Abstract
Direct lineage conversion is a promising approach for disease modeling and regenerative medicine. Cell divisions play a key role in reprogramming of somatic cells to pluripotency, however their role in direct lineage conversion is not clear. Here we used transdifferentiation of fibroblasts into neuronal cells by forced expression of defined transcription factors as a model system to study the role of cellular division in the direct conversion process. We have shown that conversion occurs in the presence of the cell cycle inhibitors aphidicolin or mimosine. Moreover, overexpression of the cell cycle activator cMyc negatively influences the process of direct conversion. Overall, our results suggest that cell divisions are not essential for the direct conversion of fibroblasts into neuronal cells.
Abbreviations
| iPS cells | = | induced pluripotent stem cells |
| ES cells | = | embryonic stem cells |
| BAM | = | Brn2, Ascl1 and Myt1l |
| BAM+M | = | Brn2, Ascl1, Myt1l and cMyc |
| DOX | = | doxycycline, BrdU, bromodeoxyuridine |
| MEF | = | mouse embryonic fibroblasts |
Introduction
Reprogramming of the cell fate using defined transcription factors clearly showed genome plasticity of somatic cells.Citation1 Moreover, efficient protocols are available to guide the differentiation of induced pluripotent stem (iPS) cells and embryonic stem (ES) cells into different types of somatic cells. As a further development, protocols for the conversion (transdifferentiation) of one set of differentiated lineage-restricted cells directly into another were published. For instance, overexpression of different sets of transcription factors or microRNAs was shown to convert fibroblasts into macrophages,Citation2 cardiomyocytes,Citation3 neuronal cells,Citation4,5 blood cells,Citation6 and hepatocytes.Citation7 The principle of both, reprogramming of somatic cells to pluripotent state and direct conversion to specific cell types is similar and based on forced overexpression of defined master-genes (transcription factors and/or microRNAs), which activate downstream gene networks specific for a desired cell type. However, the mechanisms of these processes have not been completely studied yet.
Using generation of iPS cells as a model it was found that the reprogramming process is generally slow, inefficient, and is based on a stochastic model, but could be improved by expression of additional transcription factors or supplementation with small molecular substances.Citation8,9 Wide scale epigenetic changes, such as X chromosome reactivation and changes in histone modification and genome methylation pattern are required to establish and consolidate the pluripotent state of reprogrammed cells.Citation10 These global epigenetic changes are usually associated with cell cycle progression,Citation11 leading to the hypothesis that acceleration of cell divisions favors faster reprogramming. Indeed, upregulation of genes responsible for cell cycle progression, such as cMyc, or knockdown of cell cycle regulators (e.g, p53) facilitates reprogramming.Citation12,13 Moreover, it was shown that reprogramming to pluripotent state consists of 2 independent components, one responsible for the gain of pluripotent properties and another for activation of cell cycle progression.Citation8 When quiescent cells, such as post-mitotic neurons, are used as a source for iPS cell generation, additional factors are required to obtain stem-like cells.Citation14 Thus, cell divisions play a key role in reprogramming of cells to pluripotent state.
The situation is different when one set of differentiated cells is converted into another. First, the scale of epigenetic changes required for the direct conversion depends on the original and desired cell types and usually is less than in the case of iPS cell generation. In addition, the transdifferentiation process occurs fast and efficient and could even result in nearly 100% conversion rates in some cases.Citation15 Second, in contrast to stem cells, which require active proliferation to maintain their properties, differentiated cells could exit the cell cycle and stay in G0 phase. It was already shown that conversion between closely related cell types such as B-cells and macrophages or astroglia and neuronal cells could occur in the absence of cell divisions.Citation16,17 Moreover, reprogramming of mouse fibroblasts into neuronal cells generally occurs with a maximum of one cell division.Citation4,18 However, whether cell divisions is a limiting factor for the conversion of distinct cell types has not been yet investigated. Here we used transdifferentiation of mouse embryonic fibroblasts (MEFs) into neuronal cells by forced expression of defined transcription factors as a model system to study the role of cellular division in the direct conversion process. We also exploit this system to investigate effects of the cell cycle activator cMyc on the efficiency of transdifferentiation.
Results
Direct conversion of fibroblasts into neurons provides a useful system to study the role of cell division in the transdifferentiation process
Cells of many types actively proliferate when cultured in vitro. If cell cycle progression is inhibited, these cells might undergo differentiationCitation19 or apoptosis.Citation20 Thus, it might be complicated to distinguish between effects of cell cycle arrest on the transdifferentiation process and effects of cell cycle arrest on the subsequent culture of obtained cells. To overcome this limitation we decided to use direct conversion of somatic cells into quiescent postmitotic neurons as a model system.
We used lentiviral transduction with 3 transcription factors (Brn2, Ascl1 and Myt1l, BAM) under the control of a DOX-inducible promoter to convert mouse embryonic fibroblasts into neurons ().Citation4 We assessed the dynamics of activation of neuronal markers, β-III tubulin (Tuj1) and neurofilament 200 (NF200), during the transdifferentiation process. First Tuj1-positive cells appeared in culture at day 5 after viral transduction (). The number of Tuj1-positive cells increased by day 9, and their morphology changed from fibroblast- to neuron-like with long branches. At this time point (day 9) some of the Tuj1-positive cells showed expression of NF200. The amount of NF200 positive cells increased during the next week of cultivation and at day 19 we observed a number of Tuj1/NF200 double-positive neuron-like cells (). Such cells formed mature networks and survived in culture for at least one month. Importantly, we did not find any Tuj1- or NF200-positive cells in not transfected control cells at any time point (data not shown).
Figure 1. Dynamics of conversion of MEFs into neurons by exogenous BAM expression. (A) Experimental design. MEFs were plated at day -2, transduced with BAM+RT viruses at day -1 and cultured in the presence of DOX starting from day 0 until day 5–19. (B) Immunostaining with Tuj1 (yellow) and NF200 (red) antibodies in the targeted cells 5–19 d after viral transduction. (C) Same as B for neuronal markers Tuj1 (yellow) and MAP2 (red) antibodies. (D) Immunostaining with NF200 (red) and collagen-1 (green) antibodies of the targeted cells 19 d after viral transduction. Cell nuclei (DAPI) are shown in blue.
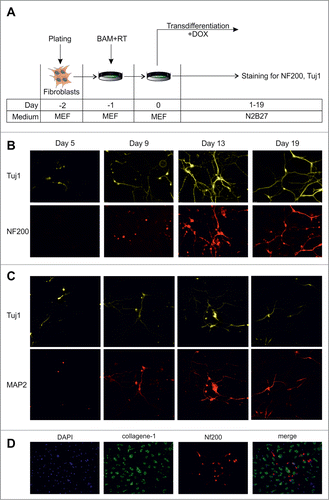
We also assessed the kinetics of activation of another neuronal marker, MAP2 during transdifferentiation (). The first MAP2-positive cells appeared between day 5 and 7 and were detected at subsequent time points. We noted that, similarly to NF200, the MAP2 signal was highly overlapping with the Tuj1 signal.
We found that the fibroblast marker, collagen-1, was down regulated in Tuj1-positive cells as early as 5 d after transduction (data not shown). In addition, 19 days after transduction we found cells expressing collagen-1 or NF200, but not both of them together (). Overall, these results show rapid conversion of fibroblasts into neuronal cells by defined transcription factors.
Transdifferentiation does not require cell divisions
We used aphidicolin (reversible inhibitor of DNA polymerase α) or mimosine (reversible inhibitor of DNA replication via a decrease in cellular dNTP concentrations) to study the role of cellular division in the process of direct conversion of fibroblasts to neuronal cells. Using the bromodeoxyuridine (BrdU) incorporation assay as an indicator of cellular divisions, we first assessed the efficiency of the aforementioned substances as cell cycle inhibiting agents in fibroblast cells (). We found that 24 hs incubation of fibroblasts in BrdU-containing medium resulted in almost no BrdU-positive cells in the presence of aphidicoline or mimosine (number of BrdU-positive cells was <0.1%), whereas among non-treated fibroblasts up to 60% of cells were BrdU-positive.
Figure 2. (See previous page) Direct conversion of MEFs into neuronal cells by BAM in the presence of cell division inhibitors. (A) BrdU incorporation in MEFs treated with cell division inhibitors aphidicolin or mimosine for 24 hr. (B) Number of MEFs survived after 2–10 d of culture in the presence of inhibitors. Aph – aphidicolin, Mim – mimosine, Ctr - untreated cells. X-axis indicates experimental time-frame starting from plating of MEFs (day -2) until day 10. Y-axis indicates the number of survived cells. The arrow shows the moment when medium was supplemented with cytostatics (day 0). The culture medium used at each time point of the experiment is shown in boxes under the plot. (C) Experimental design. MEFs were plated at day -2, transduced with BAM+RT viruses at day -1 and cultured in the presence of DOX and aphidicolin starting from day 0 until day 11. (D) Immunostaining with Tuj1 (yellow) and MAP2 (red) antibodies in the aphidicolin treated cells 5–11 d after viral transduction. (E) Tuj1/NF200 double-stained neurons generated from MEFs in the presence of aphidicolin 10 d after viral transduction. (F) Quantification of synapsin (Syn1) and Tuj1 expression (Log10 scale) in the aphidicolin treated cells 5–11 d after viral transduction. MEFs: control fibroblasts not transduced with viruses and cultured for 11 d in N2B27 media without aphidicolin *P < 0.05 in comparison to control MEFs.
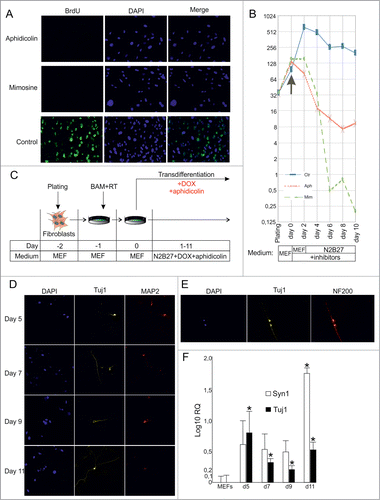
We have shown before () that activation of the neuronal markers, NF200 and Tuj1, could be detected in fibroblasts earliest 9–10 days after viral transduction. We cultivated MEF cells in the presence of aphidicolin or mimosine for 2, 4, 6, 8 or 10 days in the N2B27 medium (medium used during transdifferentiation) and quantified the number of survived cells to investigate the toxic effect of these cytostatics (). We observed a gradual decrease of cell viability when fibroblasts were cultured in the presence of aphidicolin. In case of mimosine the gradual decrease of viability observed during the first 4 days of culture was followed by a sharp drop at day 6 resulting in almost no cells survived at day 10. Interestingly, we observed a slight decrease of proliferative activity of control MEF starting from day 2–4, probably due to contact inhibition and cultivation in serum-free N2B27 medium ().
We then employed aphidicolin to prevent cell divisions during the transdifferentiation process. MEFs were transduced with BAM viruses and cultured in the presence of DOX and aphidicolin (). Starting from day 2–4, we observed a massive cell death, most probably due to the toxic effect of aphidicolin. Immunocytochemical analysis of survived cells showed Tuj1 and MAP2 activation in cells treated with aphidicolin (). We were able to detect first Tuj1/MAP2 double-positive cells as early as at day 5 (), indicating that conversion of fibroblasts into Tuj1/MAP2 double-positive cells may occur without cell divisions. Moreover, we were able to detect rare Tuj1/NF200 double-positive cells with neuronal-like morphology among survived cells at day 10 (). Consistent with this, upregulation of Tuj1 and synapsin was observed in neurons treated with aphidicolin (). Importantly, expression of the mature neuron marker synapsin at day 11 was more than 80 times higher in transduced cells treated with aphidicolin than in not transduced cells.
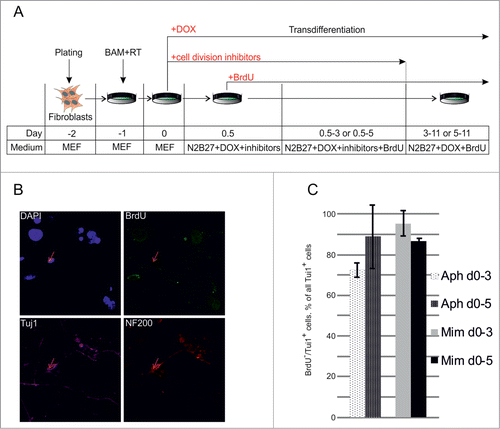
The highly variable rates of cell death prevented the comparison of transdifferentiation efficiencies between neuronal cells obtained in the presence and absence of aphidicolin. To improve the efficiency of our experimental system we decided to apply inhibitors only for a short period of time. Considering the rapid decrease of cell viability after 4–6 days of incubation with cytostatics, we decided to perform treatment of MEFs with aphidicolin or mimosine only during the first 3–5 days after viral transduction followed by incubation in cytostatics-free medium during the subsequent 5–7 days. Using this strategy, we were able to improve cell viability and obtained many survived cells at day 11 after transduction with BAM viruses. Some of these cells showed typical neuronal morphology and were positive for the expression of the neuronal markers, Tuj1 and NF200 (data not shown). To exclude the possibility that these cells underwent cell divisions during the last 5–7 days when they were incubated in cytostatics-free medium we repeated the experiment supplementing the culture medium with BrdU (). Taking in consideration that DOX-inducible genes require 12–24 hs to reach high expression levelsCitation21 and to avoid detection of cells that were in the S-phase at the beginning of the experiment we started the BrdU treatment 12 hours after the addition of the inhibitors. Starting from this time point cell culture medium was continuously supplemented with BrdU. Eleven days after viral transduction we detected some NF200/Tuj1 double-positive neuronal cells, which incorporated BrdU, indicating that the transdifferentiation process of these cells was accompanied with cell divisions. However more than 70% of Tuj1/NF200 double-positive cells observed in culture were BrdU-negative, indicating a division-free transdifferentiation process (, C). We did not observe significant differences between the numbers of BrdU-negative neuronal cells obtained using aphidicolin and mimosine, suggesting that inhibitor-specific effects (such as binding of the DNA-polymerase in case of aphidicolin or reduction in cellular dNTP concentrations in case of mimosine) do not influence the transdifferentiation process (). The identification of BrdU-negative Tuj1/NF200 double-positive neuron-like cells suggests a direct conversion of fibroblasts into neuronal cells in the absence of cell divisions.
Figure 3. Direct conversion of MEFs to neurons without cell divisions. (A) Experimental design. MEFs were plated at day -2, transduced with BAM+RT viruses at day -1 and cultured in the presence of DOX from day 0 until day 11. Cells were treated with aphidicolin or mimosine starting from day 0 for 3 or 5 d. BrdU was first added 12 hours after cell division inhibitors (day 0.5) and was present in medium until the end of experiment. (B) Immunostaining with antibodies against Tuj1, NF200 and BrdU at day 11 of transdifferentiation, performed in the presence of aphidicolin from day 0 to day 5. Arrows show an example of cells treated with aphidicolin and positive for Tuj1 and NF200, but negative for BrdU. (C) Quantification of NF200+/Tuj1+/BrdU− cells obtained in the transdifferentiation experiments, performed in the presence of aphidicoline or mimosine during days 0–3 or 0–5 of transdifferentiation. BrdU− cells account for more than 70% of all NF200+/Tuj1+ neuronal-like cells. Aph – aphidicolin, Mim – mimosine. Number of experiments N = 3.
Aphidicolin blocks conversion of fibroblasts to iPS cells
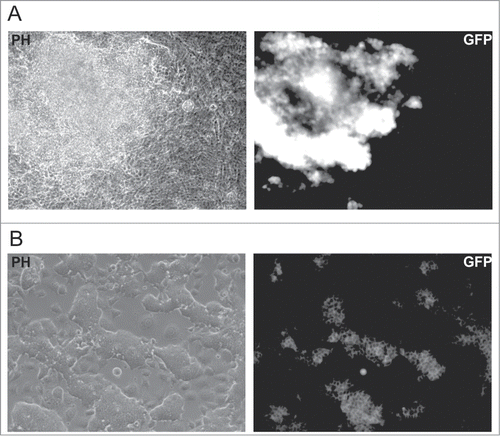
To check whether iPS cells could be generated from quiescent fibroblasts, we used OG2 MEFs carrying GFP under promoter of Oct4.Citation22 We treated OG2 MEFs with lentiviral vectors carrying DOX-inducible Oct4, Sox2, cMyc and Klf4 factors (OKSM) and reversed transactivator and cultured cells for 8 days in the presence of DOX and aphidicolin and then 3 more weeks in aphidicolin-free medium without DOX. During the experiment, the cells were routinely monitored for the activation of the Oct4::GFP reporter, however no GFP-positive cells were observed. In contrast, control MEFs cultured in similar conditions but without aphidicolin produced a number of GFP-positive colonies at days 10–15 after infection (). The efficiency of colony formation was estimated as ∼0,01%. Most of the colonies grew rapidly, maintained expression of GFP and showed a phenotype similar to the phenotype of ES cells during subsequent cultivation (). To check whether the effect of aphidicolin on iPS cell generation is due to block of cell proliferation and not cytotoxicity, we cultured generated iPS cells in presence of aphidicolin for 2–6 days. Starting from day 3, we observed significant cell death and almost all cells died at day 6. Thus, we can conclude that the presence of aphidicolin prevents generation of iPS cells from MEFs. However, it is not clear whether this effect of aphidicolin reflects the inability of quiescent fibroblasts to become iPS cells or cytotoxicity of aphidicolin on generated iPS cells.
Figure 4. Generation of iPS cells from OG2 MEFs. (A) Morphology (PH) and GFP fluorescence of the iPS cells produced in the absence of aphidicolin at day 12 after viral transduction of OG2 MEFs, carrying GFP under the promoter of Oct4. (B) Morphology and GFP fluorescence of an iPS cell culture obtained from a single clone after 4 weeks expansion.
Forced expression of cMyc negatively influences the transdifferentiation process
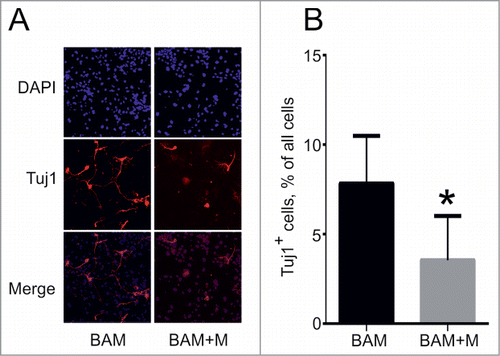
We next investigated how cell cycle activation affects the transdifferentiation process. Consistent with previously published data,Citation23 overexpression of cMyc shortened the fibroblast doubling time by ∼1.7-fold as compared to control cells (data not shown). However, the dynamics of neuronal cell formation remained unchanged when the BAM cocktail of transdifferentiation factors was supplemented with cMyc (BAM+M). We observed formation of first cells with neuronal-like morphology around day 5. The number of such cells increased until day 18, also the morphology of cells became more complex, including formation of networks (). We compared the number of Tuj1-positive cells in culture at day 18 after viral transduction with BAM and GFP (BAM+G) and BAM+M lentiviruses. Almost a twofold decrease in the percentage of Tuj1-positive cells was observed when cMyc was added to the BAM cocktail (), indicating that forced expression of cMyc negatively influences the transdifferentiation process induced by BAM. To exclude the possibility that the amount of Tuj1-positive cells was reduced due to a toxic effect of cMyc on generated neurons we performed transdifferentiation in the absence of cMyc, and treated obtained cells with cMyc or GFP at day 14 after DOX addition. We observed no significant changes (P-value = 0.28) between cells treated with cMyc or GFP at day 28 after DOX addition, indicating that cMyc affects the process of transdifferentiation, but not the survival of the obtained neurons.
Figure 5. Effect of cMyc overexpression on direct conversion of MEFs into neuronal cells. (A) Phenotype of Tuj1+ neuronal cells (red) obtained from MEFs 18 d after transduction with BAM and BAM+M lentiviruses. (B) Percentage of Tuj1+ cells out of all cells obtained at day 18. BAM+M – MEFs transduced with BAM and cMyc viruses, BAM – MEFs transduced with BAM viruses only. Number of experiments for each condition: N = 6. Data are presented as means ± SEM (standard error of means). *P < 0.05
Discussion
Here we have shown that the direct conversion of fibroblasts into neuronal cells is possible without cell divisions. This observation is in agreement with the finding that astroglia cells could be converted to neurons without cell divisions.Citation16 In addition, it was recently shown that conversion of preB-cells to macrophages does not require cell divisions.Citation17 However, in both studies transdifferentiation occurs between closely related cell types and at least in case of the preB-cells to macrophages conversion takes place in the absence of significant DNA methylation changes.Citation24 Our data indicates that cell division is not a sine qua non also for more distinct cell types, such as fibroblasts and neurons.
During development the process of DNA replication remodels chromatin structure and allows access to previously inactive regulatory domains, thereby changing the expression patterns of specific genes in dividing cells.Citation11 It is still unclear to what extent chromatin remodeling could occur without cell divisions. Experiments with hybrid cells conducted by Blau et al.Citation25 suggested the possibility of broad epigenetic changes at a quiescent heterokaryon stage. In agreement with this, weCitation26 and othersCitation27 have shown that the molecular changes that determine the success and direction (or dominance) of reprogramming occur during the first hours after cell fusion, even before the first cell division.
On the other hand, experiments conducted with both hybrid and iPS cells show that the newly acquired gene expression profiles of reprogrammed cells should be fixed over a number of cell divisions.Citation28,29 Recently, Lahn and colleagues proposed the classification of genes in two categories. Genes of the first category are trans-silenced and could be activated by exogenous transcription factors without cell division. Genes of the second category are cis-silenced and require cell cycle progression for their activation.Citation27 In our experiments, we identified an activation of neuronal markers, NF200, MAP2 and Tuj1, in quiescent fibroblasts transduced with defined transcription factors. However, it might be that cell divisions are required for the activation of cis-silenced genes.
The successful conversion of fibroblasts into neuronal cells in the absence of cell divisions encouraged us to investigate the effect of the cell cycle activation on the efficiency of the transdifferentiation process. We demonstrated that ectopic expression of cMyc decreases the yield of neurons, indicating that active cell cycle progression blocks the transdifferentiation process. In contrast, a positive effect of proliferation activators has been shown in iPS cell generation experiments.Citation8,30 This difference in the cMyc effects might be due to different regulation of the neuronal and pluripotent cell cycles. Active proliferation is required for maintaining the pluripotency of ES cells,Citation31 whereas cell cycle exit plays a major role in neural cell specification and differentiation.Citation32 Consistently, the reprogramming of neurons to iPS cells could be achieved only after forced cell proliferation by p53 suppression.Citation12,13
On the other hand, it is not excluded that cMyc affects the transdifferentiation efficiency not only as a cell cycle activator. For instance, it was shown that cMyc blocks myogenic differentiation and this effect of cMyc does not depend on the process of cell transformation.Citation33,34 Thus, we can conclude that expression of cMyc negatively influences the process of direct conversion, but the detailed mechanisms of this phenomenon remain to be elucidated.
Altogether, our results indicate that cell cycle adjustment may be essential during the conversion process; however, proliferation itself is not required for the switch between cell types.
Materials and Methods
Plasmids and lentiviruses
To convert mouse embryonic fibroblasts (MEFs) into neuronal cells or iPS cells lentiviruses, encoding Ascl1, Brn2, Myt1l, M2RTta, cMyc or single polycistronic cassette carrying Oct4, Sox2, cMyc and Klf4 (OKSM) factors were generated. 293T cells were transfected with envelope-encoding pMD2G (5 mg), packaging pCMV-dR8.74PAX2 (5 mg), and either Ascl1 (Addgene #27150), Brn2 (Addgene #27151), Myt1l (Addgene #27152), M2RTta (Addgene #20342), cMyc Citation35 or OKSM (Addgene #20328)- encoding LVTHM-based plasmids (20 mg) by the calcium-phosphate method. Lentiviruses were collected from the cell culture supernatant and processed as described elsewhere.Citation36
Fibroblast isolation and viral transduction
Unless specified, all cell culture products were from Invitrogen (Darmstadt, Germany). MEFs were routinely maintained in high glucose DMEM supplemented with 10% FBS (Sigma-Aldrich, St. Louis, MO), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 1x non-essential amino acids, 50 μM β-mercaptoethanol (MEF medium). To isolate MEFs naturally mated NMRI (Charles River, Germany), ISR (ICG, Novosibirsk) or OG2Citation22 (ICG, Novosibirsk) female mice were checked for the presence of a vaginal plug and thereafter sacrificed at day 13 of pregnancy (E13). Isolated embryos were freed from head and visceral tissues, minced, subsequently trypsinized and plated on tissue culture dishes (passage 0).
For the transdifferentiation experiments MEFs (passage 2) were seeded at 2.5 × 104 cells/cm2 in a 24-well plate (day -2) and transduced 24 hs later (day -1) with the Ascl1, Brn2, Myt1l and M2RTta lentiviruses (BAM, see above) in MEF medium. When indicated, the virus cocktail was supplemented with cMyc or GFP (BAM+M and BAM+G). 16–20 hs after transduction (day 0) the medium was changed to N2B27 medium (mixture 1:1 of N2-medium [DMEM/F12 supplemented with 1x N2, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.005% BSA, 25 μM β-mercaptoethanol] and B27 medium [NBM supplemented with 1x B27 (without RA), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 25 μM β-mercaptoethanol]). The transdifferentiation process was started by addition of doxycycline (DOX, Sigma-Aldrich, 2ug/ml) at day 0. Cells were further cultivated in the presence of DOX in N2B27 medium until immunostaining was performed. Aphidicolin (Sigma-Aldrich, 5 ug/ml), mimosine (Sigma-Aldrich, 400 nM), or BrdU (Millipore, 10uM) were added to the culture medium when indicated.
iPS cells were generated from OG2 MEFs as described elsewhere.Citation35 Briefly, 105 OG2 MEFs were infected with OKSM and M2RTta lentiviruses and cultured in DMEM supplemented with 10% FBS (Sigma-Aldrich, St. Louis, MO), 5% KSR, 1,000 U/mL LIF, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 1x non-essential amino acids, 50 μM β-mercaptoethanol. The culture medium was supplemented with DOX (2ug/ml) and aphidicolin (5 ug/ml) when indicated.
Immunocytochemistry
Cells were washed with PBS and fixed with 4% formaldehyde (Fluka, Germany) for 10 min at room temperature. After fixation, the cells were permeabilized with 0.1% Triton X-100 (Fluka) in PBS for 20 min at room temperature, rinsed with PBS, blocked with 1% bovine serum albumin (BSA; Sigma, USA) in PBS for 30 min and incubated for 1 h at room temperature with primary antibodies in the following combinations: anti-b-III tubulin (Tuj1, mouse, 1:500; Covance, France), anti-NF200 (rabbit, 1:500; Sigma-Aldrich), anti-MAP2 (rabbit, 1:500, Almabion) and anti-BrdU (rat, 1:1500,Biozol). For BrdU staining, DNA was denatured in 2N HCl for 20 min at 37°C before primary antibodies were applied. After 1 h incubation with primary antibody, cells were washed 3 times with PBS and incubated for 1 h at room temperature with the Alexa488-, Alexa647- (Life technologies), Cy3-, or Cy5 (Jackson ImmunoResearch Laboratories) conjugated secondary antibodies (1:1000) Thereafter, the cells were washed 3 times with PBS, counterstained with 4,6-diamidino-2-phenylindole (DAPI, Sigma, Germany), mounted in a glycerol solution containing 1,4 diazobicyclo-[2.2.3] octane (DABCO; Sigma, USA) and examined either on a fluorescent microscope Leica DM6000 with 5x, 10x, 20x, and 40x air objectives or a confocal Leica TCS SP5 microscope with 20× air and 63× oil immersion objectives. Images were acquired and processed using the Leica software.
Quantitative reverse transcriptase–PCR (qPCR)
Total RNA was isolated using the Quick-RNA™ MicroPrep Kit (Zymo Research). 0.125–0.25 ug of DNase treated RNA was reverse transcribed using a RevertAid First Strand cDNA Synthesis kit (Thermo Scientific). Quantitative RT-PCR analysis was performed in triplicate using 1/10 of the reverse transcription reaction in an StepOnePlus™ Real-Time PCR System (Applied Biosystems®) with SYBR Green I qPCR mix (Syntol). PCR primer sequences are available on request.
Statisctical analysis
The total number of Tuj1-positive cells with a neuronal morphology, defined as cells having a circular, 3-dimensional appearance that extend thin processes at least 3 times longer than their cell body, was quantified in ×20 visual fields. The total number of cells in corresponding fields was quantified according to DAPI signal. The percentage of neuronal cells was calculated as a ratio between Tuj1-positive cells and the total number of cells. The resulting number was averaged for 30 randomly selected fields. This calculation was repeated in 6 independent experiments. The same method was used to calculate the efficiency of BrdU incorporation. Statistical analysis was performed using Wilcoxon- and t-tests.
Statistical analysis of qPCR was performed using REST software. P < 0.05 was considered as a significant difference in all statistical tests.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The study was supported by RFBR grant No. 13-04-00441, SB RAS grant No. 136, in part by RSF grant No. 14-15-00131, and BMBF/WTZ grant No. 01DJ3012 and C. Zeiss (Optec), IALS, and DAAD (A/10/85864) fellowships to VF.
References
- Takahashi K, Yamanaka S. Induction of Pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663-76; PMID:16904174; http://dx.doi.org/10.1016/j.cell.2006.07.024
- Feng R, Desbordes SC, Xie HF, Tillo ES, Pixley F, Stanley ER, Graf T. PUA and C/EBP alpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A 2008; 105:6057-62; PMID:18424555; http://dx.doi.org/10.1073/pnas.0711961105
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010; 142:375-86; PMID:20691899; http://dx.doi.org/10.1016/j.cell.2010.07.002
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010; 463:1035-41; PMID:20107439; http://dx.doi.org/10.1038/nature08797
- Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 2012; 11:100-9; PMID:22683203; http://dx.doi.org/10.1016/j.stem.2012.05.018
- Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 2010; 468:521-6; PMID:21057492; http://dx.doi.org/10.1038/nature09591
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 2011; 475:390-3; PMID:21716291; http://dx.doi.org/10.1038/nature10263
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 2009; 462:595-601; PMID:19898493; http://dx.doi.org/10.1038/nature08592
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 2013; 502:65-70; PMID:24048479; http://dx.doi.org/10.1038/nature12587
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 2007; 1:55-70; PMID:18371336; http://dx.doi.org/10.1016/j.stem.2007.05.014
- Probst A V, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 2009; 10:192-206; PMID:19234478; http://dx.doi.org/10.1038/nrm2640
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009; 460:1132-5; PMID:19668191; http://dx.doi.org/10.1038/nature08235
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 2009; 460:1145-8; PMID:19668190; http://dx.doi.org/10.1038/nature08285
- Kim J, Lengner CJ, Kirak O, Hanna J, Cassady JP, Lodato MA, Wu S, Faddah DA, Steine EJ, Gao Q, et al. Reprogramming of postnatal neurons into induced pluripotent stem cells by defined factors. Stem Cells 2011; 29:992-1000; PMID:21563275; http://dx.doi.org/10.1002/stem.641
- Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, Herms S, Wernet P, Kögler G, Müller FJ, et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods 2012; 9:575-8; PMID:22484851; http://dx.doi.org/10.1038/nmeth.1972
- Heinrich C, Blum R, Gascon S, Masserdotti G, Tripathi P, Sanchez R, Tiedt S, Schroeder T, Gotz M, Berninger B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. Plos Biol 2010; 8:e1000373; PMID:20502524; http://dx.doi.org/10.1371/journal.pbio.1000373
- Di Tullio A, Graf T. C/EBP alpha bypasses cell cycle-dependency during immune cell transdifferentiation. Cell Cycle 2012; 11:2739-46; PMID:22771961; http://dx.doi.org/10.4161/cc.21119
- Marro S, Pang ZP, Yang N, Tsai MC, Qu K, Chang HY, Sudhof TC, Wernig M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 2011; 9:374-82; PMID:21962918; http://dx.doi.org/10.1016/j.stem.2011.09.002
- Hindley C, Philpott A. Co-ordination of cell cycle and differentiation in the developing nervous system. Biochem J 2012; 444:375-82; PMID:22642576; http://dx.doi.org/10.1042/BJ20112040
- Pucci B, Kasten M, Giordano A. Cell cycle and apoptosis. Neoplasia 2000; 2:291-9; PMID:11005563; http://dx.doi.org/10.1038/sj.neo.7900101
- Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science 1995; 268:1766-9; PMID:7792603; http://dx.doi.org/10.1126/science.7792603
- Yao H, Jiang Y, Zhang Y, Liu W, Huang B, Wang X, Gao S. Establishment of novel embryonic stem (ES) cell lines from OG2/rtTA blastocysts. J Genet Genomics 2011; 38:289-95; PMID:21777853; http://dx.doi.org/10.1016/j.jgg.2011.05.007
- Karn J, Watson JV, Lowe AD, Green SM, Vedeckis W. Regulation of cell cycle duration by c-myc levels. Oncogene 1989; 4:773-87; PMID:2660073
- Rodriguez-Ubreva J, Ciudad L, Gomez-Cabrero D, Parra M, Bussmann LH, di Tullio A, Kallin EM, Tegner J, Graf T, Ballestar E. Pre-B cell to macrophage transdifferentiation without significant promoter DNA methylation changes. Nucleic Acids Res 2012; 40:1954-68; PMID:22086955; http://dx.doi.org/10.1093/nar/gkr1015
- Palermo A, Doyonnas R, Bhutani N, Pomerantz J, Alkan O, Blau HM. Nuclear reprogramming in heterokaryons is rapid, extensive, and bidirectional. Faseb J 2009; 23:1431-40; PMID:19141533; http://dx.doi.org/10.1096/fj.08-122903
- Gridina MM, Serov OL. Bidirectional reprogramming of mouse embryonic stem cell/fibroblast hybrid cells is initiated at the heterokaryon stage. Cell Tissue Res 2010; 342:377-89; PMID:21103994; http://dx.doi.org/10.1007/s00441-010-1085-2
- Foshay KM, Looney TJ, Chari S, Mao FF, Lee JH, Zhang L, Fernandes CJ, Baker SW, Clift KL, Gaetz J, et al. Embryonic stem cells induce pluripotency in somatic cell fusion through biphasic reprogramming. Mol Cell 2012; 46:159-70; PMID:22445485; http://dx.doi.org/10.1016/j.molcel.2012.02.013
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010; 467:285-U60; PMID:20644535; http://dx.doi.org/10.1038/nature09342
- Serov OL, Matveeva NM, Khabarova AA. Reprogramming mediated by cell fusion technology. Int Rev Cell Mol Biol 2011; 291:155-90; PMID:22017976; http://dx.doi.org/10.1016/B978-0-12-386035-4.00005-7
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2008; 2:10-2; PMID:18371415; http://dx.doi.org/10.1016/j.stem.2007.12.001
- Neganova I, Zhang X, Atkinson S, Lako M. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene 2009; 28:20-30; PMID:18806832; http://dx.doi.org/10.1038/onc.2008.358
- Galderisi U, Jori FP, Giordano A. Cell cycle regulation and neural differentiation. Oncogene 2003; 22:5208-19; PMID:12910258; http://dx.doi.org/10.1038/sj.onc.1206558
- Miner JH, Wold BJ. c-myc inhibition of MyoD and myogenin-initiated myogenic differentiation. Mol Cell Biol 1991; 11:2842-51; PMID:1850105
- La Rocca SA, Crouch DH, Gillespie DA. c-Myc inhibits myogenic differentiation and myoD expression by a mechanism which can be dissociated from cell transformation. Oncogene 1994; 9:3499-508; PMID:7970710
- Liskovykh M, Chuykin I, Ranjan A, Safina D, Popova E, Tolkunova E, Mosienko V, Minina JM, Zhdanova NS, Mullins JJ, et al. Derivation, characterization, and stable transfection of induced pluripotent stem cells from Fischer344 rats. PLoS One 2011; 6:e27345; PMID:22076153; http://dx.doi.org/10.1371/journal.pone.0027345
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol 2003; 77:8957-61; PMID:12885912; http://dx.doi.org/10.1128/JVI.77.16.8957-8951.2003
