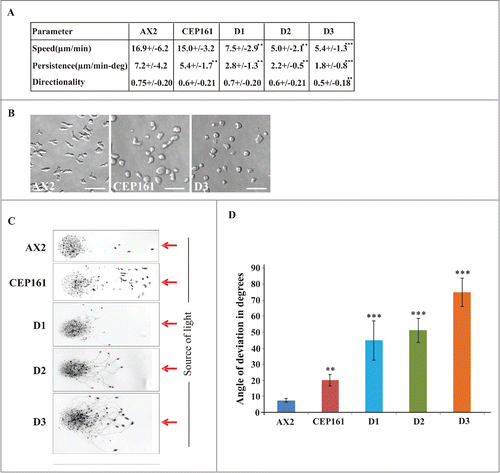
During development, multicellular migrating pseudoplasmodia (slugs) are formed.Citation28 D. discoideum slugs are polar with a tip at the anterior end consisting of prestalk cells, which sense the light that helps to control their migration and phototactic turning. AX2 and CEP161 slugs migrated in a nearly straight path toward the light source. D1, D2 and D3 deviated from this path and migrated in an angle toward light. Also, the path length was shorter, in particular for D1 and D2 ().
Identification of CEP161 as an interacting partner of the Hippo homolog Hrk-svk and its influence on the kinase activity of Hrk-svk
To identify further interaction partners of CEP161 we carried out immunoprecipitation experiments using GFP-tagged CEP161. In our mass spectrometry analysis we identified severin kinase SvkA (DDB_G0286359) as a potential interacting partner. SvkA was described as a homolog of human MST3, MST4 and YSK1 kinases.Citation20 Our detailed sequence analysis showed that it is related to hippo of Drosophila (25.1% identity, 35.7% similarity) and Hippo related Stk3 (MST1) of human (34.7% identity; 52.0% similarity). Particularly high homology was observed in the serine/threonine protein kinase domain of the proteins (). Given the significance of SvkA to our study and its relation to Hippo, we have renamed it as Hippo related kinase-svk (Hrk-svk). To confirm the interaction we performed GST pull down assays with GST-CEP161-D2 (residues 1-763) and GST-CEP161-D3 proteins (residues 763-1114). The D2 protein encompasses the γ-TuRC domain and a coiled coil domain, D3 encodes 2 coiled coil domains ( Bi). Hrk-svk interacted with the D2 and D3 proteins when the precipitate was probed with Hrk-svk polyclonal antibodies (). The interaction was further tested by using GST fusions of Hrk-svk for pull downs of CEP161. Only Hrk-svk-E1 (residues 1-323) which encompasses the serine threonine kinase domain (residues 1-323) interacted with the CEP161 polypeptides whereas Hrk-svk-E2 (residues 290–478) did not ( Bii, D). We next tested whether Hrk-svk phosphorylates CEP161. For this we used Hrk-svk-E1 which contains the catalytic domain and has kinase activity. It phosphorylates itself and myelin basic protein (MBP) (Arasada et al., 2006; our unpublished observations). We observed autophosphorylation of Hrk-svk-E1 and phosphorylation of MBP. When we added CEP161-D2, increasing amounts of the polypeptide inhibited increasingly the phosphorylation of MBP as well as autophosphorylation ().
Figure 5 (See previous page). DdCEP161 as a novel interacting partner of DdHrk-svk. (A) Sequence alignment for the Serine/Threonine protein kinase domain of Human (Hs) MST2 (NP_006272.2), Drosophila (Dm) Hippo (NP_611427.1) and Dictyostelium (Dd) Hrk-svk (DDB_G0286359). The numbers indicate the position of the amino acid sequence of the STK domain in the respective proteins. Color code: Red background, identical residues; yellow background, similar residues. (B) GST-tagged polypeptides for (i) CEP161 and (ii) Hrk-svk. (C) CEP161 interacts with Hrk-svk. Pull down assay with GST-tagged residues 1-763 and 763-1114 of CEP161. (D) Pull down assay with GST-tagged residues 290-478 and 1-323 of Hrk-svk. AX2 cell lysates were used for the pull downs in C and D. The blots were probed with the indicated antibodies. The GST-fusions were revealed by Ponceau S staining. (E) Phosphorylation assay. GST-CEP161-D2 negatively regulates the auto- and trans-phosphorylation abilities of GST-Hrk-svk-E1. MBP, myelin basic protein. (F) Immunofluorescence analysis showing the colocalization of CEP161 with GFP-tagged Hrk-svk in AX2 cells. CEP161 was detected by mAb K83-632-4, nuclei were stained with DAPI. Size bar, 5 μm.
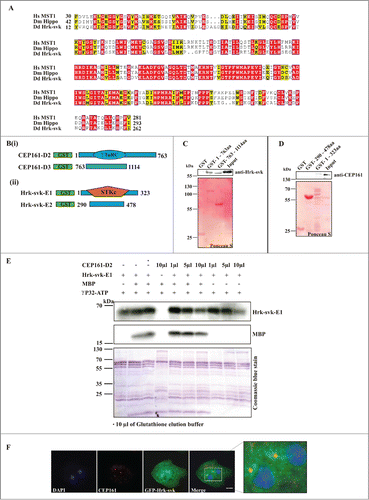
To investigate the localization of Hrk-svk we expressed it as GFP-tagged protein. Previously we have reported for GFP-Hrk-svk a cytosolic localization and enrichment at the centrosome (Rohlfs et al., 2007). When we stained GFP-Hrk-svk expressing cells for CEP161 using mAb K83-632-4 we detected CEP161 staining in the strongly GFP positive region near the nucleus showing a yellow puncta in the overlay which indicates a colocalization ().
Discussion
CEP161 is a novel component of the D. discoideum centrosome harboring a gamma-tubulin ring complex (γ-TuRC) domain and 4 predicted coiled coil domains and presumably forms an elongated molecule which is the characteristics of a pericentriolar matrix protein. CEP161 is present at the centrosome where it remains throughout the cell cycle (data not shown). For this association, the N-terminal part of the protein encompassing the γ-TuRC domain is required. The interaction site with its centrosomal binding partner CP250 is also located in this region. In a proteomic analysis, we identified Hrk-svk as interaction partner for CEP161. This interaction appears to interfere with the kinase activity of Hrk-svk since in in vitro experiments increasing amounts of a CEP161 polypeptide led to a decrease in the phosphorylation activity of Hrk-svk. Hrk-svk is the homolog of D. melanogaster Hippo and its serine/threonine kinase domain shows high similarity to Hippo homologs in higher organisms.
The analysis of cells ectopically expressing C-terminally truncated polypeptides revealed an involvement of CEP161 in growth and development. These are properties in which Hippo signaling is typically involved. Furthermore we observed changes in the adhesion properties and in the motile behavior of individual cells as well as of the multicellular slug. In D. discoideum regulation of the adhesive properties and of cell migration have been revealed as key functions of the Hippo pathway.Citation21 Cells deficient for the binding partner CP250 exhibited similar defects in growth, development and motility.Citation24 Whether these phenotypes are also linked to Hippo signaling is unclear at present. Ectopic expression of full length CEP161 caused a mislocalization of the centrosome. In ∼40% of the cells it was located >900 nm away from the nucleus, whereas ectopic expression of the D1 and D2 proteins led to an altered nucleus centrosome ratio. From these data it appears that the N-terminal part of CEP161 is important for interactions with pathways that impact on cell physiology and centrosome biology. It harbors the CP250 and the centrosome binding site and has also the ability to interact with and inhibit the Hrk-svk kinase. Taking into account the inhibitory effect of CEP161-D2 on the Hrk-svk kinase activity it might well be that the phenotypes which we have described like enhanced growth and increased size of multicellular structures are caused by inhibition of the Hippo signaling pathway (). Interestingly, several of these phenotypes were detected in the Hrk-svk mutant and in mutants deficient for other D. discoideum components of the Hippo signaling pathway as well. All of them had altered growth characteristics, development was affected to different degrees and motility of single cells as well as phototactic migration were altered ().Citation20-22,29
Table 1. Phenotypes of D. discoideum mutants with defects in components of the Hippo pathway
Figure 6. Model for the regulation of Hippo signaling by CEP161. In D. discoideum, CEP161 interacts with Hrk-svk and reduces its auto- and trans-phosphorylation ability presumably leading to an inhibition of the pathway. CEP161 influence on Hippo pathway components is shown in various colors, with black pointed arrows for direct interactions and red blunt lines indicating inhibitory interactions and blue arrows point to the affected functions.
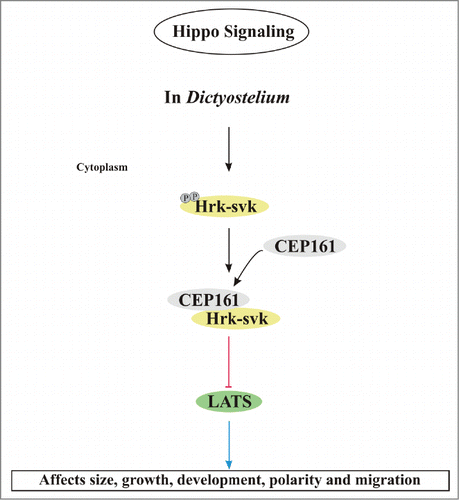
A link of Hippo signaling and the centrosome was recently suggested by findings that MST1 signaling controls centrosome duplication and that in D. discoideum deletion of the LATS homolog NdrC leads to centrosomal abnormalities.Citation22,30 We observed an impact of CEP161 mutant proteins on the nucleus centrosome distance and the nucleus centrosome ratio.
Taken together, we have identified a further component of the D. discoideum centrosome which interacts with the pericentrosomal D. discoideum CP250 and is the ortholog of the mammalian centrosomal CDK5RAP2. We provide evidence for a further interaction with Hippo-related kinase Hrk-svk which presumably takes place at the centrosome and leads to an inactivation of the kinase. Thus, the centrosome may also be a player in the Hippo pathway and determine through this mechanism cell proliferation and further cellular properties.
Materials and methods
Dictyostelium strains, cell culture, and vector construction
Strain AX2–214 is the parent of all strains generated in this study. Growth and development were done as described.Citation31 For expression of CEP161 GFP fusion proteins, sequences encompassing amino acids 1-1114 corresponding to nucleotides 1-3340 (GFP-CEP161-D1), amino acids 1-763 corresponding to nucleotides 1-2290 (GFP-CEP161-D2) and amino acids 763-1114 corresponding to nucleotides 2290-3340 (GFP-CEP161-D3) were cloned into pBsr GFP N2.Citation24 Expression was under the control of the constitutively active actin15 promoter. The plasmids were introduced into AX2 cells by electroporation following standard procedures.Citation32 Selection of the transformants was with blasticidin (3.5 μg/ml). Cells expressing GFP-tagged proteins were identified by immunofluorescence analysis and western blotting. Mutant analysis was done as described.Citation31,33,34 Cell size was determined using cells that had been treated with 20 mM EDTA in Soerensen phosphate buffer (17 mM Na/K-phosphate buffer, pH 6.0) in order to obtain perfectly round cells. For expression as glutathione S-transferase (GST)-fusion proteins, cDNA sequences encoding amino acids 1-763 (GST-CEP161-D2) and amino acids 763-1114 (GST-CEP161-D3) were cloned into pGEX4T (GE Healthcare). Yeast-two-hybrid experiments were carried out as described.Citation24
Generation of antibodies
Mouse monoclonal antibodies were generated against CEP161-D2 (amino acids 1-763) as described.Citation35,36 For immunization of mice the GST-part was removed by thrombin cleavage. The identity of the CEP161 polypeptide was confirmed by mass spectrometry. mAb K83-632-4 was used in this study. It recognized the bacterially produced recombinant protein and the endogenous protein in immunofluorescence analysis. Rabbit polyclonal antibodies specific for CEP161 were generated against a GST fusion protein containing the amino acids 763-1114 (Pineda, Berlin, Germany) and affinity purified. They recognized the recombinant and the endogenous protein in protein gel blots of whole cell lysates.
Immunofluorescence analysis
Immunofluorescence analysis of D. discoideum cells was carried out as described.Citation35 Antibodies used were mouse monoclonal antibodies mAb 47-16-8 directed against α-actinin,Citation36 mAb 33-294 against the cell adhesion molecule csA,Citation37 mAb K3-184-2 against GFP,Citation34 mAb 190-340-8 against comitinCitation25 and mAb K83-632-4 against CEP161 (this study). The appropriate secondary antibodies were Cy3-conjugated sheep anti-mouse IgG (Sigma, St. Louis, MO) and Alexa 568- or 647-conjugated goat anti-mouse or donkey anti-mouse and Alexa 647 donkey anti-rabbit IgG or Alexa 568-coupled goat anti-rabbit IgG (Molecular Probes, Eugene, OR). Nuclei were stained with DAPI (Invitrogen). The coverslips were mounted and subjected to immunofluorescence microscopy using a Leica TCS SP5 microscope equipped with a HyD detector.
Mutant analysis
Analysis of D. discoideum nuclear and centrosome abnormalities
The centrosome was stained with mAb K83-632-4 for CEP161, DNA was stained with DAPI. Fixation was with methanol. Confocal microscope images were taken and nuclei and centrosome number and the distance between centrosome and nucleus determined. The centrosome-nucleus distance was measured using the scale bar in the LAS-AF-LITE software for confocal microscopy imaging.
Phagocytosis assay
For analysis of phagocytosis, cells were collected from Petri dishes, an equivalent amount of TRITC labeled yeast was added to the cells, the cells were fixed with methanol after a 15 min incubation time, and the number of ingested yeast determined.
D. discoideum cell migration studies
This analysis was done as previously described.Citation24,38 Briefly, aggregation competent cells were plated in a chamber (ibidi GmbH-Martinsried, Germany) and random motility was followed. Images were recorded at intervals of 6 s using a Leica DM-IL inverse microscope (Deerfield, IL; 40× objective) and a conventional CCD video camera and analyzed using Dynamic Image Analysis Software (DIAS, Soll Technologies, Iowa City, IA).
D. discoideum cell adhesion assay
To analyze cell-substrate adhesion a substrate detachment assay was carried out. A total of 1×10Citation6 cells in growth medium was added per well (24 well plates, Costar) and incubated for 4 hours at 22°C. Then the plates were shaken on a gyratory shaker at 60, 120 and 200 rpm for one hour each. The number of detached cells was counted in a hemocytometer. The total number of cells was determined after resuspension of all cells and the percentage of detached cells calculated.
Phototaxis assay
Phototaxis assays were essentially performed as described earlier.Citation39 The filters exhibiting the stained slime trails were used to determine the distance traveled by the slugs toward the source of light from the point of application. The angle of deviation of the slugs was measured with the software Kreiswinkelmesser.
Pull down and immunoprecipitation assays
For pull down and immunoprecipitation experiments D. discoideum cells were lysed in 50 mM (10 mM for immunoprecipitation assay) Tris/HCl, pH 7.4, 150 mM NaCl, 0.5% NP40, supplemented with protease inhibitor cocktail (Sigma), 0.5 mM PMSF, 0.5 mM EDTA, and 1 mM Benzamidine by passing them through a 25G syringe (10–20 strokes) and incubated with agitation for 15 min at 4ºC to ensure complete cell lysis followed by a centrifugation step at 16,000 rpm for 10 min. The supernatants were either incubated with GST and GST-fusion proteins bound to Glutathione Sepharose beads, respectively, or with GFP-trap beads (ChromoTek, Martinsried, Germany). After incubation for 3 h GST beads were washed 3 times with wash buffer (50 mM Tris/HCl, pH 7.4, 150 mM NaCl, protease inhibitor cocktail, 0.5 mM PMSF, 0.5 mM EDTA, 1 mM Benzamidine). GFP-trap beads were washed with a different wash buffer (10 mM Tris/HCl, pH 7.4, 50 mM NaCl, protease inhibitor cocktail, 0.5 mM PMSF, 0.5 mM EDTA, 1 mM Benzamidine). The beads were resuspended in SDS sample buffer, incubated at 95ºC for 5 min and the proteins separated by SDS-PAGE and analyzed by western blotting. Hrk-svk was detected with polyclonal antibodies.
Phosphorylation assay
Hrk-svk activity was assayed in a reaction mixture (40 μl) containing 10 mM Tris/HCl, pH 7.5, 1 mM dithiothreitol, 1 mM EGTA, 1 mM Na3VO4, 2 mM NaF, 10 mM MgCl2, 0.05 mg/ml bovine serum albumin, 0.1 mM ATP containing 2–5 μCi of [32P]ATP, 0.01% NaN3, and 1–4 μM substrate. The reaction was initiated by addition of either the substrate or the kinase or together with CEP161 to the reaction mixture and carried out at 30 °C. The GST-tagged proteins CEP161-D2 and Hrk-svk-E1 were used for the assay. Glutathione was used to elute the GST-tagged proteins. The glutathione elution buffer (100 mM Tris, pH 8.0, 20 mM L-Glutathione (Sigma G4251)) was used for control. Myelin basic protein (MBP) (2–4 μM final concentration) was used as substrate. The phosphorylation was terminated after 30 min by the addition of 20 μl of 3× SDS sample buffer and boiling for 5 min. Proteins were separated by SDS-PAGE and electrophoresis was terminated before the running front reached the lower buffer chamber and the gel was cut just above the running front to remove the lower gel strip which contains most of the non-incorporated radioactive ATP. Protein bands were visualized by staining with Coomassie brilliant blue and, after drying of the gels, labeled proteins were detected by autoradiography on Amersham hyper films.
Oligonucleotide sequences
Oligonucleotides used for PCR (Polymerase Chain Reaction) were purchased from Sigma-Genosys in Steinheim.
Cep161 1F GGATCCATGAATGGATGGGGAGAAAGTGACG
Cep161 2320R GGATCCGTAGATCTTCCTTCTCATCGAATAGC
Cep161 2293F GGATCCTTGCTATTGGATGAGAAGGAAGATCTAC
Cep161 3342R CCCGGGGATATCAGGATTTAAAGTTATTAAAGATGAAG
Cep161 3314F GGATCCTCATCTTTAATAACTTTAAATCCTGATATC
Cep161 4143R CCCGGGTTTTATTTGTTGTTTAAGTAAATTTAATTGTTTG
Cep161 3295R GAGCTAAAGAGATGGTCATGGTTCTTTTGGTTGTGG
Cep161 2290R CCTTCTCATCCAATAGCAATTACTGTTGTTGTTGATTAGCC
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Tobias Lamkemeyer, Proteomics Facility of CECAD, for mass spectrometry analysis, Maria Stumpf for help with phosphorylation assays, Julia Janzen for help with cloning, Berthold Gaßen for providing antibodies and Ramesh Rijal for help with RT-PCR. SK was a member of the CECAD Graduate School.
Funding
This work was supported by the CMMC, CECAD and DFG (Schl 204/11-1).
References
- Bornens M. The Centrosome in Cells and Organisms. Science 2012; 335:422-6; PMID:22282802; http://dx.doi.org/10.1091/10.1126/science.1209037
- Keryer G, Di Fiore B , Celati C, Lechtreck KF, Mogensen M, Delouve A, Lavia P, Bornens M, Tassin A. Part of Ran is associated with AKAP450 at the Centrosome : involvement in microtubule-organizing activity. Mol Biol Cell 2003; 14:4260-71; PMID:14517334; http://dx.doi.org/10.1091/10.1091/mbc.E02
- Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol 2005; 21:411-34; PMID:16212501; http://dx.doi.org/10.1091/10.1146/annurev.cellbio.21.122303.120418
- Ueda M, Schliwa M, Euteneuer U. Unusual centrosome cycle in Dictyostelium: correlation of dynamic behavior and structural changes. Mol Biol Cell 1999; 10:151-60; PMID:9880333; http://dx.doi.org/10.1091/mbc.10.1.151
- Reinders Y, Schulz I, Gräf R, Sickmann A. Identification of novel centrosomal proteins in dictyostelium discoideum by comparative proteomic approaches. J Proteome Res 2006; 5:589-98; PMID:16512674; http://dx.doi.org/10.1021/pr050350q
- Andersen JS, Wilkinson CJ, Mayor T. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003; 426:570-4; PMID:14654843; http://dx.doi.org/10.1091/10.1038/nature02163.1
- Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol 2005; 15:303-11; PMID:15953548; http://dx.doi.org/10.1091/10.1016/j.tcb.2005.04.008
- Badano JL, Teslovich TM, Katsanis N. The centrosome in human genetic disease. Nat Rev Genet 2005; 6:194-205; PMID:15738963; http://dx.doi.org/10.1091/10.1038/nrg1557
- Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 2014; 156:893-906; PMID:24581491; http://dx.doi.org/10.1091/10.1016/j.cell.2013.12.043
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 2003; 114:457-67; PMID:12941274; http://dx.doi.org/10.1091/10.1016/S0092-8674(03)00557-9
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol 2003; 5:914-20; PMID:14502294; http://dx.doi.org/10.1091/10.1038/ncb1050
- Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, Kim MC, Jeong WI, Calvisi DF, Kim JM, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci U S A 2010; 107:8248-53; PMID:20404163; http://dx.doi.org/10.1091/10.1073/pnas.0912203107
- Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A 2010; 107:1437-42; PMID:20080689; http://dx.doi.org/10.1091/10.1073/pnas.0911427107
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development 2011; 138:9-22; PMID:21138973; http://dx.doi.org/10.1091/10.1242/dev.045500
- Pantalacci S, Tapon N, Léopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol 2003; 5:921-7; PMID:14502295; http://dx.doi.org/10.1091/10.1038/ncb1051
- Tapon N, Harvey KF, Bell DW, Wahrer DCR, Schiripo T A, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 2002; 110:467-78; PMID:12202036
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 1995; 9:534-46; PMID:7698644
- Kango-Singh M. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 2002; 129:5719-30; PMID:12421711; http://dx.doi.org/10.1091/10.1242/dev.00168
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005; 122:421-34; PMID:16096061; http://dx.doi.org/10.1091/10.1016/j.cell.2005.06.007
- Rohlfs M, Arasada R, Batsios P, Janzen J, Schleicher M. The Ste20-like kinase SvkA of Dictyostelium discoideum is essential for late stages of cytokinesis. J Cell Sci 2007; 120:4345-54; PMID:18042625; http://dx.doi.org/10.1091/10.1242/jcs.012179
- Artemenko Y, Batsios P, Borleis J, Gagnon Z, Lee J, Rohlfs M. Tumor suppressor Hippo / MST1 kinase mediates chemotaxis by regulating spreading and adhesion. Proc Natl Acad Sci U S A 2012; 109:13632-7; PMID:22847424; http://dx.doi.org/10.1091/10.1073/pnas.1211304109
- Müller-Taubenberger A, Kastner PM, Schleicher M, Bolourani P, Weeks G. Regulation of a LATS-homolog by Ras GTPases is important for the control of cell division. BMC Cell Biol 2014; 15:25-34; PMID:24986648; http://dx.doi.org/10.1091/10.1186/1471-2121-15-25
- Sebé-Pedrós A, Zheng Y, Ruiz-Trillo I, Pan D. Premetazoan Origin of the Hippo Signaling Pathway. Cell Rep 2012; 1:13-20; PMID:22832104; http://dx.doi.org/10.1091/10.1016/j.celrep.2011.11.004
- Blau-Wasser R, Euteneuer U, Xiong H, Gassen B, Schleicher M, Noegel AA, Robert U, Johnson W. CP250 , a novel acidic coiled coil protein of the Dictyostelium centrosome, affects growth, chemotaxis and the nuclear envelope. Mol Biol Cell 2009; 20:4348-61; PMID:19692569; http://dx.doi.org/10.1091/10.1091/mbc.E09
- Weiner OH, Murphy J, Griffiths G, Schleicher M, Noegel AA. The actin-binding protein comitin (p24) is a component of the Golgi apparatus. J Cell Biol 1993; 123:23-34; PMID:8408201; http://dx.doi.org/10.1091/10.1083/jcb.123.1.23
- Choi YK, Liu P, Sze SK, Dai C, Qi RZ. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol 2010; 191:1089-95; PMID:21135143; http://dx.doi.org/10.1091/10.1083/jcb.201007030
- Faix J, Gerisch G, Noegel AA. Constitutive overexpression of the contact site A glycoprotein enables growth-phase cells of Dictyostelium discoideum to aggregate. EMBO J 1990; 9:2709-16; PMID:2390970
- Wallraff E, Wallraff HG. Migration and bidirectional phototaxis in Dictyostelium discoideum slugs lacking the actin cross-linking 120 kDa gelation factor. J Exp Biol 1997; 200:3213-20; PMID:9364027
- Arasada R, Son H, Ramalingam N, Eichinger L, Schleicher M, Rohlfs M. Characterization of the Ste20-like kinase Krs1 of Dictyostelium discoideum. Eur J Cell Biol 2006; 85:1059-68; PMID:16842885; http://dx.doi.org/10.1091/10.1016/j.ejcb.2006.05.013
- Hergovich A, Kohler RS, Schmitz D, Vichalkovski A, Cornils H, Hemmings BA. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr Biol 2009; 19:1692-702. PMID:19836237; http://dx.doi.org/10.1091/10.1016/j.cub.2009.09.020
- Khurana B, Khurana T, Khaire N, Noegel AA. Functions of LIM proteins in cell polarity and chemotactic motility. EMBO J 2002; 21:5331-42; PMID:12374734; http://dx.doi.org/10.1091/10.1093/emboj/cdf550
- Faix J, Kreppel L, Shaulsky G, Schleicher M, Kimmel AR. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res 2004; 32:e143-9; PMID:15507682; http://dx.doi.org/10.1091/10.1093/nar/gnh136
- Noegel AA, Rivero F, Albrecht R, Janssen KP, Köhler J, Parent CA, Schleicher M. Assessing the role of the ASP56/CAP homologue of Dictyostelium discoideum and the requirements for subcellular localization. J Cell Sci 1999; 112:3195-203; PMID:10504325
- Noegel AA, Blau-Wasser R, Sultana H, Müller R, Israel L, Schleicher M, Patel H, Weijer CJ. The cyclase-associated protein CAP as regulator of cell polarity and cAMP signaling in Dictyostelium. Mol Biol Cell 2004; 15:934-45; PMID:14595119; http://dx.doi.org/10.1091/10.1091/mbc.E03-05-0269
- Xiong H, Rivero F, Euteneuer U, Mondal S, Mana-Capelli S, Larochelle D, Vogel A, Gassen B, Noegel AA. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic 2008; 9:708-24; PMID:18266910; http://dx.doi.org/10.1091/10.1111/j.1600-0854.2008.00721.x
- Schleicher M, Gerisch G, Isenberg G. New actin-binding proteins from Dictyostelium discoideum. EMBO J 1984; 3:2095-100; PMID:16453551
- Bertholdt G, Stadler J, Bozzaro S, Fichtner B, Gerisch G. Carbohydrate and other epitopes of the contact site A glycoprotein of Dictyostelium discoideum as characterized by monoclonal antibodies. Cell Differ 1985; 16:187-202; PMID:2408765; http://dx.doi.org/10.1016/0045-6039(85)90516-0
- Müller R, Herr C, Sukumaran SK, Omosigho NN, Plomann M, Riyahi TY, Stumpf M, Swaminathan K, Tsangarides M, Yiannakou K, et al. The cytohesin paralog Sec7 of Dictyostelium discoideum is required for phagocytosis and cell motility. Cell Commun Signal 2013; 11:54-69; PMID:23915312; http://dx.doi.org/10.1091/10.1186/1478-811X-11-54
- Khaire N, Müller R, Blau-Wasser R, Eichinger L, Schleicher M, Rief M, Holak TA, Noegel AA. Filamin-regulated F-actin assembly is essential for morphogenesis and controls phototaxis in Dictyostelium. J Biol Chem 2007; 282:1948-55; PMID:17121815; http://dx.doi.org/10.1091/10.1074/jbc.M610262200
- Kastner PM, Schleicher M, Müller-Taubenberger A. The NDR family kinase NdrA of Dictyostelium localizes to the centrosome and is required for efficient phagocytosis. Traffic 2011; 12:301-12. PMID:21134080; http://dx.doi.org/10.1091/10.1111/j.1600-0854.2010.01147.x
