Abstract
Breast cancer stem cells (BCSCs) have the greatest potential to maintain tumorigenesis in all subtypes of tumor cells and were regarded as the key drivers of tumor. Recent evidence has demonstrated that BCSCs contributed to a high degree of resistance to therapy. However, how BCSCs self renewal and tumorigenicity are maintained remains obscure. Herein, our study illustrated that overexpression of let-7a reduced cell proliferation and mammosphere formation ability of breast cancer stem cells(BCSCs) in a KRas-dependent manner through different pathways in vitro and in vivo. To be specific, we provided the evidence that let-7a was decreased, and reversely the expression of KRas was increased with moderate expression in early stages (I/II) and high expression in advanced stages (III/IV) in breast cancer specimens. In addition, the negative correlation between let-7a and KRas was clearly observed. In vitro, we found that let-7a inhibited mammosphere-forming efficiency and the mammosphere-size via NF-κB and MAPK/ERK pathway, respectively. The inhibitory effect of let-7a on mammosphere formation efficiency and the size of mammospheres was abolished after the depletion of KRas. On the contrary, enforced expression of KRas rescued the effect of let-7a. In vivo, let-7a inhibited the growth of tumors, whereas the negative effect of let-7a was rescued after overexpressing KRas. Taken together, our findings suggested that let-7a played a tumor suppressive role in a KRas-dependent manner.
Abbreviations
| BCSCs | = | breast cancer stem cells |
| MFE | = | mammosphere-forming efficiency |
Introduction
Breast cancer stem cells (BCSCs), which exist only as a minority proportion in breast tumor cells, have been regarded as the only cells with unlimited proliferation and tumorigenic potential; therefore, being capable of driving tumor growth, progression and metastasis due to the characteristics: self-renewal and differentiation.Citation1-3 It is reported that BCSCs play a key role in radio and chemo therapy resistance. Based on these observations, the BCSCs have become the foundation for new preventive and therapeutic strategies in breast cancer.Citation1,4-6 CSCs have been identified and isolated using putative cell surface markers.Citation7–9 It is reported that BCSCs have been enriched in CD44+/CD24−/low, SP, ALDH+ subpopulations.Citation3,10–15 CSCs typically have the capacity to grow as mammospheres, which can be serially passaged in vitro.Citation15,16 Based on this characteristic, isolating spherical clusters of self-replicating cells from suspension cultures is also used as a sorting method of tumorigenic CSCs. However, how BCSCs self renewal and tumorigenicity are maintained is still poorly understood.
Let-7, the first identified tumor suppressing miRNA, consists of 12 subtypes in its family.Citation17 The let-7 family can regulate many oncogenes to inhibit tumor progression by binding to complementary sequences in the 3′UTRs of the target mRNAs, including Ras, c-Myc and HMGA2.Citation18-21 In addition, reduced expression of let-7 has been found in lung cancer, colon cancer, ovary caner and breast cancer.Citation21,22 Many studies suggested reduced expression of miRNAs in stem cells was associated with the ability of cell self-renewal and differentiation.Citation23,24 However, there is still lack of understanding how miRNAs regulate the properties of CSCs.
Ras proteins control multiple intracellular signaling networks involved in cell proliferation, differentiation, apoptosis and cell migration.Citation25-27 Previous study showed let-7 is complementary to multiple sequences in the 3′UTR of human RAS genes, and represses its expression.Citation28 NF-κB can act as downstream of Ras pathway.Citation29 In general, a key regulatory step in the activation of NF-κB involves the phosphorylation of IKK complex, including heterodimers of IKKα and IKKβ, which subsequently results in ubiquitination and degradation of IκB and finally leads to the nuclear translocation of NF-κB, where it activates the transcription of various genes.Citation30,31 Moreover, Ras/MEK/ERK pathway involves in the activation of many transcription factors correlation with tumorigenesis.Citation32,33 The factors which inhibit steps in the NF-κB and MAPK/ERK pathway are regarded as potential targets for treating cancer.
Our study showed let-7a is reduced in human breast cancer patient samples and KRas is increased along with the cancer progression, and the negative correlation between let-7a and KRas was clearly observed. Let-7a regulates mammosphere formation capacity and tumor formation through Ras/NF-κB pathway and Ras/MAPK/ERK pathway in a KRas-dependent manner.
Results
The negative correlation between let-7a and KRas in breast cancer
We first examined the protein level of KRas by performing immunohistochemical (IHC) staining in breast specimens (n = 53). The results showed that the expression level of KRas was significantly increased in cancerous tissues with moderate expression in early stages (I/II) and high expression in advanced stages (III/IV) (). We further evaluated the abundance of KRAS mRNA in these specimens by qRT-PCR analysis. The mRNA level of KRAS was significantly increased in breast cancer tissues (n = 38) compared with normal tissues (n = 15), which is consistent with the increased protein expression (). Next, We analyzed the expression level of let-7a in breast cancer specimens (n = 15) and matched normal-like control from the same patient (n = 15) by quantitative RT-PCR(qRT-PCR) analysis. As shown in , let-7a was decreased significantly in breast cancer specimens compared with normal tissues (P < 0.05) (). Taken together, these observations suggested that the level of let-7a was reduced in human breast cancer patient samples and KRas was increased along with the cancer progression.
Figure 1. Let-7a was downregulated in breast cancer and inversely correlated with KRas levels. (A) Representative examples of immunohistochemical staining for KRas in each of the clinical stages of breast cancers as indicated. (B) Quantification of KRas relative immunostaining intensity for each clinical stage of breast cancers. Data is shown as mean ± SEM for N as indicated in the figure in parenthesis as shown. (C) Relative expression levels of KRas mRNA in breast specimens. KRas abundance was normalized to 18S rRNA. Data is shown as mean ± SEM for N as indicated in the figure in parethesis as shown. (D) Relative expression levels of let-7a in human breast specimens. Let-7a(miRNA) levels were measured by TaqMan stem-loop qRT-PCR. U6 was used as the control. (E) Let-7a and KRas were inversely correlated in human breast cancers(r = −0.5355, p = 0.0023).
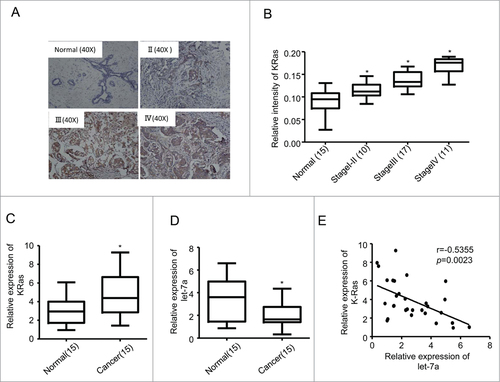
We subsequently analyzed the relationship between KRas and let-7a. As shown in , when KRas mRNA levels were plotted against let-7a expression, significant inverse correlations were observed in clinical human breast cancer specimens.
Let-7a inhibited mammosphere formation capacity in a KRas-dependent manner in breast cancer cells
To explore the function of let-7a upon mammosphere formation in mammary cell lines, we established stable cell lines (MCF-7 and SKBR3) by lentiviral transduction. As shown in , the expression of let-7a in these cell lines were higher than that in scramble control cells. To determine the functional significance of let-7a in regulating the mammosphere formation capacity of breast cancer cells, we conducted mammosphere formation assays and found that overexpression of let-7a in MCF-7 and SKBR3 cells inhibited the mammosphere number and size ().
Figure 2. Let-7a inhibited mammosphere formation capacity in a KRas-dependent manner in breast cancer cells. (A) let-7a was detected after transfecting with lenti- let-7a and lenti-scramble, the fold-changes were showed as indicated(data are mean ± SD; n = 3). (B) Images showing mammosphere formation in MCF7 cells and SKBR3 cells transfected as indicated in (A). The cells were cultured with mammosphere culture medium containing EGF, bFGF and B27 supplement. Scale bar=100μm. (C and D) The efficiency of mammosphere-formation and the size of mammospheres were measured after 10 d in MCF7 cells and SKBR3 cells transfected lenti-let-7a and lenti-scramble or vector control (data are mean ± SD; n = 3, #P< 0.05, relative to the values in the respective controls). (E and F) MCF-7 and SKBR3 cells were infected with lenti-let-7 and/or KRas-siRNA. The number of mammospheres was counted, the percentage of mammosphere-forming cells was determined (data are mean ± SD; n = 3, #P < 0.05, relative to the values in the respective controls).
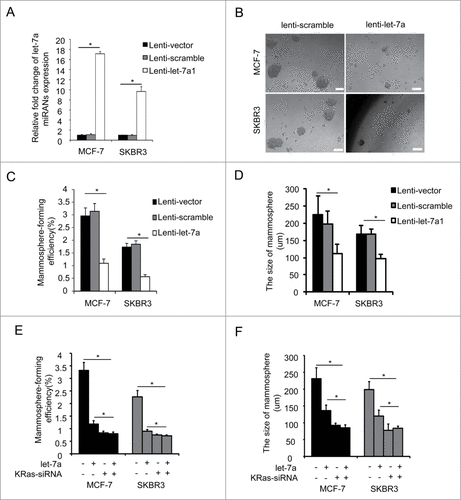
The inverse correlation between let-7a and KRas raised a possibility that let-7a may functionally interact with KRas signaling. To explore this possibility, we overexpressed let-7a with simultaneous depletion of KRas in MCF-7 and SKBR3 cells. Let-7a inhibited mammosphere-forming efficiency and the size of mammospheres in the presence of endogenous KRas, while the mammospere formation capacity in MCF-7 and SKBR3 cell lines cotransfected with let-7a and KRas-siRNA was not decreased compared to the cell lines transfected with KRas-siRNA alone(). This result suggested the suppressive effect of let-7a on mammospere formation capacity was abolished after the depletion of KRas, and the suppressive role of let-7a was dependent on KRas.
Let-7a-mediated suppression of KRAS expression and transactivation requires let-7a binding to the 3′UTR
NF-κB pathway promotes tumor growth and progression, and acts as a direct downstream target of Ras pathway [29]. The phosphorylation of IKKα and IKKβ results in ubiquitination and degradation of IκB, and finally leads to the nuclear translocation of NF-κB, where it activates the transcription of various genes.Citation30,31 Ras/MEK/ERK pathway involves in the activation of many transcription factors, which leads to the uncontrolled cancer cell growth and promotes tumorigenesis.Citation32,33 Our studies showed that let-7a inhibited mammosphere formation capacity in breast cancer cells in a KRas-dependent manner. To validate the mechanism of let-7a-regulated signaling pathways, we detected the expression of KRas and the phosphorylation levels of IKKα/β, IκBα and ERK in let-7a overexpressing MCF-7 and SKBR3 cells. The expression of KRas and the phosphorylation levels of IKKα/β, IκBα and ERK decreased markedly (), indicating that let-7a repressed the activation of RAS/NF-κB and RAS/MAPK/ERK pathway.
Figure 3. KRas is a direct target of let-7a. (A) The protein expression levels were determined in MCF-7 and SKBR3 cells transfected lenti-let-7a or lenti-scramble by western blot. (B-D) Luciferase activities were analyzed in 2 breast cell lines 36h after cotransfection of let-7a with either mutant or wild-type KRas luciferase reporter vectors.
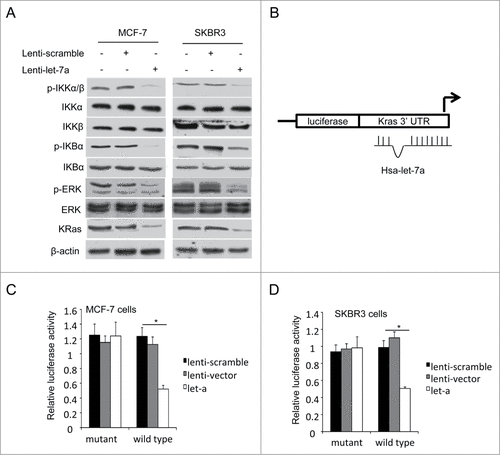
Bioinformatics analysis indicated that KRas was also a potential target of let-7a (). To validate this, we performed luciferase assays using reporter containing wild-type KRas 3′-UTR and mutant defective for let-7a binding. As shown in, overexpression of let-7a inhibited KRas wild-type, but not mutant luciferase reporter activities in MCF-7 and SKBR3 cells, indicating that let-7a specifically targets KRas through binding its 3′ UTR directly.
Let-7a regulates NF-κB and MAPK/ERK pathway in a KRas-dependent manner in breast cancer cells
Previous studies showed let-7a did not directly bind to the complementary sequences in the 3′UTRs of the genes in NF-κB pathway and MAPK/ERK pathway.Citation30,34 KRas was the target gene of let-7 as well as upstream of NF-κB and MAPK/ERK pathway. Based on these observations, we hypothesized that let-7a repressed NF-κB and MAPK/ERK pathway in a KRas-dependent manner. To explore this possibility, we first overexpressed let-7a with simultaneous depletion of KRas in MCF-7 and SKBR3 cells. Let-7a repressed the phosphorylation of IKKα/β, IκBα and ERK in the presence of endogenous KRas, while the phosphorylation of IKKα/β, IκBα and ERK in MCF-7 and SKBR3 cell lines cotransfected with let-7a and KRas-siRNA was not decreased compared to the cell lines transfected with KRas-siRNA alone().
Figure 4. Let-7a regulates NF-κB and MAPK/ERK pathway in a KRas-dependent manner in breast cancer cells (A and B) The protein expression levels were determined in MCF-7 and SKBR3 cells transfected lenti- let-7a and/or KRas-siRNA by western blot. (C and D) The protein expression levels were determined in MCF-7 and SKBR3 cells transfected lenti- let-7a and/or KRas by western blot.
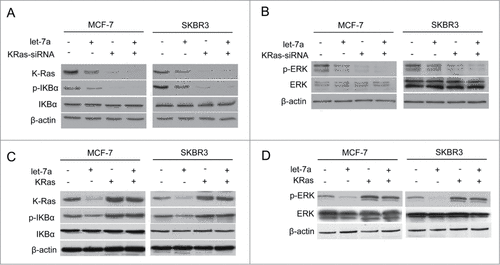
This result suggested that let-7a-mediated repression of NF-κB and MAPK/ERK pathway is dependent on the presence of KRas. Consistent with this observation, overexpression of KRas in MCF-7 and SKBR3 cells rescued the loss of p-IKKα/β, p-IκBα and p-ERK induced by let-7a overexpression (). Taken together, these data indicated that KRas was required for let-7a-mediated suppression of NF-κB and MAPK/ERK pathway.
Furthermore, after treating MCF-7 and SKBR3 cells with SN50Citation35,36 (a highly selective inhibitor of NF-κB pathway) and/or PD98059 (an inhibitor of ERK), we observed the phosphorylation levels of IKKα/β, IκB and ERK decreased (Fig. S1A and B).
Let-7a/Ras/NF-κB axis inhibits mammosphere initiation
The number of mammospheres reflects the quantity of cells capable of self-renewal in vitro, while the size of mammospheres measures the self-renewal capacity of each mammosphere-generating cell.Citation24,37 To explore whether let-7a regulated the number of mammospheres and the size of mammospheres through Ras/NF-κB axis, we transfected with lenti-let-7a and/or KRas-siRNA in MCF-7 and SKBR3 cells followed by SN50 treatment. As shown in , the mammosphere formation efficiency and the size of mammospheres were decreased significantly after transfecting with lenti-let-7a and/or KRas-siRNA, while in the precence of SN50, only the number of mammospheres remarkably decreased (; Fig. S2A-E).
Figure 5 (See previous page). Let-7a/Ras/NF-κB axis inhibits mammosphere initiation. (A) Representative images of mammospheres in MCF-7 cell transfected or treated as indicated. Scale bar=100μm. (B and C) MCF-7 cells were infected with lenti-let-7 and/or KRas-siRNA, KRas or treated with 50μM SN50. The number of mammospheres was counted, the percentage of mammosphere-forming cells was determined (data are mean ± SD; n = 3, #P< 0.05, relative to the values in the respective controls). (D and E) The primary mammospheres of MCF-7 cells transfected lenti-let-7 and/or KRas-siRNA, KRas or treated with 50μM SN50 were dissociated into single cells and grown as secondary mammospheres. The size of mammospheres was measured after 10d (data are mean ± SD; n = 3, #P < 0.05, relative to the values in the respective controls). (F-I) FACS analysis was performed to identify the cells expressing ALDH activity. The proportion of ALDH+cells was measured(data are mean ± SD; n = 3, #P < 0.05, relative to the values in the respective controls). Representative images of the proportion of ALDH+cells in MCF-7 cell transfected or treated as indicated.
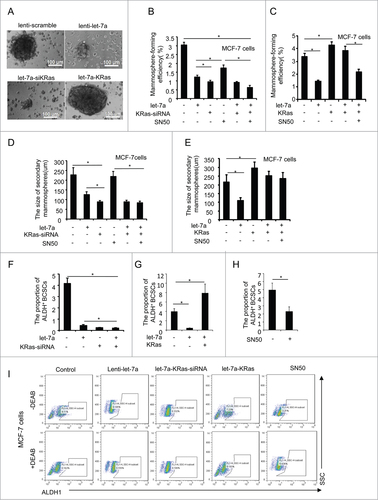
Subsequently, to observe the potential of sencondary mammospheres, the primary mammospheres of MCF-7 and SKBR3 cells as previous describe were dissociated into single cells and cultured in the mammosphere culture medium. However, these cells did not generate secondary mammospheres as efficiently as cells from mammospheres of untreated MCF-7 and SKBR3 cells, and the size of secondary mammospheres still was not decreased significantly after SN50 treatment (; Fig. S2F-G).
As showed previously, the depletion of KRas with simultaneous overexpression of let-7a abolished the inhibitory effect of let-7a on both mammosphere formation efficiency and the size of mammospheres. On the contrary, enforced expression of KRas rescued the let-7a-mediated inhibition of mammosphere formation capacity (). Taken together, these results indicated that let-7a inhibited mammosphere formation efficiency through suppressing Ras/NF-κB axis, whereas the size of mammosphere was regulated through other underlying mechanism via KRas.
FACS analysis was performed to further characterize the effects of let-7a on the proportion of BCSCs. As shown in , the proportion of ALDH+ BCSCs was decreased after overexpression of let-7a, KRas knockdown and treating with SN50, whereas enforced expression of KRas increased the proportion of ALDH+ BCSCs in MCF-7 cells transfected with let-7a.
Let-7a affects the size of mammospheres through regulating Ras/MAPK/ERK pathway
Cell proliferation during mammosphere growth determines the size of the mammospheres. In order to determine whether the pathways involved in cell proliferation could regulate the size of mammospheres, we transfected with lenti-let-7a and/or KRas-siRNA in MCF-7 and SKBR3 cells followed by SN50 treatment. Let-7a, KRas-siRNA or SN50 suppressed the cell proliferation, whereas overexpression of KRas rescued the inhibitory effect of let-7a on cell viability (; Fig. S3A-B). Our previous study showed let-7a regulated mammosphere size via KRas (), whereas NF-κB pathway was not involved in the regulation of the size of mammospheres.
Figure 6. Let-7a affects the size of mammospheres through regulating Ras/MAPK/ERK pathway. (A and B) MCF-7 were infected with lenti-let-7 and/or KRas-siRNA, KRas or treated with 50 μM SN50. Relative cell viability was estimated at indicated time by a MTT assay in which each condition was replicated 3 times(data are mean ± SD, proliferation of untreated cells was defined as 100%). (C) MCF7 cells were treated with PD98059 (25μM). OD value was estimated at indicated time by a MTT assay in which was replicated 3 times(data are mean ± SD). (D and E) MCF-7 cells were transfected with lenti-let-7, and/or KRas-siRNA, KRas or treated with PD98059. The size of mammospheres was measured after 10d (data are mean ± SD; n = 3, #P< 0.05, relative to the values in the respective controls). (F) MCF-7 cells were treated with PD98059. The number of mammospheres was counted, the percentage of mammosphere-forming cells was determined (data are mean ± SD; n = 3, #P < 0.05, relative to the values in the respective controls). (G) Schematic overview of the effect of let-7a on mammosphere formation capacity.
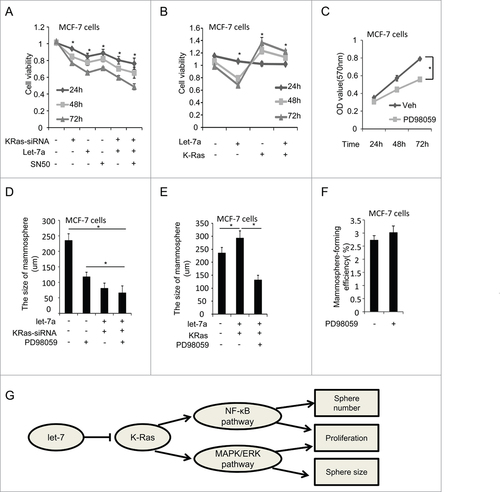
Next, we explored the role of MAPK/ERK pathway in the regulation of the size of mammospheres. After inhibiting ERK by PD98059, the cell viability was decreased and the size of mammospheres were reduced significantly (; Fig. S3C). However, we did not observe clear changes on the number of mammospheres (; Fig. S3E), suggesting MAPK/ERK pathway was also considered downstream of Ras and involved in the regulation of cell growth the size of mammospheres.
The size of mammosphers was decreased by ERK inhibitor in MCF-7 and SKBR3 cells (; Fig. S3D). After overexpression of KRas, the inhibitory effect of let-7a was rescued, whereas the size of mammosphers was still repressed by ERK inhibitor (; Fig. S3D). These findings suggested let-7a regulated the size of mammosphere through Ras/MAPK/ERK pathway ().
Let-7a inhibits tumor formation via KRas in vivo
We next determined the effect of let-7a on tumor formation. We implanted MCF-7 cells (2×106 cells per site) transfected with lenti-let-7a and KRas, or cotransfection with lenti-let-7a and KRas subcutaneously into immune-deficient mice in the presence of long-release estradiol pellets and monitored the tumor growth (). Four out of 6 mice inoculated with MCF-7 cells transfected let-7a formed smaller tumors. Moreover, overexpression of KRas rescued the inhibitory effect of let-7a on tumor formation.
Figure 7. Let-7a inhibits tumor formation via KRas in vivo. MCF-7 cells transfected with lenti-let-7 and/or KRas(2×106 cells per site) mixed with matrigel (1:1) were injected into the mammary fat pad of immunodeficient mice supplemented with long-release estradiol pellets. Tumor size was measured every 4d until the tumors were harvested. The data was shown as mean ± SEM for N > 6 separate tumors for each group. (A) Images of tumors dissected from the mice. (B) The tumor size (mm3) versus days of post injection. (C) Tumor was weighted after resection at the end of experiment. (D) mRNA levels of let-7a, KRas, IκBα and ERK were determined by qRT-PCR in tumors. (E and F) IHC staining detected the protein level of KRas, p-IκBa and p-ERK in tumor tissues derived from mice. Data for quantified IHC was shown as mean ± SEM for N = 4 tumors in each group.
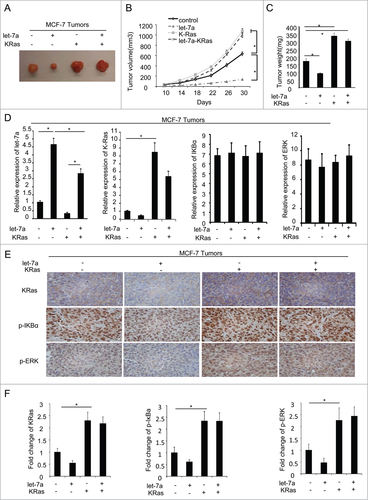
qRT-PCR analysis was conducted to determine the mRNA level of let-7a, KRas, IκBα and ERK (), and IHC was performed to detect the protein level of KRas, p-IκBα and p-ERK in tumor specimens(). Consistent with the findings in vitro, the results suggested let-7a inhibited the phosphorylation of IκBα and ERK through inhibiting KRas in vivo.
Discussion
BCSCs are not only correlated with tumorigenesis, but also responsible for tumor propagation, metastasis, recurrence and therapy resistance.Citation1,4,24 Based on this, BCSCs are regarded as novel therapeutic targets.Citation38,39 BCSCs are enriched in the ALDH+ cell population, which has the potential to generate mammospheres under anchorage-independent conditions in mammosphere culture medium.Citation15,31 However, there is a still limited understanding that how mammosphere formation is regulated and which molecular mechanisms are involved in the process of BCSCs self-renewal.
Let-7 miRNA, a tumor suppressor, regulates cell proliferation and differentiation.Citation40,41 Recently, its decreased expression has been linked to the increased tumorigenicity and poor prognosis.Citation42 Let-7a has been reported to directly suppress Ras through binding to its 3′UTR.Citation28 In the present study, we provided the evidence that let-7a was downregulated, and reversely the expression level of KRas was increased with moderate expression in early stages (I/II) and high expression in advanced stages (III/IV) in breast cancer specimens. The negative correlation between let-7a and KRas was clearly observed.
In breast cancer cells, we demonstrated that the negative effects of let-7a on mammosphere-formation capacity, which can be represented by two aspects, the number of mammospheres which reflects the quantity of cells capable of self-renewal in vitro, and the size of mammospheres which measures the self-renewal capacity of each mammosphere-generating cell and can be determined by cell proliferation during mammosphere growth.Citation24,37 In our study, let-7a regulated the mammosphere-forming efficiency and mammsphere-size in a KRas-dependent manner. Moreover, the proportion of ALDH+ BCSCs was decreased in MCF-7 cells transfected let-7a. The depletion of KRas with simultaneous overexpression of let-7a alleviated the inhibitory effect of let-7a, whereas the negative effect of let-7a was rescued after enforced expression of KRas. After inhibiting the NF-κB pathway by using SN50, the phosphorylation level of IKKα/β and IκB increased, which further leaded to the decrease of mamosphere-forming efficiency as well as the proportion of ALDH+ BCSCs, instead of the size of mammosphere. These illustrated NF-κB pathway affected mammosphere initiation but did not primarily influence cell proliferation during mammosphere growth.
It is reported that cell proliferation during mammosphere growth determined the size of mammospheres.Citation24,31 We found the proliferation decreased significantly in MCF-7 and SKBR3 cell lines transfected with lenti-let-7, KRas-siRNA or treated with SN50, whereas overexpression of KRas rescued the inhibitory effect of let-7a on cell viability. However, the size of mammospheres was not regulated by NF-κB pathway, which suggested let-7a regulated the size of mammospheres via some underlying molecular pathways involved in KRas. MAPK/ERK pathway, known as downstream of Ras, was involved in the activation of many transcription factors and cell proliferation.Citation43,44 Incorrect regulation of ERK pathway is common in tumors.Citation33,45-47 In the end, we proved that let-7a regulated the phosphorylation levels of ERK, the cell proliferation and the size of mammospheres via KRas/MAPK/ERK pathway.
In conclusion, let-7a mediated inhibition of mammosphere formation capacity and cell survival depended on KRas. Moreover, let-7a regulated the number of mammospheres and the size of mammospheres through KRas/NF-κB pathway and KRas/MEK/ERK pathway respectively. As a consequence, it is of great importance to develop drugs targeted at the molecules involved in these two different pathways to ameliorate the curative effects of breast cancer patients.
Materials and Methods
Cell culture
Breast cancer cell lines MCF7 and SKBR3 were purchased from the American Type Culture Collection (ATCC). Cells were cultured in 90% DMEM with 10% (vol/vol) fetal bovine serum (Invitrogen) at 37°C under 5% CO2. Mammospheres were generated as described below.Citation15
Mammosphere culture
Cells were plated as single cell son ultralow attachment plates (Corning) at a density of 5000 viable cells/ml in primary culture and a density of 1000 cells/mL in MCF-7and SKBR3 cell lines. Cells were cultured in mammosphere culture medium, which consisted of serum-free DMEM-F12 (BioWhittaker), supplemented with B27 (1:50, Invitrogen), 10 ng/mL EGF (BD Biosciences), 20 ng/mL bFGF (PeproTech), 0.4% bovine serum albumin (Sigma), and 4 mg/mL insulin (Sigma) and heparin (Stem Cell Technologies). In some experiments, cells were treated with NF-κB inhibitor SN50 (Calbiochem) and ERK inhibitor PD98059 (Calbiochem), at the indicated concentrations for 10 d. To culture secondary mammospheres in vitro, primary mammospheres were collected by gentle centrifugation (800rpm) after 10 days, and dissociated enzymatically (10 min in 0.05% trypsin, 0.53 mM EDTA-4Na; Invitrogen) and mechanically into a single-cell suspension. Single cells were then cultured in suspension to generate mammospheres of the next generation. The percentage of wells with mammospheres and the size of mammospheres were analyzed at indicated times.
Constructs and transfection
Oligonucleotides encoding let-7a pre-miRNA48or siRNA targeting KRASCitation49,50 were synthesized according to previously published sequences. The KRas cDNA clone was purchased from OriGene Technologies and cloned into 5′-NotI and 3′-ClaI restriction enzyme sites of MSCV-IRES-GFP (Addgen) retroviral vector. The transduction was performed as described previously.Citation24,51,52 The expression of let-7a and KRas were detected by qRT-PCR and Western blot analysis as described below.
Luciferase reporter assay
The 3′UTR of pGL3-KRas luciferase reporter construct, including the predicted binding sites for let-7a, was purchased from Addgen. The 3′UTR of mutant KRas was constructed, carrying a mutated sequence in the let-7a binding region. Cells were cultured in 24-well plates and co-transfected with a complex of let-7a and the luciferase reporter construct using lipofectamine 2000 into MCF-7 or SKBR3 cells, according to the manufacturers' guidelines. β-Gal was co-transfected in each sample as an internal control for transfection efficiency. At 36h post transfection, luciferase assay was conducted as previously described.Citation53
Western blotting
Whole-cell lysates (60μg) were separated by 10% SDS-PAGE, and the proteins were transferred to nitrocellulose membrane. Western blotting was performed with antibodies against IKKα/β, IκBα, p-IKKα/β, p-IκBα, KRas, ERK, p-ERK, and β-actin, and detection was performed with HRP-conjugated antisera and chemiluminescence. All the antibodies were purchased from Santa Cruz Biotechnology.
Aldefluor assay and flow cytometry
The ALDEFLUOR™ fluorescent reagent system (Stem Cell Technologies, Canada) was used to identify the cells expressing ALDH activity by using LSRII flow cytometer (BD Biosciences) as previously described.Citation54 Briefly, cells were trypsinized to single cells at a concentration of 0.5–1.0×106 and subsequently suspended in ALDEFLUOR assay buffer containing ALDH substrate and then incubated for 45 minutes at 37°C. In all experiments, the ALDEFLUOR-stained cells treated with diethylaminobenzaldehyde(DEAB), a specific ALDH inhibitor, served as ALDH-negative controls. The Flow cytometer setup and data acquisition were followed by Aldehyde Dehydrogenase Based Cell Detection Kit (ALDEFLUOR™, 01700). DEAB-treated cells served as control to set the ALDHhigh regions. FACS data were analyzed with Flowjo software 7.2.5 (Treestar, Oregon, USA).
Quantitative analysis of microRNAs and mRNAs
Total RNA was extracted from clinical specimens of breast cancer using TRIzol reagent (Invitrogen) according to the manufacturer's protocols. The TaqMan stem-loop realtime RT-PCR was used to assess the expression of microRNAs, U6 was used as the control. For quantitative analysis of mRNA expression, total RNA was reversely transcribed to cDNA using the Primescript RT reagent kit (TaKaRa) according to the manufacturer's instructions. qRT-PCR was carried out in Bio-Rad CFX96 Real-Time PCR Detection System with iQ SYBR Green Supermix (Bio-Rad). The real-time data were normalized based on the real-time PCR detection of 18S rRNA in the various samples. Relative miRNA expression data were analyzed using the ΔΔCT method.
Assessment of Cell Viability by MTT Assays
Initially, cells were seeded at the concentration of 4×103cells/well cells in triplicates in a 96 well format. Cells were then treated with SN50(50μM) or PD98059 (25μM) for 24hrs, 48hrs and 72hrs. Cell viability was measured by MTT cell viability assay.
Immunohistochemistry (IHC) staining
Paraffin-embedded tissue sections (human origin and mouse origin) were used to determine KRas, p-IκBα, p-ERK (Santa Cruz). Immunohistochemical analysis of human breast cancer and tumor xenograft tissue was carried out as describedCitation55,56
Tumor xenograft in nude mice
MCF-7-control, MCF-7-lenti-let-7a, MCF-7-KRas, MCF-7-let-7a-KRas cells (2×106 cells per site) were injected into the mammary fat pad of 6-week-old BALB/c femal nude mice (Shanghai Slaccas). Long-release estradiol pellets (Innovative Research of America) were implanted the day before inoculation. Tumor growth was measured using a digital caliper every 4d for 4–5 weeks and their volumes were calculated using a standard formula: width2 × length × 0.52.Citation57 Tumor weight was measured when mice were sacrificed on day 30 after cell implantation. All animal experiments were approved by our Institutional Animal Care and Use Committee.
Disclosure of Potential Conflicts of Interest
The authors declare that they have no competing or conflicting interests.
1030547_Suplemental_files.zip
Download Zip (3.5 MB)Acknowledgments
The authors thank Ke Chen for advising on the revision of the article. The authors acknowledge the assistants at the Department of Oncology and the staff of Center for Translational Medicine, the first affiliated hospital, College of Medicine, Xi'an Jiaotong University for their technical assistance.
Funding
This study was supported by the Natural Science Foundation of China, approved ID: 81272418, ID: 81402506.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature 2001; 414:105–11; PMID:11689955; http://dx.doi.org/10.1038/35102167
- Charafe-Jauffret E, Monville F, Ginestier C, Dontu G, Birnbaum D, Wicha MS. Cancer stem cells in breast: current opinion and future challenges. Pathobiology 2008; 75:75–84; PMID:18544962; http://dx.doi.org/10.1159/000123845
- Ricardo S, Vieira AF, Gerhard R, Leitao D, Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F, Paredes J. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J Clin Pathol 2011; 64:937–46; PMID:21680574; http://dx.doi.org/10.1136/jcp.2011.090456
- Velasco-Velazquez MA, Popov VM, Lisanti MP, Pestell RG. The role of breast cancer stem cells in metastasis and therapeutic implications. Am J Pathol 2011; 179:2–11; PMID:21640330; http://dx.doi.org/10.1016/j.ajpath.2011.03.005
- Chuthapisith S, Eremin J, El-Sheemey M, Eremin O. Breast cancer chemoresistance: emerging importance of cancer stem cells. Surgical Oncol 2010; 19:27–32; PMID:19251410; http://dx.doi.org/10.1016/j.suronc.2009.01.004
- Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44(+) breast cancer-initiating cells to radiation. J Natl Cancer I 2006; 98:1777–85; PMID:17179479
- Jaggupilli A, Elkord E. Significance of CD44 and CD24 as cancer stem cell markers: an enduring ambiguity. Clin Dev Immunol 2012; 2012:708036; PMID:22693526; http://dx.doi.org/10.1155/2012/708036
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells–perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res 2006; 66:9339–44; PMID:16990346; http://dx.doi.org/10.1158/0008-5472.CAN-06-3126
- Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K, Los M. Cancer stem cell markers in common cancers - therapeutic implications. Trend Mol Med 2008; 14:450–60; PMID:18775674; http://dx.doi.org/10.1016/j.molmed.2008.08.003
- Snyder EL, Bailey D, Shipitsin M, Polyak K, Loda M. Identification of CD44v6(+)/CD24- breast carcinoma cells in primary human tumors by quantum dot-conjugated antibodies. Lab Invest J Tech Method Pathol 2009; 89:857–66; PMID:19488035; http://dx.doi.org/10.1038/labinvest.2009.54
- Bauerschmitz GJ, Ranki T, Kangasniemi L, Ribacka C, Eriksson M, Porten M, Herrmann I, Ristimaki A, Virkkunen P, Tarkkanen M, et al. Tissue-specific promoters active in CD44+CD24−/low breast cancer cells. Cancer Res 2008; 68:5533–9; PMID:18632604; http://dx.doi.org/10.1158/0008-5472.CAN-07-5288
- Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24−/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res 2005; 11:1154–9; PMID:15709183
- Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, Goodell MA, Brenner MK. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A 2004; 101:14228–33; PMID:15381773; http://dx.doi.org/10.1073/pnas.0400067101
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007; 1:555–67; PMID:18371393; http://dx.doi.org/10.1016/j.stem.2007.08.014
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 2010; 140:62–73; PMID:20074520; http://dx.doi.org/10.1016/j.cell.2009.12.007
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 2011; 147:759–72; PMID:22078877; http://dx.doi.org/10.1016/j.cell.2011.09.048
- Wang X, Cao LEI, Wang Y, Wang X, Liu N, You Y. Regulation of let-7 and its target oncogenes (Review). 2012; 955–60; PMID:22783372
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215–33; PMID:19167326; http://dx.doi.org/10.1016/j.cell.2009.01.002
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007; 315:1576–9; PMID:17322030; http://dx.doi.org/10.1126/science.1137999
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Gene Dev 2009; 23:1743–8; PMID:19574298; http://dx.doi.org/10.1101/gad.1812509
- Zhao Y, Deng C, Lu W, Xiao J, Ma D, Guo M, Recker RR, Gatalica Z, Wang Z, Xiao GG. let-7 microRNAs induce tamoxifen sensitivity by downregulation of estrogen receptor α signaling in breast cancer. Mol Med 2011; 17:1233–41; PMID:21826373; http://dx.doi.org/10.2119/molmed.2010.00225
- Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih Ie M, Zhang Y, Wood W 3rd, Becker KG, Morin PJ. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PloS one 2008; 3:e2436; PMID:18560586; http://dx.doi.org/10.1371/journal.pone.0002436
- Yu Z, Li Y, Fan H, Liu Z, Pestell RG. miRNAs regulate stem cell self-renewal and differentiation. Frontier Genet 2012; 3:191; PMID:23056008; http://dx.doi.org/10.3389/fgene.2012.00191
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007; 131:1109–23; PMID:18083101; http://dx.doi.org/10.1016/j.cell.2007.10.054
- Zamkova M, Khromova N, Kopnin BP, Kopnin P. Ras-induced ROS upregulation affecting cell proliferation is connected with cell type-specific alterations of HSF1/SESN3/p21Cip1/WAF1 pathways. Cell Cycle 2013; 12:826–36; PMID:23388456; http://dx.doi.org/10.4161/cc.23723
- Takahashi C, Bronson RT, Socolovsky M, Contreras B, Lee KY, Jacks T, Noda M, Kucherlapati R, Ewen ME. Rb and N-ras function together to control differentiation in the mouse. Mol Cell Biol 2003; 23:5256–68; PMID:12861012; http://dx.doi.org/10.1128/MCB.23.15.5256-5268.2003
- Basu Roy UK, Henkhaus RS, Loupakis F, Cremolini C, Gerner EW, Ignatenko NA. Caveolin-1 is a novel regulator of K-RAS-dependent migration in colon carcinogenesis. Intl J Cancer 2013; 133:43–57; PMID:23280667; http://dx.doi.org/10.1002/ijc.28001
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell 2005; 120:635–47; PMID:15766527; http://dx.doi.org/10.1016/j.cell.2005.01.014
- Chen Y, Wu J, Ghosh G. KappaB-Ras binds to the unique insert within the ankyrin repeat domain of IkappaBbeta and regulates cytoplasmic retention of IkappaBbeta × NF-kappaB complexes. J Biol Chem 2003; 278:23101–6; PMID:12672800; http://dx.doi.org/10.1074/jbc.M301021200
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009; 139:693–706; PMID:19878981; http://dx.doi.org/10.1016/j.cell.2009.10.014
- Hinohara K, Kobayashi S, Kanauchi H, Shimizu S, Nishioka K, Tsuji E, Tada K, Umezawa K, Mori M, Ogawa T, et al. ErbB receptor tyrosine kinase/NF-kappaB signaling controls mammosphere formation in human breast cancer. Proc Natl Acad Sci U S A 2012; 109:6584–9; PMID:22492965; http://dx.doi.org/10.1073/pnas.1113271109
- Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007; 26:3291–310; PMID:17496923; http://dx.doi.org/10.1038/sj.onc.1210422
- McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochimica Biophys Acta 2007; 1773:1263–84; PMID:17126425; http://dx.doi.org/10.1016/j.bbamcr.2006.10.001
- Choudhury SN, Li Y. miR-21 and let-7 in the Ras and NF-κB Pathways. MicroRNA 2012; 1:65–9; PMID:25048092; http://dx.doi.org/10.2174/2211536611201010065
- Bonini SA, Ferrari-Toninelli G, Uberti D, Montinaro M, Buizza L, Lanni C, Grilli M, Memo M. Nuclear factor kappaB-dependent neurite remodeling is mediated by Notch pathway. J Neuroscience 2011; 31:11697–705; PMID:21832199; http://dx.doi.org/10.1523/JNEUROSCI.1113-11.2011
- Bravo SB, Pampin S, Cameselle-Teijeiro J, Carneiro C, Dominguez F, Barreiro F, Alvarez CV. TGF-β-induced apoptosis in human thyrocytes is mediated by p27kip1 reduction and is overridden in neoplastic thyrocytes by NF-kappaB activation. Oncogene 2003; 22:7819–30; PMID:14586408; http://dx.doi.org/10.1038/sj.onc.1207029
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Gene Dev 2003; 17:1253–70; PMID:12756227; http://dx.doi.org/10.1101/gad.1061803
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009; 138:645–59; PMID:19682730; http://dx.doi.org/10.1016/j.cell.2009.06.034
- Morrison BJ, Schmidt CW, Lakhani SR, Reynolds BA, Lopez JA. Breast cancer stem cells: implications for therapy of breast cancer. Breast Cancer Res 2008; 10:210; PMID:18671830; http://dx.doi.org/10.1186/bcr2111
- Ding XC, Slack FJ, Großhans H. The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation. Cell cycle (Georgetown, Tex) 2008; 7:3083–90.
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834–8; PMID:15944708; http://dx.doi.org/10.1038/nature03702
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nature reviews Genetics 2009; 10:704–14; PMID:19763153; http://dx.doi.org/10.1038/nrg2634
- Whitmarsh AJ. Regulation of gene transcription by mitogen-activated protein kinase signaling pathways. Biochimica Biophy Acta 2007; 1773:1285–98; PMID:17196680; http://dx.doi.org/10.1016/j.bbamcr.2006.11.011
- Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev 2004; 68:320–44; PMID:15187187; http://dx.doi.org/10.1128/MMBR.68.2.320-344.2004
- Steelman LS, Chappell WH, Abrams SL, Kempf RC, Long J, Laidler P, Mijatovic S, Maksimovic-Ivanic D, Stivala F, Mazzarino MC, et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging 2011; 3:192–222; PMID:21422497
- Deschenes-Simard X, Gaumont-Leclerc MF, Bourdeau V, Lessard F, Moiseeva O, Forest V, Igelmann S, Mallette FA, Saba-El-Leil MK, Meloche S, et al. Tumor suppressor activity of the ERK/MAPK pathway by promoting selective protein degradation. Gene Dev 2013; 27:900–15; PMID:23599344; http://dx.doi.org/10.1101/gad.203984.112
- Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opinion Therapeutic Targets 2012; 16:103–19; PMID:22239440; http://dx.doi.org/10.1517/14728222.2011.645805
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004; 64:3753–6; PMID:15172979; http://dx.doi.org/10.1158/0008-5472.CAN-04-0637
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 2009; 15:489–500; PMID:19477428; http://dx.doi.org/10.1016/j.ccr.2009.03.022
- Kwon O, Jeong SJ, Kim SO, He L, Lee HG, Jang KL, Osada H, Jung M, Kim BY, Ahn JS. Modulation of E-cadherin expression by K-Ras; involvement of DNA methyltransferase-3b. Carcinogenesis 2010; 31:1194–201; PMID:20375073
- Parikh C, Subrahmanyam R, Ren R. Oncogenic NRAS, KRAS, and HRAS exhibit different leukemogenic potentials in mice. Cancer Res 2007; 67:7139–46; PMID:17671181; http://dx.doi.org/10.1158/0008-5472.CAN-07-0778
- Guo L, Chen C, Shi M, Wang F, Chen X, Diao D, Hu M, Yu M, Qian L, Guo N. Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition. Oncogene 2013; 32:5272–82; PMID:23318420; http://dx.doi.org/10.1038/onc.2012.573
- Wang XR, Luo H, Li HL, Cao L, Wang XF, Yan W, Wang YY, Zhang JX, Jiang T, Kang CS, et al. Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro-Oncol 2013; 15:1491–501; PMID:24092860; http://dx.doi.org/10.1093/neuonc/not107
- Lin L, Liu A, Peng Z, Lin HJ, Li PK, Li C, Lin J. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res 2011; 71:7226–37; PMID:21900397; http://dx.doi.org/10.1158/0008-5472.CAN-10-4660
- Wang T, Liu Z, Guo S, Wu L, Li M, Yang J, Chen R, Xu H, Cai S, Chen H, et al. The tumor suppressive role of CAMK2N1 in castration-resistant prostate cancer. Oncotarget 2014; 5:3611–21; PMID:25003983
- Wang T, Guo SM, Liu Z, Wu LC, Li MC, Yang J, Chen RB, Liu XM, Xu H, Cai SX, et al. CAMK2N1 inhibits prostate cancer progression through androgen receptor-dependent signaling. Oncotarget 2014; 5:10293–306; PMID:25296973
- Martin HL, Smith L, Tomlinson DC. Multidrug-resistant breast cancer: current perspectives. Breast Cancer 2014; 6:1–13; PMID:24648765; http://dx.doi.org/10.2147/BCTT.S37638
