?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Investigation of the mechanisms leading to aneuploidy and polyploidy is critical to cancer research. Previous studies have provided strong evidence of the importance of tetraploidization as an early step in tumorigenesis. In cancer cells, tetraploid cells may contribute to abnormal mitotic progression, which may be associated with cytokinesis failure. Tetraploidy leads to genomic instability due to centrosome and chromosome over-replication. Until now, the mechanism by which cells maintain tetraploid status has been unknown. Here, we identified a novel D box-containing protein, FLJ25439, which displays a dynamic expression profile during mitosis/cytokinesis with the midbody as the most prominent associated structure. To understand the function of FLJ25439, we established stable cell lines overexpressing FLJ25439. FLJ25439-overexpression cells grew slower and displayed a tetraploid DNA content in comparison with diploid parental cells. They also showed aberrant mitosis and dysregulated expression of p53, pRb and p21, suggesting a defect in cell cycle progression. To explore the molecular mechanisms responsible for FLJ25439-induced tetraploidization, we conducted a comparative analysis of the global protein expression patterns of wild type and overexpressors using proteomics and bioinformatics approaches. Protein category profiling indicated that FLJ25439 is involved in pathways related to anti-apoptosis, protein folding, the cell cycle, and cytoskeleton regulation. Specifically, genotoxic-stress- and ER stress-related chaperone proteins greatly contributed to the FLJ25439 overexpression phenotypes. The results of this study pave the way to our further understanding of the role of this novel cytokinesis-related protein in protecting cells from environmental stress and tetraploid formation.
Abbreviations
| ER stress | = | endoplasmic reticulum stress |
| 2DE PAGE | = | 2-dimensional polyacrylamide gel electrophoresis |
| IEF | = | isoelectric focusing |
| DTT | = | dithiothreitol |
| SDS | = | sodium dodecyl sulfate |
| HRP | = | horseradish peroxidase |
| MALDI-TOF MS | = | matrix-assisted laser desorption ionization-time of flight mass spectrometry |
| PMF | = | peptide mass fingerprint |
Introduction
Cell division follows an ordered progression of events and is critical for the development of multicellular organisms. This process requires the temporal–spatial coordination of nuclear division (mitosis) and cytoplasmic division (cytokinesis) to ensure that each daughter cell receives a full complement of chromosomes and cellular material.Citation1 In animal cells, cytokinesis starts by assembly of a central spindle during early anaphase, a structure generated by compaction of the overlapping non-kinetochore microtubules in the spindle midzone as the chromosomes move toward the spindle poles.Citation2 The central spindle is required to both initiate and complete cytokinesis and together with the spindle asters it serves to stimulate the initiation of the cleavage furrow and determines the position of the cleavage plane by transmitting a spatial signal through a pathway involving the small GTPase RhoA, leading to the assembly of an actomyosin ring at the equatorial cell cortex.Citation3 Contraction of the actomyosin ring pulls the overlying plasma membrane toward the center of the cell where it reaches the central spindle and constricts components of the central spindle into a transiently focused structure called the midbody.Citation4
The final stage of cytokinesis occurs via a process referred to as abscission, in which midbody cleavage concludes cell division and 2 daughter cells are formed. The mechanism of regulation of abscission timing in animal cells is poorly defined, but seminal work has identified the existence of an abscission checkpoint (also termed NoCut) that depends on Aurora B functions as a sensor to control abscission timing in response to unsegregated chromatin in the cleavage plane and to protect missegregating cells against tetraploidization by furrow regression.Citation5 The midbody provides an anchor for the ingressed cleavage furrow. For example, midbody-localized centrosome protein 55kDa (CEP55) serves as a binding platform for the ESCRT complex, which is essential for abscission.Citation6 Several key mitotic regulators, such as polo-like kinase and components of the centralspindlin complex as well as of the chromosome passenger complex have been shown to redistribute to the central spindle during anaphase and then become highly organized into the midbody during cytokinesis.Citation7 Disruption of these midzone/midbody components results in the defective assembly of the midbody and failure of cytokinesis.
Faithful cytokinesis requires a complex interplay among many protein components related to the cytoskeleton, chromosome, cell cycle, and membrane trafficking factors.Citation2,8 Failure to complete cytokinesis can be associated with developmental or pathological cell division programs leading to formation of a persistent connection between the 2 daughter cells and subsequent tetraploid progenies. In mammals, unlike in organisms such as fish and amphibians, tetraploidization is poorly tolerated, and most cell types (with some exceptions such as hepatocytes, syncytiotrophoblasts, and megakaryocytes) do not tolerate significant variations from the diploid status, meaning that tetraploid as well as higher-order polyploid cells usually activate programmed death pathways as soon as they are generatedCitation9 or elicit immune responses resulting in their elimination.Citation10 Cells with tetraploid or polyploid DNA content have been hypothesized to be a genetically unstable intermediates that often progress to aneuploidy in the succeeding divisions,Citation11,12 which has been detected in many cancers as well as in some precancerous lesions such as cervical metaplasia or Barrett's esophagus.Citation13 Thus novel molecules identified to be related to tetraploid formation will likely be of use as tools in anti-tetraploidization approaches.
Using a yeast 2 hybrid system, we identified a novel destruction box (D box)-containing protein, FLJ25439, which displays a dynamic expression profile during mitosis/cytokinesis with the midbody as the most prominent associated structure. Transient overexpression of FLJ254309 causes midbody arrest at cytokinesis and stable FLJ25439 expression induces tetraploidization and aberrant mitosis leading to delayed cell cycle progression. During tetraploidization, genomic instability might result in cellular stress and subsequent deregulation of biochemical pathways. To further reveal the biological function of FLJ25439, high-throughput proteome tools using 2DE and TOF/TOF MS were used to elucidate the effects caused by FLJ25439 as well as the associated signaling pathways. MetaCore pathway analysis was applied to dissect the protein–protein interactions and global cellular mechanisms behind the differences in protein levels.Citation14,15 The characterization of FLJ25439 protein detailed here provides new insights into control of tetraploidization.
Results
Identification of FLJ25439 as a novel cytokinesis-associated protein
Previously, we carried out a yeast 2 hybrid screen to identify pinin interacting proteins from the human kidney cDNA library. A number of independent clones were isolatedCitation16 and among them, the MC25-1 clone (NCBI Accession: JN799997), encoding a hypothetical protein annotated as tetratricopeptide-repeat domain 23 like protein (TTC23L) in the Ensembl database, matched to the EST FLJ25439 cDNA from a testis library in the DNA database of Japan (DDBJ).Citation17 The human FLJ25439 cDNA was deduced to code for an as yet uncharacterized 361 amino acid protein. As analyzed by the software, the primary sequence of the FLJ25439 protein comprises 2 tetratricopeptide repeat (TPR) like motifs (aa 168–201, and aa 261–294), 3 D-boxes (aa 221–224, aa 228–231, and aa 331–334), and a putative nuclear export signal (NES, aa 217–226) (). The D-box is a 4 amino acid RxxL motif identified in some mitotic proteins, such as pole-like kinase-1 (Plk-1), aurora B kinase, and cyclin B, and this motif is associated with the APC/C machinery-dependent regulation.Citation18 Among the 3 D-boxes, the first and the second are conserved in FLJ25439 protein across the mammalian species, while the third is only present in human FLJ25439 (Fig. S1A). Sequence alignment of the 2 TPRs of FLJ25439 protein revealed that its TPRs are not completely compatible with the consensus of TPR (Fig. S1B).Citation19 Other than the TPR-like and D-box sequence, the FLJ25439 protein is not homologous to any known protein. Interestingly, phylogeny analysis with Clustalw 2 (http://www.ebi.ac.uk/Tools/msa/clustalws2/) indicates that human (H. sapiens) FLJ25439 shows a closer relationship in amino acid sequence with canine (C. familiaris) and ox (B. Taurus) than with mouse (M. musculus) and rat (R. norvegicus) (Fig. S1C), suggesting an evolutionary adaptation of this protein toward higher species.
Figure 1. Molecular characterization of FLJ25439 protein. (A) Schematic representation of FLJ25439 domain structure. FLJ25439 protein is 361 amino acids in length. Three domains were recognized based on their sequence structure. (B) Deduced amino acid sequence of FLJ25439 protein. The tetratricopepetide repeat (TPR) is highlighted in yellow, 3 destruction boxes (D-box) are highlighted in gray and the putative nuclear export signal is underlined. The peptide used for generating 2A4 monospecific antibody against FLJ25439 is underlined in red.

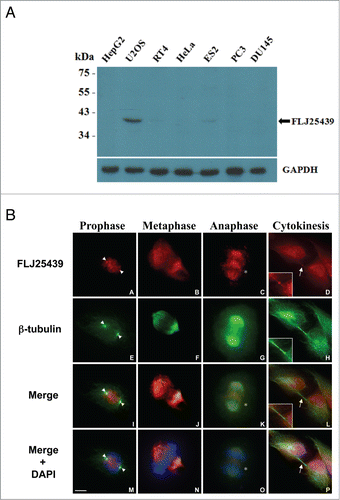
To investigate the localization and expression profile of FLJ25439 protein at the cellular level, we generated a monoclonal antibody 2A4 by immunization of mice with a synthetic peptide corresponding to amino acid residues 346–361 of FLJ25439 protein (). The 2A4 antibody is able to recognize an approximate 42 kDa endogenous protein in U2OS cells and ES2 cells, but not in other cell lines tested (HeLa, PC3 and HepG2 etc., ), suggesting a cell type-dependent expression manner. Based on the predicted size of FLJ25429 protein from cDNA cloning, the 42 kDa protein recognized by 2A4 antibody is consistent with the theoretical value of 40.80 kDa of a protein with 361aa. The specificity of the antibody to recognize FLJ25439 protein was confirmed at cellular level by imunostaining of U2OS cells in the presence of the peptide used to generate 2A4. Addition of peptide prevents cells from staining with 2A4, while 2A4 stains positively in cells without peptide treatment (Fig. S2). To examine cellular distribution of FLJ25439 protein in U2OS cells during mitosis/cytokinesis, we performed immunofluorecsence microscopy with 2A4 antibody (). In parallel, cells were also co-stained with DAPI and anti-α-tubulin antibodies to verify the position of chromosomes and the mitotic spindle. FLJ25439 displayed a highly dynamic subcellular localization during cell cycle progression; initially it was detected on split centrosomes in prophase cells (arrowheads in A,E, I, M), translocated to the mitotic spindles during metaphase ( B, F, J, M) as well as in early anaphase ( C, G, K, O), and finally localized to the midbody during cytokinesis (arrows in D, L, P). In addition to the mitotic spindle, FLJ25439 can be observed at the spindle midzone and the cleavage furrow in anaphase (stars in C, K, O), indicating FLJ25439 behaves like a mitotic passenger protein in U2OS cells. When using HeLa cells for immunofluorescence staining, however, only the midbody that stained positive for Cep55 (Fig. S3L), a marker of cytokinesis midbody, was positive for FLJ25439 (Fig. S3F and X), whereas interphase cells (Fig. S3A) and the centrosome (stained positively with γ-tubulin, Fig. S3G) were negative for FLJ25439 staining, indicating FLJ25439 expression is restricted in certain cell cycle phase which corresponds well with the Western data () that FLJ25439 are barely detected in interphase HeLa cells. Given the fact that central spindle and midbody are considered to be the essential structures required for the initiation and completion of cytokinesis,Citation4 we surmise that FLJ 25439, based on its cellular location, is functionally relevant to cell cycle progression and cytokinesis.
Figure 2. Expression profile and subcellular localization of FLJ25439 protein. (A) Western blot demonstrating cell type-variation in expression pattern of FLJ25439 protein. FLJ25439 protein was recognized as a 41 kDa protein by 2A4 antibody protein gel blot. Among cell lines tested, U2OS cells express enough FLJ25439 protein to be readily detected while other cells express little or no endogenous FLJ25439. (B) Immunofluorescent micrographs of FLJ25439 protein localization during cell cycle progression. U2OS cells were fixed and immunostained with antibodies to FLJ25439 (red), ß-tubulin (green) and nuclei were counter stained with DAPI. FLJ25439 is observed at split centrosomes (arrowheads in A, E, I, M) in prophase, localized with the mitotic spindle during mitosis, distributed in the central spindle (stars in C, K, O) in anaphase and targeted to the midbody (arrows in D, L, P) in cytokinesis. The insets are an enlarged version of the region indicated by the arrows. Bar, 5 μm.
FLJ25439 overexpression elicits cytokinetic arrest and retards cell proliferation
To gain insight into the function of FLJ25439 protein, we used a gain-of-function approach to overexpress FLJ25439 protein in HeLa cells. The reason that we carried out overexpression rather than depletion to examine FLJ25439 protein function is because FLJ2543l9 protein is expressed at very low levels in HeLa cells, making overexpression a logical rationale for experimental manipulation. To thoroughly investigate the role of FLJ25439 in cell cycle progression, we generated stable HeLa clones overexpressing HA-tagged FLJ25439 protein by means of retroviral infection. Among the 4 clones selected, the 1–16 clone, referred to as HeLa(1-16), expressed relatively higher level exogenous FLJ25439 protein () and was chosen for subsequent experiments. HeLa cells were found to pair together after transient transfection of EGFP-tagged FLJ25439, and exogenous FLJ25439, as revealed by GFP, can be observed at the midbody of transfected cells undergoing cell division (arrow in upper panel). Quantitation of EGFP-FLJ25439 expressing cells indicated that more than 40% vs. <5% EGFP only-transfected cells were arrested in cell division with prominent midbody localization ( lower panel), suggesting that FLJ25439 overexpression interferes with the cell division cycle and may arrest cells at cytokinesis. Using immunofluoresence microscopy analysis, we analyzed distribution of exogenously expressed FLJ25439 protein during mitosis/cytokinesis in HeLa(1-16) cells (Fig. S4) and found that its cellular distribution was strikingly similar to that of endogenous proteins in U2OS cells at various cell cycle phases (). During cell cycle progression, HA-tagged FLJ25439 proteins co-localized with γ-tubulin at centrosomes in prophase, associated with the microtubule spindle in the metaphase, co-distributed with the central spindle microtubules in anaphase and telophase, and were present in the midbody during cytokinesis (arrows in Fig. S4F). Thus it appears the relative expression levels of FLJ25439 may determine its cellular distribution during mitosis/cytokinesis and, at cytokinesis, the midbody can be considered the most obviously recognizable organelle with which FLJ25439 associates in our tested cell lines.
Figure 3. FLJ25439 overexpression induces midbody arrest and retards mitosis during cell cycle progression. (A) Western blot identification of stable HeLa clones expressing exogenous HA-FLJ25439 fusion protein. HeLa cells were transfected for stable expression of HA-Flag-FLJ25439 protein, and stably expressing HA-tagged FLJ25439 protein was selected based on immunoblotting of cell lysate with anti-HA or 2A4 antibodies. Among cell lines tested HeLa(1-16) expresses relatively higher level of exogenous FLJ25439 protein. (B) Representative image of HeLa cells transfected with EGFP-FLJ25439 fusion protein (upper panel) and percentage of cells with midbody was scored (lower panel). Note that a large amount of cells transfected with EGFP-FLJ25439 display cytokinetic defect with prominent midbody (white arrows in D, F) decorated with EGFP compared to EGFP vector only-transfected cells. Data are mean ± S.D (n = 4, 100 cells each). **, P < 0.05. Bar, 5 μm. (C) Sequential images of HeLa (upper panel) and HeLa(1-16) (lower panel) synchronized by release from double thymidine block were captured by time lapse microscope. Arrows indicate cells undergoing mitosis. Red arrows track HeLa (1-16) that show increased time in mitosis compared to its normal counterparts (yellow arrows in HeLa or orange arrows in HeLa(1-16)). (D) Quantitative analysis of cell cycle phase of HeLa (1-16) vs HeLa wild type as a function of time (minutes). Pro-M, prometaphase; M, metaphase; A, anaphase; T, telophase. Data are mean ± S.D (n = 11). (E) HeLa(1-16) is less sensitive (IC50 = 647 μM) to oxidative stress than HeLa (IC50 = 106 μM). Cell viability was assayed by MTT assay. Data are mean ± SD (n = 3).
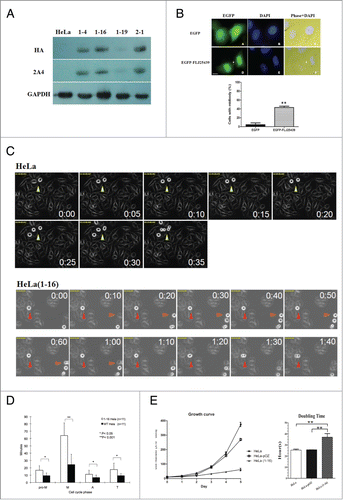
Using phase microscopy observation, we observed that the size and growth rate of HeLa(1-16) cells as well as of other stable HeLa clones expressing exogenous FLJ25439 were different from those of the parental cells. When stained with wheat germ agglutinin (WGA) and DAPI to visualize the cell boundary and nuclei, respectively, cells were bigger in terms of nuclear size and cell volume (, right panel). To analyze growth rate, we grew the same number of HeLa(1-16), HeLa-pOZ (vector-only transfected HeLa cells) or parental HeLa cells and compared their growth after one to 5 days, respectively. Compared with the parental or HeLa-pOZ cells, which displayed exponential growth day-by-day, HeLa(1-16) grew in a linear fashion without a significant increase in cell number (, left panel). In general the doubling time of HeLa(1-16) and other stable clones (data not shown) was 37 hours on average while HeLa-pOZ, like parental HeLa, divided at a shorter interval of 25 hours ( right panel). Subsequent experiments, therefore, used HeLa only as a comparison control for HeLa(1-16). Taken together, overexpression of FLJ25439, a novel midbody-associated protein, perturbs cell cycle progression and may lead to defects in cell growth.
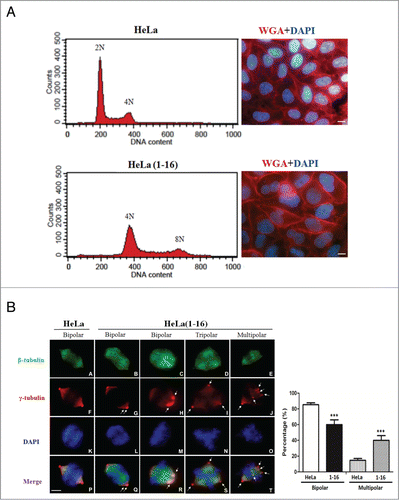
Figure 4. FLJ25439 overexpression elicits tetraploidization and aberrant mitoses. (A) Representative flow cytomerty analysis of DNA content of HeLa compared to HeLa(1-16). The DNA histogram showed HeLa(1-16) harbored tetraploid DNA complement, while the parental HeLa cells are diploid. Both the nucleus (stained blue by DAPI) and cell (stained red by WGA) size of HeLa(1-16) cells are larger than those of the parental HeLa. (B) HeLa(1-16) display multipolar mitosis during cell cycle progression. Cells were synchronized by release from thymidine block and immunostained (left panel) with β-tubulin (green), γ-tubulin (red) and counter stained with DAPI (blue). Bipolar or multipolar mitoses were scored for HeLa(1-16) compared to HeLa (right panel). Data are mean ± SD (n = 3, each 100 cells). Arrows indicate centrosomes. Bar, 5 μm. ***, P < 0.01.
FLJ25439 stable expression induces HeLa tetraploidization
Given that failure in cytokinesis leads to tetraploidization and increased cell size is the most obvious and consistent consequence of an increase in ploidy, we attempted to analyze HeLa(1-16) DNA content by flow cytometry. For cell cycle profile analysis, cells were fixed and stained with propidium iodide to measure their DNA content. Based on the fluorescence parameter used for DNA analysis, parental HeLa had a diploid (2N) complement of chromosomes. Interestingly, HeLa(1-16) did not exhibit a peak profile consistent with the diploid complement, it rather displayed a DNA complement equivalent to tetraploid (4N) cells with some cells displaying an octoploid (8N) DNA complement ( left panel). In general tetraploid cells are characterized by 4N DNA content in the G1 phase of the cell cycle and by an 8N DNA content in G2 and M phase. Based on our cytometry analysis we conclude that HeLa(1-16) and other FLJ25439 expressing stable clones (data not shown) exhibit bona fide tetraploidy traits. As revealed by both flow cytometry analysis and western blot, the tetraploid DNA contents of HeLa(1-16) were stable for at least 40 generations.
To examine how tetraploidization might have occurred, we tracked the onset and process of mitosis in synchronous cell populations by time-lapse microscopy (). The HeLa(1-16) and parental HeLa cells were synchronized at the G1/S boundary by double thymidine administration (65% vs 52% efficiency for HeLa and HeLa(1-16), respectively), and the synchronized cells were released in fresh medium for 10 hours to observe cells at mitosis. Cytokinesis failure can result from events during various stages of this process: specification of the cleavage plane, formation and then ingression of contractile ring and finally cell abscission. To investigate which of these events is relevant, we started by following round-up cells, which represent prometaphase stage, and monitored their morphological changes along with time until 2 daughter cells were clearly paired together (telophase). Using this method, we found that HeLa(1-16) (red arrowheads in lower panel and supplement movie 1) took a much longer time (64.1 ± 17.4 min, ) than parental HeLa (yellow arrowheads in upper panel and supplement movie 2) to reach anaphase (24.5 ± 14.0 min, ) but both cell types spent a similar amount of time, about 6.8∼11.4 min, to progress from anaphase to telophase (). Although using our current cellular model, which is not tagged with fluorescent labeling in live imaging, we were unable to trace whether there is any delay in abscission (abscised cells vs. interphase cells), results from EGFP-FLJ25439 overexpression () clearly demonstrated that FLJ25439 functions in abscission control through its association with the midbody. Based on the time-lapse imaging of HeLa(1-16) and transient transfection data, we conclude that FLJ 25439 overexpression perturbs anaphase onset and arrests/delays cell division at cytokinesis.
Tetraploidy constitutes a metastable state because tetraploid cells tend to lose chromosomes progressively during aberrant dipolar mitosesCitation12 and/or undergo multipolar mitosis during which chromosomes are distributed among the daughter cells in a near-random manner.Citation20 To examine the mitotic figures occurring in HeLa(1-16) during cell division, we also observed tetraploid cells in metaphase where the abnormal mitotic spindle can be identified (). HeLa(1-16) or parental HeLa were synchronized by double thymidine block and examined for mitotic spindles and centrosomes by antibodies against α-tubulin and γ-tubulin, respectively ( left panel). Events in which numbers of mitotic cells with either bipolar (with classical metaphase plates organized by 2 centrosomes), tripolar or multipolar (with more than 3 or 4 centrosomes and abnormal Y- or X-shaped metaphase figure) spindles were quantified (n = 3, each 100 cells, , right panel) and the results showed that HeLa(1-16) cells contain significantly more multipolar spindles than their parental counterparts, while, conversely, parental HeLa cells contain more bipolar spindles than HeLa(1-16) cells. Interestingly, though bipolar in nature, HeLa(1-16), unlike parental HeLa cells, contain supernumerary (more than 2) centrosomes clustered in spindle poles (arrows in , left panel, G and H), suggesting aberrant mitosis in HeLa(1-16). Aberrant mitoses in HeLa(1-16), therefore, may explain why HeLa(1-16) grew slowly during in vitro imaging () and routine culture ().
Identification of differential protein profiles of HeLa(1-16) vs. parental HeLa by proteomic analysis
FLJ25439 stable clones can generate tetraploidy in HeLa cells and the appearance of polyploidy cells is usually associated with cellular stress which should result in protein alterations in quality or quantity.Citation20 Consistent maintenance of a tetraploidy state also leads to profound changes in cellular physiology. To explore the global changes in protein profiles between cells transfected with or without FLJ25439, we conducted a proteomic analysis. As shown in , more than 1898 protein spots appeared in the 2-DE reference maps derived from the samples. Among these spots, 17 proteins with significant and meaningful changes were further identified by a PMF analysis as indicated by the Arabic numerals on the silver-stained gels. Results in all 3 repeats were consistent and reproducible. All the differentially expressed proteins were unambiguously identified by MALDI-TOF/TOF MS analyses. A typical example is shown in Fig. S5, in which GRP78 was characterized with a maximum of 34 matching peptides representing 54% sequence coverage. The results of the spectrometric analyses and protein functions are summarized in . As expected, a group of chaperones and the proteins involved in modulation of ER stress, protein folding and oxidative stress were revealed, including protein glucose regulated protein (GRP) 75, GRP78, GRP94, HSC7C, TCPB, and PRDX6.
Table 1. List of significantly different protein spots between mock and FLJ35439 transfected HeLa cells
Figure 5. Expression profiling and network analysis of HeLa(1-16) with FLJ25439 overexpression. (A) Comparison of 2-DE protein profiles of samples from HeLa and HeLa (1-16) groups. The protein spots found to be significantly different in volume are indicated by Arabic numbers. (B) Protein expression levels of proteins with significant changes in volume were validated by western blotting analysis. β-actin was used as an internal control and the relative expression to β-actin is shown at the bottom. (C) Top-ranked GO processes from GeneGoMetaCore pathway analysis. Pathways were ranked according to P values; bars represent the inverse log of the P value.
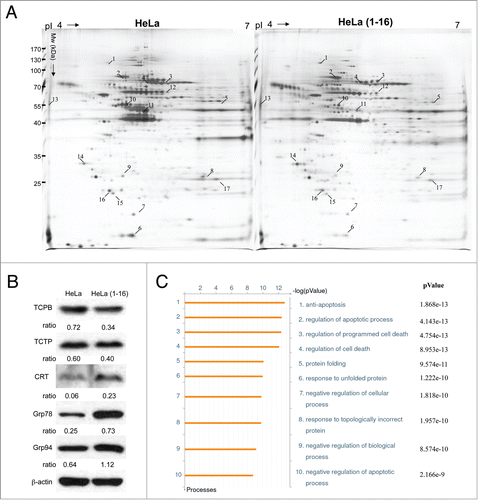
Western blot analysis was performed by using antibodies against several proteins to confirm the changes in protein expression levels revealed by proteomic tools. We detected a remarkable increase in levels of calreticulin (CRT), GRP78 and GRP94, whereas TCPB and TCTP were noticeably decreased in FLJ25439 overexpressed HeLa(1-16) as compared with the control (). The plasmid pOZ-transfected cells showed no inducing in GRP78 level, suggesting that the vector would not markedly affect cell physiology in our cell model (Fig. S6). These findings are in line with our results of the 2-DE protein expression profiles on HeLa(1-16) and the control.
Functional network analysis
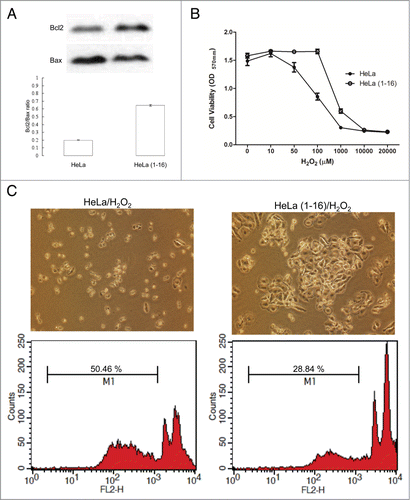
To further reveal the relationship of the differentially expressed proteins elucidated by the 2-DE analysis and their significance in the mechanisms involved in tetraploidization induced by FLJ25439 stable expression, proteins were analyzed by applying the MetaCore analytical tool. The network was generated using the shortest-path algorithm to map interactions and a biological process was assigned to each network. As demonstrated in , the top 10 enrichment GO pathways indicated that the differentially expressed proteins after stable FLJ25439 expression were primarily involved in the following cellular networks; anti-apoptosis (P = 1.868 × 10−13), regulation of apoptotic process (P = 4.143 × 10−13), regulation of programmed cell death (P = 4.754 × 10−13), regulation of cell death (P = 8.953 × 10−13) and protein folding (P = 9.574 × 10−11). To further validate the results revealed by the bioinformatic results, the signal transduction pathways inducing cell apoptosis were determined via immunoblot analysis. It has been suggested that the Bcl-2/Bax ratio may be a key indicator of susceptibility of cells to apoptosis. Therefore, we evaluated the expression level of Bcl-2 and Bax. As expected, FLJ25439 overexpressing cells had a higher Bcl-2/Bax ratio, suggesting that FLJ25439 would inhibit apoptosis and enhance cell survival (). Based on the 2-DE analysis, FLJ25439 overexpression would upregulate the expression levels of several proteins associated with modulating oxidative and ER stress. We next studied whether FLJ25439 stable clones could protect cells from oxidative stress by exposing HeLa(1-16) or control cells to various concentrations of H2O2 for 24 h and MTT assays were performed to evaluate the cell viability (). The results indicated that 100 μM of H2O2 obviously reduced control HeLa cell viability, while FLJ25439 stable clones could survive under administration of higher concentration of H2O2. Moreover, control HeLa cells obviously detached from culture dishes and became round shape while HeLa(1-16) were intact under hydrogen peroxide treatment (). Again, the proportion of apoptotic cells (M1) was promoted in control cells compared to FLJ25439 stable clones by the flow cytometric analysis (). These findings suggest that FLJ25439 overexpression may attenuate sensitivity to cellular stress.
Figure 6. FLJ25439 overexpression renders HeLa(1-16) resistant to oxidative stress. (A) The protein levels of Bcl-2 and Bax were determined by protein gel blotting assays. Density ratio of Bcl2 over Bax was measured by a densitometer. (B) HeLa(1-16) is less sensitive (IC50= 647 μM) to oxidative stress than HeLa (IC50 = 106 μM). Cell viability was assayed by MTT assay. Data are mean ± SD (n = 3). (C) Cell images and flow cytometric analysis for HeLa and HeLa(1-16) under hydrogen peroxide exposure. M1 represents percentage of apoptotic cells.
Tetraploids caused by FLJ25439 modulate cell-cycle gene expression
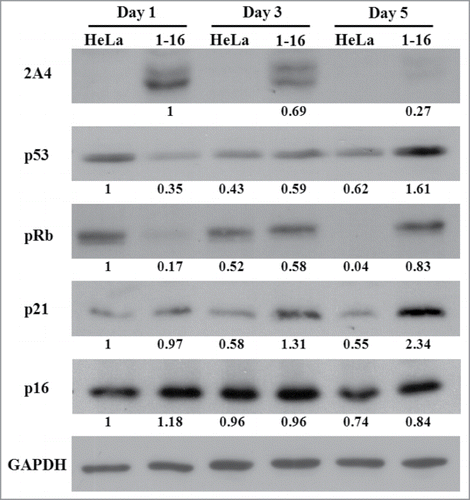
Since inactivation of p53 was reported to be permissive for tetraploidization and facilitate survival of tetraploid cells,Citation9,21 we further attempted to elucidate the relationship of FLJ25439 expression with regard to p53 axis-associated cell cycle proteins. Western blot was performed on extracts from HeLa(1-16) and control cells cultured for different periods of time and expression levels of FLJ25439, determined by 2A4 antibody, as well as those of p53, p21, pRb and p16 were quantified (). Surprisingly we found that in HeLa(1-16), exogenously expressed FLJ25439 was decreased markedly from day 1 to day 5, while p53, p21 and pRb expression levels were significantly increased to various degrees. In contrast with HeLa(1-16), control cells expressed decreased levels of p53, p21 and pRb upon continuous culture with little change in p16 expression. Thus it appears that FLJ25439 not only affects stress response chaperone expression but also its expression inversely correlates with that of p53-associated signaling pathway proteins.
Discussion
A better understanding of cytokinesis is fundamentally important for biological research, and also highly relevant for medical research because failed cytokinesis generates tetraploid cells, which are genomically unstable and can promote tumorigenesis.Citation12 Here, we report the identification of FLJ25439, a novel D-box-containing protein which is a cytokinesis-associated molecule whose expression levels are extremely low in most of the cell types tested. FLJ25439 overexpression renders HeLa arrest at cytokinesis and delays cell growth/cell cycle progression through tetraploidization induction. Based on proteomic analysis, stable FLJ25439-expressing HeLa cells are less vulnerable to oxidative stress due to enhanced expression of oxidative and ER stress proteins.
There are several features of FLJ25439 in terms of sequence structure and subcellular location that make it an attractive protein for cell cycle research. First, FLJ25439 contains 3 D-boxes in sequence near its C-terminal half (). Destruction box is a signature motif frequently found in substrates of anaphase-promoting complex/cyclosome (APC/C). The APC/C, RING finger type ubiquitin ligase, is a key ubiquitin ligase that targets a number of cytokinesis-relevant substrates for destruction, including anillin, PLK1, Aurora A/B and controls several transitions in the cell cycle.Citation18 It will be of interest to investigate whether FLJ25439 is an APC/C substrate and how it might be regulated at different phases during cell cycle progression. Secondly, 2 TPR-like motifs occur in the FLJ25439 sequence which displays minor imperfection when compared to the TRP consensus (Fig. S1B). The TPR motif, a repeating 34 amino acid sequence, was originally identified in yeast cell division cycle (CDC)-associated proteins. TPR is predicted to be responsible for protein-protein interaction and is conserved in evolution.Citation22 In addition to cell cycle regulation, TPR-containing proteins participate in various cellular activities including mRNA transcription/splicing, protein trafficking and chaperoning.Citation19,23 It is plausible that FLJ25439 acts in cell-cycle regulation by interacting with proteins engaging in the above-mentioned cellular activity via the TPR motif. Third, we found FLJ25439 protein is located at the midbody at the final stage of cell division, while it may distribute in centrosomes and/or central spindles in different phases during cell cycle progression ( and Figs. S3 and S4). FLJ25439 protein thus exhibits an expression profile with a dynamic distribution pattern during mitosis/cytokinesis. Interestingly, however, FLJ25439 was not identified in a proteomic analysis of isolated intercellular bridges sample in which more than 100 different protein components were detected and many of which are essential for completion of cytokinesis.Citation8
The midbody is a transient structure that connects 2 daughter cells at the end of cytokinesis, with the principal function being to localize the site of abscission, which physically separates 2 daughter cells.Citation4 The midbody forms from the central spindle—a bipolar microtubule array that assembles between separating sister chromatids during anaphase and most central spindle components persist on the midbody until abscission.Citation7 Conversion of the central spindle to midbodies correlates positively with furrow ingression and the central spindle and midbody are therefore considered to be the essential structures required for the initiation and completion of cytokinesis. Thus proteins associated with the central spindle and/or midbody represent prime candidates for the regulation of cytokinetic stages. Indeed FLJ52439, when overexpressed transiently in HeLa cells, elicits midbody arrest at telophase and impairs cytokinesis (), while stable expression renders cellular tetraploidization as well as aberrant mitoses (), leading to retardation of cell cycle progression ().
One prominent mechanism of tumorigenesis involves an initial event of tetraploidization. Several possible mechanisms, including cytokinesis failure of tumor-relevant tetraploidization have been proposed. One scenario suggests that tetraploidy may arise from illicit cell-to-cell fusion among somatic cells triggered by virally encoded fusogenic proteins. Another scenario involves failure to complete mitosis (mitotic slippage), resulting from cells with defects in cytokinesis but that exhibit normal karyokinesis, such that CHK1-haploinsufficient cells with replicated chromosomes proceed directly from G2 to G1-S phase.Citation24 The third proposed pathway to tetraploidy is a scenario in which telomere damage promotes tetraploidization through endoreplication (DNA replication without mitosis), leading to incomplete cytokinesis.Citation25 Though similar in effect to generation of tetraploid cells, the detailed mechanism of tetraploidization induction by FLJ25439 overexpression remains largely elusive and warrants careful examination.
It has been shown that tetraploid cells exhibit an enhanced rate of spontaneous apoptosis that can be inhibited by p53 depletion or by knockdown of proapoptotic p53 target genes, indicating differential apoptosis regulation in tetraploid cells in a p53-dependent manner.Citation9 In addition, tetraploid cells were, unexpectedly, more resistant to DNA damaging agents than their diploid counterparts. That said, inactivation of p53 is permissive for tetraploidization (because p53-proficient cells usually die from apoptosis as they become tetraploid) and facilitate survival of tetraploid cells.Citation9,21 In our study, we observed that p53 expression increases after prolonged culture while exogenously expressed FLJ25439 decreases over time in the HeLa(1-16) stable tetraploid cell line (). There is also an inverse correlation between expression of p21, pRb and FLJ25439 during prolonged culture (). Although the mechanism for suppressing apoptosis in tetraploid/polyploid cells remains unclear, there is some evidence that genotoxic stress resistance appears to be linked to the p53-p21-pRb pathwayCitation26 in which p21, a cyclin-dependent kinase inhibitor, can inhibit Chk1, a key component of the DNA damage response pathway, thereby blocking apoptosis in response to DNA damage. Based on our analysis of relative expression of p53-p21-pRb vs. FLJ25439 in HeLa(1-16) culture compared to parental HeLa (), we conclude that FLJ25439 expression may override the p53 expression-induced apoptosis occurring in stable tetraploidization and enhanced p21 expression may therefore confer HeLa(1-16) resistance to oxidative stress (). However, we cannot exclude the possibility that increasing gene copy numbers by FLJ25439-induced tetraploidization could function as a buffer to protect cells against genotoxic damage.
Other than tetraploid nuclei, supernumerary centrosomes have been noted in the early stages of many solid tumors.Citation27 While the role of supernumerary centrosomes in tumor biology is likely to be multifaceted, it may impose a fitness cost on the growth of mature cancers because of the potential for multipolar mitosis. To circumvent this problem, many cancer cells develop mechanisms that suppress multipolar mitosis, the best studied being clustering of supernumerary centrosomes into 2 groups at spindle poles enabling bipolar mitosis.Citation28,29 Consistent with tetraploid nuclei identified in HeLa(1-16), we observed these cells displayed supernumerary centrosomes during mitosis with a marked increase in multipolar mitosis compared to parental HeLa () and prominent centrosome clustering in bipolar mitosis of HeLa(1-16). Centrosome clustering in tumor cells is incompletely understood; however, it is expected to be dependent, to a significant degree, on microtubule associated proteins (MAPs) and motors that organize the spindle poles.Citation29,30 Additional data also showed multipolar mitoses are more frequent when p53 is downregulated and the product of the Mos oncogene is upregulated.Citation20 Although HeLa(1-16) is susceptible to increased risk of genomic instability due to aberrant multipolar mitoses occurring during cell cycle progression (), we surmise that FLJ25439 overexpression may act as a factor, having an effect similar to MAPs but opposite to Mos oncogene, to promote coalescence of supernumerary centrosomes that allow for normal bipolar mitoses of tetraploid cells. Further experiments such as using RNAi against FLJ25439 are needed to demonstrate the role of FLJ25439 in centrosome clustering.
It has been shown that global transcriptome changes associated with tetraploidy are subtle and there is a minor (20–40%) increase in the expression of several p53 target genes.Citation9 Also multiple studies comparing the genome-wide expression patterns between isolated tetraploid/polyploid and diploid mammalian cells found very few differentially expressed genes,Citation9,31 suggesting that tetraploid/polyploidization does not induce specific transcriptional reprogramming events, but rather increases the transcriptional and translational output of the same set of gene products required for a cell type-specific function. For this reason, we set up experiments to detect the global protein expression profile of HeLa(1-16) using a proteomic approach with 2-DE coupled with MALDI-MS. In this study 17 protein spots displayed significant changes in HeLa cells after FLJ25439 stable expression (). GO network analysis demonstrated that most of these proteins are involved in anti-apoptosis, protein folding response and response to oxidative stress (). Particularly, there is a marked trend of elevated expression of ER stress proteins. The most abundant and well-characterized ER chaperone proteins including GRP 78 and CRT can be readily recognized in our list. ER stress triggers an evolutionarily conserved quality control mechanism, the unfolded protein response (UPR), which restores ER homeostasis by enhancing protein-folding capacity and degrading the misfolded ER proteins.Citation32 During tumorigenesis, for example, ER chaperones serve a host of important roles in maintaining ER homeostasis contributing to cancer cell survival and progression. Recently Senovilla et al.Citation10 reported that tetraploid cancer cells become immunogenic because of a constitutive ER stress response. Given that FLJ25439 overexpression causes cytokinesis defect, aberrant mitoses and subsequent stable tetraploidization followed by ER stress response and the ER stress response elicited by tetraploidization promotes tetraploid cell survival, we conclude that tetraploidization induced by FLJ25439 overexpression confers HeLa(1-16) resistance to oxidation and apoptosis through elevated expression of ER stress proteins.
In conclusion we identified a novel cytokinesis-associated protein, FLJ25439, whose distribution and expression levels correlate intimately with organelle/proteins engaged in cell cycle progression. Overexpression of FLJ25439 promotes cellular tetraploidization and induces subsequent ER stress response which protects tetraploids from apoptotic assault and maintains their survival. Further characterization of the role FLJ25439 plays in cell cycle regulation will provide a new direction for understanding hyperploidy formation and prevention.
Materials and Methods
Cell culture and transfection
HeLa and U2Os cells were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS) and antibiotics (50 U penicillin-streptomycin) at 37°C in a 5% atmosphere inside a humidified incubator. Transient transfection was performed using calcium-phosphate based on standard protocol. For G1/S boundary synchronization, the cells were blocked at G1/S phase by double thymidine administration, and the mitotic cells were collected after the synchronized cells were released in fresh medium at 10 h. Unless stated otherwise, cells were treated with the following reagents at the indicated concentration: taxol (0–500 nM, Sigma-Aldrich) and H2O2 (0–20 mM, Sigma-Aldrich). Fresh medium and drugs were replenished every 24 h.
Plasmid construction
The FLJ25439 open reading frame clone was established by PCR amplification. The forward primer CCGGATCCATGCAAGCCAGCCCCATC, and the reverse primer CCGGATCCCCTTGTCCTGTTTTTCTCCATTTTG were applied in the PCR amplification. The PCR products were digested with Bam HI and cloned into pUC18 vector. For FLJ25439/pEGFPC1 construction, FLJ25439/pUC18 plasmid was digested with Bam H I and the desired DNA fragment was introduced to pEGFP C1 vector at BglII sites. For pOZ retroviral expression vector construction, the open reading frame of FLJ25439 cDNA with the NotI and XhoI sites from FLJ25439/pUC18 was ligated to the corresponding sites in pOZ vector.
Antibody generation
The BALB/c mice are immunized by using standard immunization protocols. Synthetic peptides: EEFCKWLVQNGEKQDK corresponding to amino acid residues 346–361 in FLJ25439 protein, were used in mice immunization. The mice were intraperitoneally immunized 3 times with 100 μg synthetic peptides. One month after immunization, the mice were sacrificed, and splenocytes were isolated. The PEG is used for the fusion of splenocytoes and myeloma to generate hybridoma. After fusion, the hybridoma cells were grown in a selection medium containing hypoxanthine, aminopterin and thymidine (HAT) and screened for cells that produce the desired antibody.
Immunofluorescence microscopy
Cells were rinsed in PBS and fixed in chilled methanol for 5 minutes, or 4% paraformaldehyde and permeabilized with 0.5% triton X-100 for 10 minutes. Immunofluorescence was performed as describedCitation21 using the following primary antibodies: mouse anti-HA, mouse anti-FLAG, mouse anti-γ-tubulin GTU-88 (1:500, Sigam-Aldrich), mouse anti-cep55 clone 11A5, and rabbit anti-β-tubulin (1:200, Sigam-Aldrich). TRITC or FITC conjugated goat anti-mouse or rabbit secondary antibodies (1:200) were from Jackson Laboratory. Cells were mounted in 50% glycerol containing 0.4% n-propylgallate with or without DAPI. Imagines were captured using an Axioscope 2 microscope equipped with epifluorescence (Carl Zeiss).
Time-lapse microscopy
Cells were seeded onto 24-well plates and imaged using an Axiovert 200 M inverted fluorescent microscope (Carl Zeiss) equipped with a Coolsnap camera and a Chamlide TC temperature, humidity and CO2 control system. Data acquisition was performed at 5 min/frame.
Western blot analysis
Cells with various treatments were harvested in a lysis buffer and measured the concentrations with the Bradford Protein Assay Kit (AMRESCO). Proteins (30 μg) were separated on 10% SDS–PAGE gels and transferred to PVDF membranes. The blots were incubated with primary antibodies and then secondary antibodies conjugated to horseradish peroxidase. The signal was detected by enhanced chemiluminescence reagent. The density of western-blot bands was quantified by GeneTools software as previous reported.Citation33
2-DE and image analysis
Cell lysis was diluted in IPG sample buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, 65 mM DTT, and 1% IPG buffer to a volume of 350 μL. After rehydration for 12 h, IEF was conducted automatically with a total of 90 kVh. Following IEF separation and equilibration, electrophoresis was carried out on 10% acrylamide gels (Bio-Rad, Hercules, CA, USA) at 30 mA. Proteins were visualized by silver staining and then scanned using Imagescanner (GE Healthcare, Taiwan). Protein spots were quantified using the Prodigy SameSpots software (Nonlinear Dynamics, Newcastle, UK). The expression levels of protein spots changed more than 1.5 fold between cells transfected with FLJ25439 and mock control should be considered as statistical significance (P value <0.05).Citation34 All experiments were biologically repeated 3 times to confirm the reproducibility.
In-gel enzymatic digestion and MALDI-TOF/ TOF MS analysis
Silver-stained spots were excised and in gel-digested with trypsin according to procedures described previously.Citation19 The MALDI-TOF analysis was performed on an Ultraflex mass spectrometer (Bruker-Daltonik). Monoisotopic peptide masses were assigned and used for Swiss-Prot sequence database searches (SwissProt 2013_9, 540958 sequences; Taxonomy: Homo sapiens, 20272 sequences) with the BioTools 3.2 software (Bruker-Daltonik) and the Mascot search engine (http://www.matrixscience.com). Search parameters were set as follows: a maximum allowed peptide mass error of 50 ppm, and consideration of one incomplete cleavage per peptide. For the MS/MS, the 3 most intense precursor ions with a signal/noise ratio >25 were selected after exclusion of the common background signal. The MS/MS mode was operated at 1 keV and products of metastable decomposition at elevated laser power were detected. MS data were acquired with external calibration and MS/MS data were using the default instrument calibration.Citation14
Network analysis using MetaCore
MetaCore software (vers. 5.1 build 16271, GeneGo, St. Joseph, MI) was used to elucidate the ontological classes and relevant pathways represented among the proteins identified by 2-DE and the peptide mass fingerprint. Based on the gene ontological categorization, we used 2 algorithms for the network analysis: (a) an analysis network algorithm to deduce scoring processes regulated by differentially expressed proteins and (b) the shortest path algorithm to build a network consisting of the smallest possible number of direct interactions between differentially expressed proteins. The statistical relevance of the ontological matches was calculated as the P value, which is the probability of a match occurring by chance, given the size of the database. The P value was calculated using the formula:
where N is the total number of nodes in the MetaCore database, R is the number of network objects corresponding to genes and proteins in the list, n is the total number of nodes in each small network generated from the list, and r is the number of nodes with data in each small network generated from the list.Citation14
Statistical analysis
All values are presented as the mean ± standard deviation (SD) of a minimum of 3 replicate tests obtained from 3 individual experiments. A Kruskal–Wallis (nonparametric ANOVA) test was carried out using SPSS software (SPSS, Inc., Chicago, IL, USA) when multiple comparisons were made. Differences were considered as being significant at P < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1010906_Supplementary_Materials.zip
Download Zip (12.8 MB)Funding
This work was supported in part by the Chang Gung Memorial hospital (CMRPD1B0432 to P.O. and CMRPD1D0341 to P.T.L.) and National Science Council of Taiwan, ROC (NSC101-2320-B-182-003-MY3 to P.O. and NSC102-2628-B-182-003-MY3 to P.T.L.)
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: from parts list to mechanisms. Annu Rev Biochem 2006; 75:543-66; PMID:16756502; http://dx.doi.org/10.1146/annurev.biochem.74.082803.133425
- Glotzer M. The molecular requirements for cytokinesis. Science 2005; 307:1735-9; PMID:15774750
- Glotzer M. Cleavage furrow positioning. J Cell Biol 2004; 164:347-51; PMID:14757750
- Steigemann P, Gerlich DW. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol 2009; 19:606-16; PMID:19733077; http://dx.doi.org/10.1016/j.tcb.2009.07.008
- Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, Gerlich DW. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 2009; 136:473-84; PMID:19203582; http://dx.doi.org/10.1016/j.cell.2008.12.020
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 2007; 316:1908-12; PMID:17556548; http://dx.doi.org/10.1126/science.1143422
- Hu CK, Coughlin M, Mitchison TJ. Midbody assembly and its regulation during cytokinesis. Mol Biol Cell 2012; 23:1024-34; PMID:22278743; http://dx.doi.org/10.1091/mbc.E11-08-0721
- Skop AR, Liu H, Yates J 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 2004; 305:61-6; PMID:15166316; http://dx.doi.org/10.1126/science.1097931
- Castedo M, Coquelle A, Vivet S, Vitale I, Kauffmann A, Dessen P, Pequignot MO, Casares N, Valent A, Mouhamad S, et al. Apoptosis regulation in tetraploid cancer cells. EMBO J 2006; 25:2584-95; PMID:16675948; http://dx.doi.org/10.1038/sj.emboj.7601127
- Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, Galluzzi L, Adjemian S, Kepp O, Niso-Santano M, et al. An immunosurveillance mechanism controls cancer cell ploidy. Science 2012; 337:1678-84; PMID:23019653; http://dx.doi.org/10.1126/science.1224922
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005; 437:1043-7; PMID:16222300; http://dx.doi.org/10.1038/nature04217
- Ganem NJ, Pellman D. Limiting the proliferation of polyploid cells. Cell 2007; 131:437-40; PMID:17981108; http://dx.doi.org/10.1016/j.cell.2007.10.024
- Galipeau PC, Cowan DS, Sanchez CA, Barrett MT, Emond MJ, Levine DS, Rabinovitch PS, Reid BJ. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett's esophagus. Proc Natl Acad Sci USA 1996; 93:7081-4; PMID:8692948
- Pan TL, Wang PW, Huang CC, Yeh CT, Hu TH, Yu JS. Network analysis and proteomic identification of vimentin as a key regulator associated with invasion and metastasis in human hepatocellular carcinoma cells. J Proteomics 2012; 75:4676-92; PMID:22387118; http://dx.doi.org/10.1016/j.jprot.2012.02.017
- Pan TL, Wang PW, Hung YC, Huang CH, Rau KM. Proteomic analysis reveals tanshinone IIA enhances apoptosis of advanced cervix carcinoma CaSki cells through mitochondria intrinsic and endoplasmic reticulum stress pathways. Proteomics 2013; 13:3411-23; PMID:24167031; http://dx.doi.org/10.1002/pmic.201300274
- Chang WL, Lee DC, Leu S, Huang YM, Lu MC, Ouyang P. Molecular characterization of a novel nucleolar protein, pNO40. Biochem Biophy Res Commun 2003; 307:569-77; PMID:12893261; http://dx.doi.org/10.1016/S0006-291X(03)01208-7
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet 2004; 36:40-5; PMID:14702039; http://dx.doi.org/10.1038/ng1285
- Li M, Zhang P. The function of APC/CCdh1 in cell cycle and beyond. Cell Div 2009; 4:2; PMID:19152694; http://dx.doi.org/10.1186/1747-1028-4-2
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci 2003; 28:655-62; PMID:14659697; http://dx.doi.org/10.1016/j.tibs.2003.10.007
- Vitale I, Senovilla L, Jemaa M, Michaud M, Galluzzi L, Kepp O, Nanty L, Criollo A, Rello-Varona S, Manic G, et al. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J 2010; 29:1272-84; PMID:20186124; http://dx.doi.org/10.1038/emboj.2010.11
- Senovilla L, Vitale I, Galluzzi L, Vivet S, Joza N, Younes AB, Rello-Varona S, Castedo M, Kroemer G. p53 represses the polyploidization of primary mammary epithelial cells by activating apoptosis. Cell Cycle 2009; 8:1380-5; PMID:19342895; http://dx.doi.org/10.4161/cc.8.9.8305
- Sikorski RS, Boguski MS, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 1990; 60:307-17; PMID:2404612; http://dx.doi.org/10.1016/0092-8674(90)90745-Z
- Smith DF. Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones 2004; 9:109-21; PMID:15497498
- Wilsker D, Chung JH, Bunz F. Chk1 suppresses bypass of mitosis and tetraploidization in p53-deficient cancer cells. Cell Cycle 2012; 11:1564-72; PMID:22433954; http://dx.doi.org/10.4161/cc.19944
- Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell 2010; 141:81-93; PMID:20371347; http://dx.doi.org/10.1016/j.cell.2010.01.031
- Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol 2001; 21:1066-76; PMID:11158294; http://dx.doi.org/10.1128/MCB.21.4.1066-1076.2001
- Reid BJ, Barrett MT, Galipeau PC, Sanchez CA, Neshat K, Cowan DS, Levine DS. Barrett's esophagus: ordering the events that lead to cancer. Eur J Cancer Prev 1996; 5(Suppl. 2): 57-65; PMID:9061296; http://dx.doi.org/10.1097/00008469-199612002-00009
- Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol 2001; 11:18-21; PMID:11146294; http://dx.doi.org/10.1016/S0962-8924(00)01872-9
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer 2002; 2:815-25; PMID:12415252; http://dx.doi.org/10.1038/nrc924
- Karsenti E, Vernos I. The mitotic spindle: a self-made machine. Science 2001; 294:543-47; PMID:11641489; http://dx.doi.org/10.1126/science.1063488
- Lu P, Prost S, Caldwell H, Tugwood JD, Betton GR, Harrison DJ. Microarray analysis of gene expression of mouse hepatocytes of different ploidy. Mamm Genome 2007; 18:617-26; PMID:17726633; http://dx.doi.org/10.1007/s00335-007-9048-y
- Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 2013; 32:805-18; PMID:22508478; http://dx.doi.org/10.1038/onc.2012.130
- Pan TL, Wang PW, Leu YL, Wu TH, Wu TS. Inhibitory effects of Scutellaria baicalensis extract on hepatic stellate cells through inducing G2/M cell cycle arrest and activating ERK-dependent apoptosis via Bax and caspase pathway. J Ethnopharmacol 2012; 139:829-37; PMID:22210104; http://dx.doi.org/10.1016/j.jep.2011.12.028
- Pan TL1, Wu TH, Wang PW, Leu YL, Sintupisut N, Huang CH, Chang FR, Wu YC. Functional proteomics reveals the protective effects of saffron ethanolic extract on hepatic ischemia-reperfusion injury. Proteomics 2013; 13:2297-311; PMID:23696413; http://dx.doi.org/10.1002/pmic.201200551