The DNA is tightly packed by chromatin, which controls the expression of genetic information encoded by the DNA sequence. The chromatin scaffold responds dynamically to physiological and pathological signals and can be altered by histone modification, DNA methylation, and nucleosome remodeling.Citation1 Histone and DNA modifications include covalent modifications of histone side chains and DNA bases, whereas nucleosome remodeling involves non-covalent changes of the composition or positioning of nucleosomes along the genomic DNA. Remodeling of nucleosomes is executed by ATP-dependent chromatin-remodeling complexes; however, it is largely unknown how these chromatin remodelers target specific genomic sites to control local nucleosome dynamics. Our recent work revealed that a long non-coding RNA sequesters the core subunit of a chromatin-remodeling complex from its genomic targets to inhibit gene reprogramming in pathologically stressed hearts.Citation2 This sheds new light on the pathogenesis of heart failure and the targeting mechanism of chromatin-remodeling complexes.
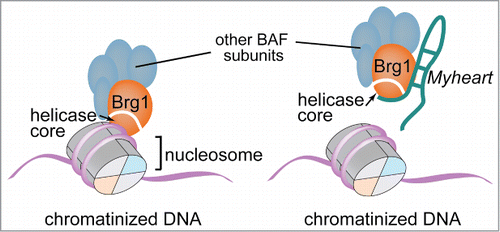
Long non-coding RNAs (LncRNA) can partner with a variety of histone- and DNA-modifying enzymes, such as Polycomb Repressive Complex, MLL/TrxG complex, histone demethylase LSD1 and DNA methyltransferase DNMT1, to control covalent modifications of chromatin.Citation3 In contrast, there were no known roles of lncRNAs in nucleosome remodeling; neither were there known functions of lncRNAs during the development of heart failure. The recent discovery of Myheart (Mhrt) provides new paradigms for lncRNA–chromatin interaction and cardiac biology. Mhrt is the first kind of lncRNA known to protect the heart from stress-induced hypertrophy and heart failure.Citation2 Mhrt maintains the heart function by inhibiting the chromatin activity of Brg1, an essential subunit of the SWI/SNF-like BAF chromatin-remodeling complex that reprograms cardiac gene expression to induce pathological hypertrophy.Citation4 Brg1 contains an evolutionarily conserved ATPase/helicase domain that belongs to the Superfamily 2 helicases.Citation5 This helicase domain doesn't bind to naked DNA but recognizes chromatinized or nucleosomal DNA. By binding to the chromatinized promoter DNA, the helicase domain tethers Brg1/BAF to its genomic DNA target sites where it remodels nucleosomes to control gene expressionCitation2 (). The helicase domain also binds to Mhrt with high affinity, enabling a competitive inhibition mechanism by which Mhrt sequesters Brg1 from its chromatinized DNA targetsCitation2 (). Therefore, Mhrt can modulate how Brg1/BAF targets to genomic loci to control nucleosome remodeling. This mechanism of Mhrt action provides a new model of lncRNA–chromatin regulation.
Figure 1. Brg1 (orange) recognizes and binds to the chromatinized/nucleosomal DNA, which wraps around the histone core (left panel). The long noncoding RNA Myheart (emerald) binds to Brg1 and sequesters it from its genomic DNA target locus (right panel).
The BAF complex occupies regulatory elements that are essential for transcription and is located near genomic regions critical for chromosome organization or DNA replication.Citation6 How BAF recognizes these specific genomic sites, however, remains elusive. The studies of Mhrt and Brg1 suggest that Brg1/BAF can recognize and bind favorably to certain nucleosome conformation constituted by DNA sequence,Citation2 which is critical for determining nucleosome positions.Citation7 The targeting specificity also requires other domains of Brg1 and subunits of the BAF complex. For instance, the bromodomain of Brg1 and Baf45 subunit of BAF can recognize acetylated and/or methylated histone signatures of chromatin. Accordingly, the DNA sequence and modified histones can formulate specific nucleosome conformations that Brg1/BAF recognizes to excise its nucleosome remodeling function. This targeting mechanism and Mhrt regulation of the SWI/SNF-like Brg1/BAF complex are likely applicable to the other families of ATP-dependent chromatin-remodeling complexes (CHD, ISWI, and INO80), which share a conserved Brg1-like ATPase/helicase component. Further studies will be crucial to elucidate how Mhrt interacts with the other chromatin remodelers to control chromatin and gene expression.
In the heart, Mhrt seizes the helicase domain and prevents Brg1 from recognizing its chromatin targets and executing remodeling function, whereas Brg1 inhibits the transcription of Mhrt by directly repressing its promoterCitation2 This Mhrt–Brg1 feedback circuit is critical for maintaining the homeostasis of cardiac epigenome and heart function—Pathological perturbation of the Mhrt–Brg1 loop results in cardiac gene reprogramming and heart failure.Citation2 Such reciprocal inhibition of Mhrt and Brg1 provides a new mechanism by which the chromatin-remodeling factor and lncRNA reach a homeostatic state to control chromatin and genomic activity. It remains to be seen whether other lncRNAs and their epigenetic partners have similar reciprocal regulation to control chromatin structure under different physiological and pathological conditions.
Funding
C-PC was supported by National Institutes of Health (NIH; HL118087, HL121197), the American Heart Association (AHA; Established Investigator Award 12EIA8960018), March of Dimes Foundation (#6-FY11-260), Indiana University (IU) School of Medicine—IU Health Strategic Research Initiative, and the IU Physician-Scientist Initiative, endowed by Lilly Endowment.
References
- Han P, et al. Circ Res 2011; 108:378-96; PMID:21293009; http://dx.doi.org/10.1161/CIRCRESAHA.110.224287
- Han P, et al. Nature 2014; 514:102-6; PMID:25119045; http://dx.doi.org/10.1038/nature13596
- Rinn JL, et al. Annu Rev Biochem 2012; 81:145-66; PMID:22663078; http://dx.doi.org/10.1146/annurev-biochem-051410-092902
- Hang CT, et al. Nature 2010; 466:62-7; PMID:20596014; http://dx.doi.org/10.1038/nature09130
- Linder P, et al. Nat Rev Mol Cell Biol 2011; 12:505-16.
- Euskirchen GM, et al. PLoS Genet 2011; 7:e1002008.
- Segal E, et al. Nature 2006; 442:772-8; PMID:16862119
