Abstract
PR homology domain-containing member 12 (PRDM12) belongs to a family of conserved transcription factors implicated in cell fate decisions. Here we show that PRDM12 is a key regulator of sensory neuronal specification in Xenopus. Modeling of human PRDM12 mutations that cause hereditary sensory and autonomic neuropathy (HSAN) revealed remarkable conservation of the mutated residues in evolution. Expression of wild-type human PRDM12 in Xenopus induced the expression of sensory neuronal markers, which was reduced using various human PRDM12 mutants. In Drosophila, we identified Hamlet as the functional PRDM12 homolog that controls nociceptive behavior in sensory neurons. Furthermore, expression analysis of human patient fibroblasts with PRDM12 mutations uncovered possible downstream target genes. Knockdown of several of these target genes including thyrotropin-releasing hormone degrading enzyme (TRHDE) in Drosophila sensory neurons resulted in altered cellular morphology and impaired nociception. These data show that PRDM12 and its functional fly homolog Hamlet are evolutionary conserved master regulators of sensory neuronal specification and play a critical role in pain perception. Our data also uncover novel pathways in multiple species that regulate evolutionary conserved nociception.
Abbreviations
| PRDM12 | = | PR homology domain-containing member 12 |
| HSAN | = | hereditary and sensory autonomic neuropathy |
| TRHDE | = | tyrotropin-releasing hormone degrading enzyme |
| RA | = | retinoic acid |
| Brn3d | = | brain 3d |
| Tlx3 | = | T-cell leukemia homeobox 3 |
| Hmx3 | = | H6 family homeobox 3 |
| Drgx | = | dorsal root ganglia homeobox |
| Sncg | = | Synuclein Gamma (Breast Cancer-Specific Protein 1) |
| En1 | = | engrailed-1 |
| RT-qPCR | = | real-time quantitative polymerase chain reaction |
| MO | = | morpholino oligonucleotide |
| ChIP | = | chromatin immunoprecipitation |
| S1PR1 | = | Sphi8ngosine-1-phosphate receptor 1 |
| CGNL1 | = | cyclin L1 |
| IL1R1 | = | interleukin 1 receptor type 1 |
| TRH(DE) | = | tryrotropin-releasing hormone degrading enzyme |
| pCMV6 | = | plasmid cytomegalovirus |
| DDK | = | DYKDDDDK epitope |
| GAPDH | = | glyceraldehyde 3-phospate dehydrogenase |
| NBT/BCIP | = | nitro blue tetrazolium / 5-bromo-4-chloro-3-indolyl-phosphate |
| SET | = | Su(var)3–9 and ‘Enhancer of zeste’ |
| PDB | = | protein data base |
| HEK293 | = | human embryonic kidney cell 293 |
| PBS | = | phosphate buffered saline |
| DAPI | = | 4′,6-diamidino-2-phenylindole |
| GFP | = | green fluorescent protein |
| BSA | = | bovine serum albumin |
| HRP | = | horseraddish peroxidase |
| ECL | = | enhanced chemiluminescence |
| FPKM | = | fragments per kilobase exon |
| FDR | = | false discovery rate |
| GEO | = | gene expression omnibus |
| PMID | = | pubmed identification. |
Introduction and Results
Chronic and acute pain affects millions of people worldwide producing an enormous financial and quality of life burden. The detection of noxious or damaging stimuli (nociception) is an ancient process that alerts living organisms to environmental dangers.Citation1-3 Harmful stimuli activate receptors on specific sensory neurons called nociceptors,Citation4 which mediate information transfer via the spinal cord to higher order processing centers resulting in protective behaviors and awareness of pain.Citation5 Pain perception is essential for an animal to thrive, and human patients that cannot sense pain, such as patients with hereditary sensory and autonomic neuropathy (HSAN), die prematurely due to multiple injuries.Citation6
We and others have shown that Prdm12, a member of the PR domain containing zinc finger transcription factor family, is selectively expressed in p1 progenitors of the developing vertebrate spinal cord where it promotes V1 interneuron cell fate. It is also expressed in specific brain regions and in cranial ganglia.Citation7,8 Through RNAseq analysis of retinoic acid (RA) treated neuralized caps derived from control and Prdm12 overexpressing embryos, about 300 differentially expressed genes were identified. As expected, the list of regulated genes includes many V1 interneuron markers. Intriguingly, among the list of upregulated genes, we also found genes encoding transcription factors playing essential roIes in sensory neurogenesis such as Islet1, Brn3d, Tlx3, Hmx3 and Drgx and sensory neuron markers such as Sncg and Nprrf2, some of which have been previously implicated in nociception.Citation9-14a shows validation of these Prdm12 target genes in RA-treated neuralized animal cap explants by RT-qPCR. To determine whether Prdm12 is required for sensory neurons, embryos were injected with a Prdm12 antisense morpholino (MO) and analyzed for the markers Sncg, strongly expressed in trigeminal ganglia, as well as Tlx3 and Islet1 expressed more broadly in cranial sensory ganglia (). All markers were reduced in Prdm12 antisense MO treated embryos (68% reduced and 30% unaffected, n = 43 for Sncg; 64% reduced and 36% unaffected, n = 51 for Tlx3; 61% reduced, 27% unaffected and 12% increased, n = 51 for Islet1). In contrast, Sncg, Tlx3 and Islet1 expression were unaffected in embryos injected with control Prdm12 mismatch morpholinos (66% unaffected, 19% increased and 15% decreased, n = 53 for Sncg; 66% unaffected, 19% increased and 15% decreased, n = 41 for Tlx3; 60% unaffected, 25% increased and 15% decreased, n = 20 for Islet1). In regions where Prdm12 is not expressed such as the dorsal spinal cord, Sncg expression was, as expected, not affected by Prdm12 morpholino injections (data not shown). These results indicate that PRDM12 has a critical role in the specification of sensory neurons during Xenopus development.
Figure 1. Prdm12 gain and loss of function affects expression of sensory neuronal markers in Xenopus. (A) Real-time qPCR analysis to assess the expression of the indicated genes in stage 28 animal cap explants isolated from Xenopus embryos injected with noggin mRNA, mouse Prdm12 mRNA and treated with retinoic acid (RA) as indicated. Expression levels (fold increase ± SD) were normalized to GAPDH and compared to the expression level of noggin-injected RA treated caps, which was arbitrarily defined as 1. (B) Lateral views of Xenopus tailbud or tadpole stage embryos injected unilaterally with Prdm12 antisense morpholinos (MO) and hybridized with the indicated antisense probes. The injected side is revealed by LacZ staining in blue. Note that all sensory markers tested are upregulated upon Prdm12 overexpression in caps, and in Prdm12 morphant embryos their expression is reduced in trigeminal and epibranchial placodes. (C) Sequence alignment of Homo sapiens (Hos) and Xenopus laevis (Xel) PRDM12 proteins. Orange and blue bars indicate the positions of the SET (PF00856) and ZnF_C2H2 (SM00355) domains, respectively. Conserved residues are boxed. Light blue boxes highlight Cys and His residues which bind the Zn ions. Residues found to be mutated in patients are highlighted with red background and red triangles. NCBI protein accessions NP_067632 and NP_001079854. Alignment was rendered using ESPript (PMID: 24753421 deciphering key features in protein structures with the new ENDscript server).
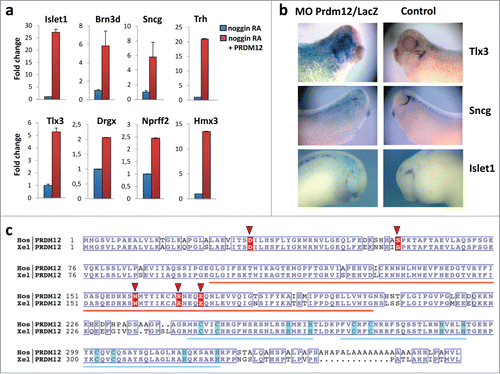
In a manuscript submitted elsewhere PRDM12 coding mutations have been identified in several human HSAN patients.14b HSAN is a group of heterogeneous diseases characterized by the absence of peripheral sensory neuronal function.Citation15 Since we found that Prdm12 controls the specification of sensory neurons in Xenopus and Prdm12 mutations may affect pain perception in humans, we performed sequence comparisons between the human and Xenopus Prdm12 proteins. Intriguingly, the identified human Prdm12 mutations, i.e. D31Y, S58fs, W160C, R168C, and E172D, occurred in evolutionary conserved residues (). Some of the residues mutated in the HSAN patients were even evolutionary conserved up to Trichoplax adhaerens, a simple multicellular Metazoa (Fig. S1). To evaluate the importance of these mutations, we first used a 3D modeling approach (). The mutated residues D31Y and S58fs could not be modeled due to the lack of structural information in the respective region. The mutated residues W160C, R168C, and E172D cluster in the PR methyltansferase domain, for which an X-ray structure is available (PDB: 3EP0). Residues R168 and E172 are both located in a loop fragment (amino acids 166–175) of the PR domain and, in the wild type protein, interact closely by forming a salt bridge between their guanidine and carboxyl groups, respectively. This contact stabilizes the loop structure whose geometry is further constrained by the nested C-terminal part of the protein (, colored in cyan), the most rigid fragment of the domain according to B-factor values. Specifically, the R168C substitution disables the ionic interaction with E172. Similarly, introduction of Asp, having a side chain that is shorter than that of Glu, at position 172 weakens the salt bridge with R168, since conformational rearrangements of the loop are limited by the C-terminus of the PR domain (). The W160C substitution, also located in PRDM12 PR domain, can potentially result in 2 significant structural effects: 1) destabilization of the hydrophobic core of the domain due to the introduction of a less hydrophobic, smaller side chain, and 2) misfolding of the domain due to a potential disulfide bond formation with the neighboring C166 (the only cysteine in the PR domain). The latter can also occur with the R168C mutation. We next examined subcellular localization of the mutant PRDM12 proteins in HEK293T transfected cells (). While wild type PRDM12 protein exhibits exclusive nuclear localization with punctate staining, W160C, R168C, and E172D mutant proteins cluster into large aggregates in the nucleus (, and Fig. S2A). The D31Y mutant lost nuclear staining and localized primarily in the cytoplasm (). Thus, the Prdm12 mutations impact the structure of the protein and its subcellular localization.
Figure 2. Human PRDM12 mutations cause structural instability and impair induction of sensory neuronal markers. (A) The structural modeling of the human PRDM12 mutations is based on the crystal structure of the human PRDM12 methyltransferase domain (PDB:3EP0). Mutated residues and the substitutions are colored in green and gray, respectively, and shown in stick representation. Protein is shown using cartoon and bonds representations. N- and C-terminal parts are colored in violet and cyan, respectively. (B) Western blot analysis using anti-flag antibodies to detect DDK-tagged wild type PRDM12 or the indicated human PRDM12 mutant proteins transiently transfected into HEK cells. Note that we failed to detect the truncation mutant S58fs. (C) Immunofluorescence analysis of HEK cells transiently transfected with DDK-tagged human wild type PRDM12 (WT) or the human R160C, D31Y, and E172D PRDM12 mutants. Arrows indicate protein aggregates of the mutant R160C and E172D protein as well as loss of nuclear staining for the mutant D31Y. The DDK-tag was visualized with anti-flag antibodies (green); nuclei are counterstained with DAPI (blue). Overlays appear in white. Representative images are shown. Magnifications 100×. (D) Real-time RT-PCR analysis of the expression of the indicated genes (±SEM) in stage 26 Xenopus animal cap explants isolated from embryos injected with noggin mRNA, together with wild type or mutated human Prdm12 mRNAs as indicated and treated with retinoic acid (RA). Data are presented as the mean ± SEM of 3 idependent experiments. Expression levels (fold change) were normalized to GAPDH and compared to the expression level of noggin-injected RA treated caps, which was defined as 1. Significance was determined by Student's t-test, where p-values were defined as: * P ≤ 0.05; **P ≤ 0.01; *** P ≤ 0.001.
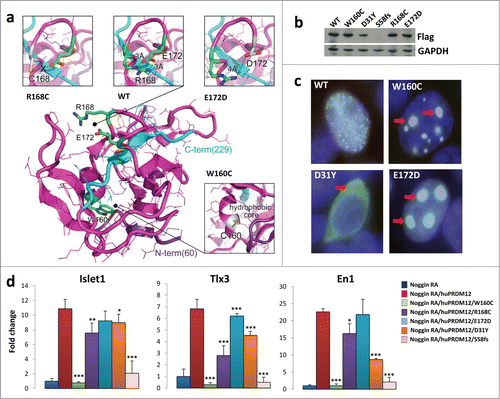
To determine whether these mutations affects PRDM12 function, we expressed the wild type and various mutated forms of human Prdm12 in Xenopus retinoic acid (RA) treated neuralized caps. Ectopic expression of human Prdm12, as its Xenopus Prdm12 ortholog,Citation8 induced the sensory neuronal markers Islet1 and Tlx3 (). The V1 interneuron marker, En1, was as expected,Citation8 also induced by human PRDM12 (). Importantly, all mutants analyzed, with the exception of E172D, resulted in a reduced ability to induce Islet1 and Tlx3 as compared to wild type human PRDM12. Among them, the frame shift mutation S58fs, which results in an early stop codon, and the W160C mutation exhibited the strongest impact on Islet1 and Tlx3 induction (). Of note, all of the human mutations tested, with the exception of E172D, also reduced the ability of PRDM12 to induce En1, the V1 interneuron marker (). This data shows that PRDM12 is highly evolutionarily conserved and that the identified mutations alter the functional properties of the transcriptional regulator PRDM12.
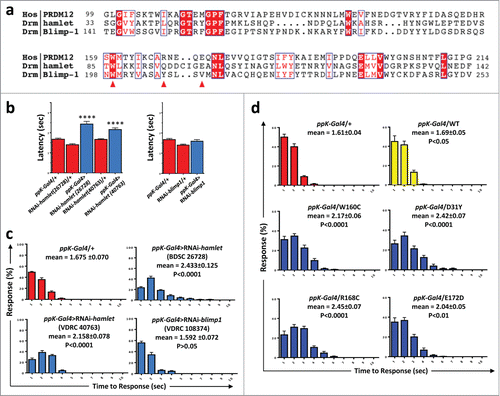
As PRDM12 controls the specification of sensory neurons in Xenopus and PRDM12 mutations may be the cause of impaired pain perception in humans, we asked whether PRDM12s function in pain perception is evolutionarily conserved. Since Xenopus is not an established model for behavioral pain studies, we turned to Drosophila genetics. We and others have previously shown strong functional conservation in nociception between flies and mammals. For example, TRPA1, the TRP channel Painless, or the calcium channel subunit straightjacket/α2δ3 are required for flies to sense and escape from noxious heat insult.Citation16-19 Most importantly, Drosophila genetics allows us to specifically knock-down genes in nociceptive sensory neurons and thus avoid any confounding developmental effects on other tissues such as the central nervous system.Citation20,21 In Drosophila, 4 PRDM family members containing a PRDM/SET domain have been found, namely, CG43347, CG13296, Hamlet and Blimp1. Among these 4, we used RNA interference to knockdown expression of Hamlet and Blimp1 () specifically in nociceptive ppK- positive type IV multidendritic sensory neurons using the ppK-Gal4 driver. Nociceptive behavior was assayed as the latency response to a local heat stimulus applied to abdominal segments A4-A6 of fly larvae. We detected increased latency responses, i.e., reduced nociception, using 2 different Hamlet UAS-RNAi lines (BDSC 26728 and VDRC 40763) as compared to controls (). Knock-down of Blimp1 did not result in any apparent nociceptive phenotype as compared to the control lines (). Similar to our data observed in hamlet RNAi knockdown lines, we also detected increased latency in the nocifensive responses of hypomorph hamlet mutant flies (Fig. S2B). Finally, we expressed the human PRDM12 mutants in nociceptive neurons using the ppK-Gal4 driver. Overexpression of all the human PRDM12 mutants tested resulted in impaired nocifensive behavior in Drosophila as compared to overexpression of the wild type, which was not different from control (). These data show that hamlet is a functionally relevant PRDM12 fly homolog that controls nociceptive behavior and that expression of human PRDM12 mutants in nociceptor neurons impairs pain perception in Drosophila.
Figure 3. Knockdown of Hamlet causes hypoalgesia in Drosophila. (A) Protein sequence alignment of human PRDM12 and the Drosophila PRDM-family members Hamlet and Blimp1 SET domains. The human W160, R168 and E172 residues mutated in patients are denoted by red arrows. Conserved (red background) and partially conserved (red letters) residues are highlighted in blue boxes. Sequences with the following accessions were used here: PRDM12 NP_067632=Q9H4Q4; Hamlet NP_724130; Blimp1 NP_647982. (B) Sensory-neuron specific knockdown of hamlet using a ppk-Gal4 driver (class IV md-da neurons) results in increased latency response times (±SEM) to a thermal stimulus of 46°C. Results were confirmed in 2 different RNAi lines (26728 and 40763). Knockdown of the closely related blimp1 gene did not affect the thermal nocifensive response (right panel). The ppK-Gal4/+ driver line is shown as a control. ****, P < 0.0001 (Kruskal-Wallis non-parametric test for median comparisons with Dunn's post-hoc multiple comparisons test). (C) Average percentage of larvae (±SEM) responding at each time point (1–10 seconds) to a 46°C noxious thermal stimulus; data corresponding to panel b. Mean response times (±SEM) and p-values are indicated for each line. (D) Average percentages of larvae (±SEM) responding at each time point (1–10 seconds) to a 46°C noxious thermal stimulus for ppK-Gal4 control flies and fly lines that were engineered to express the indicated human mutations in nociceptive neurons. Mean response times (±SEM) and p-values are indicated for each line. n ≥ 60 for all thermal nociception assays.
To identify genes downstream of PRDM12, we next performed RNA sequencing of transcriptomes from human fibroblasts derived from patients carrying the PRDM12 D31Y and E172D mutations (kindly provided by Drs. Auer-Grumbach and Senderek). Total protein levels of PRDM12 in patient fibroblasts as compared to their respective first-order and sex-matched relatives were not affected by either mutation in PRDM12 (). When we performed differential gene expression analyses using RNAseq, we identified only 19 genes whose expression was significantly affected by both PRDM12 mutations as compared to wild type fibroblasts derived from their respective parents (). Some of these hits were confirmed by RT-qPCR as well as Western blotting (, and not shown). To screen for the functional relevance of these PRDM targets in nocifensive responses, we again used nociceptor specific RNAi-mediated knockdown in fruit fly larvae. Using the automated homolog prediction databases Ensambl Compara and OrthoDB, we could assign Drosophila homologues for tetratricopeptide repeat domain 12 (TTC2), disintegrin and a metallopeptidase with thrombospondin type 1 motif 9 (ADAMTS9), ubiquitin specific peptidase 53 (USP53), mesenchyme homeo box 2 (MEOX2), WNT1 inducible signaling pathway protein (WISP1), nuclear receptor subfamily 4 group A member 1 (NR4A1; also known as nerve growth factor IB, NGFIB, or Nur77), and thyrotropin-releasing hormone degrading enzyme (TRHDE). Knocking down the Drosophila TTC12, USP53, MEOX2, and NR4A1 homologues using the nociceptor specific driver line ppK-Gal4, we detected a significant decrease in the nocifensive behavior as compared to controls (Fig. S2C). Of note, Usp53 mRNA has been found to be significantly upregulated in inflammatory painCitation17 and mutations in MEOX2 have been linked to Alzheimer disease.Citation22 TTC12 is a candidate gene for mechanical nociception (geneweaver.org) and has been associated with nicotine and alcohol dependence.Citation23-24 NR4A1 has been linked to pain responses to cold stimulation.Citation25 Moreover, NR4A1 is rapidly induced by nerve growth factor (NGF), previously shown to be required for the differentiation and survival of primary sensory fibers.Citation26 Finally, we previously identified S1PR1 as a regulator of nociception in Drosophila,Citation2 later confirmed as a mediator of nociception in mice.Citation27,28 Of note, S1PR1 was also found in the list of Prdm12 upregulated genes we identified by RNA-Seq analysis of Prdm12 overexpressing RA treated neuralized caps and the S1PR1 gene was bound by Prdm12 as detected by CHIP-Seq.Citation8
Figure 4. Identification of TRHDE as a novel pain gene. (A) Western blot analysis of fibroblasts from patients (pat.) carrying D31Y or E172D PRDM12 mutations and their sex matched parents (par.), respectively, reveal no change in expression of endogenous PRDM12 protein levels. GAPDH is shown as a loading control. (B) Heat map of the 19 genes significantly deregulated by DESeq2 in fibroblasts from both patients as compared to their respective first order relatives. (C) Western blot analysis shows a decrease in TRHDE and S1PR1 protein levels in patient (pat.) fibroblasts as compared to the respective first order relatives (par.). Tubulin is shown as a control. (D) Average percentage of larvae (±SEM) responding at each time point (1–10 seconds) to a 46°C noxious thermal stimulus testing control (ppK-Gal4/+) flies and ppK-Gal4 RNAi fly lines to knock-down gene expression of the fly TRHDE homolog (CG11951) in nociceptive neurons. Moreover, a line (BDSC18316) carrying a mutation in the fly TRHDE homolog showed increased response times to the thermal stimulus. Mean response times (±SEM) and p-values (Kruskal-Wallis non-parametric test for median comparisons with Dunn's post-hoc multiple comparisons test) are indicated for each line. (E) Confocal images of dorsal cluster (ddac) class IV neurons at abdominal segment A5 labeled by UAS-mCD8-GFP (green) driven by ppk-Gal4 (ppK-Gal4;mCD8-GFP) at the third instar larval stage. RNAi knockdown of the Drosophila TRHDE homolog (ppk-Gal4>RNAi-CG11951) exhibits a significant decrease in dendritic branching and dendritic field size as compared to control neurons. 8-bit tracing of ddaC cell body and dendrites are shown on the left of their respective images. Scale bar represents 100 μm. (F) Quantification of dendritic branch order by the centrifugal counting method reveals significantly reduced branching, especially in higher order branches. (G) The total number of dendritic branch terminals in ppk-Gal4>RNAi-CG11951 is significantly decreased compared to the control ppK-Gal4;mCD8-GFP line (same color-coded labeling as in panel f is used). Data in f and g are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (Two-way ANOVA with Bonferroni's multiple comparisons test and unpaired 2-tailed t-tests with Welch's correction).
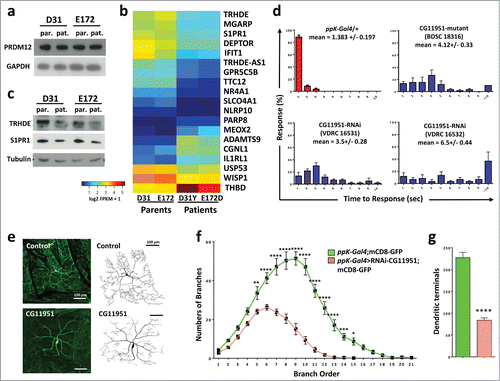
We finally assessed the role of the Drosophila CG11951, homolog of tyrotropin-releasing hormone degrading enzyme (TRHDE) in nocifensive behavior. We focused on TRHDE, because it has been reported that TRH, the substrate for TRHDE, can affect pain in human and rodents.Citation29-31 Moreover, we found that TRH is strongly upregulated in our Prdm12 overexpressing RA treated neuralized caps,Citation8 a result validated by RT-qPCR (). PpK-Gal4- driven knock-down of CG11951, a fly homolog of TRHDE, indeed resulted in a significantly impaired nocifensive response to the noxious heat stimulus as compared to their respective controls (). Importantly, 2 independent RNAi lines to knock-down TRHDE exhibited impaired nociception in Drosophila (). Moreover, a TRHDE mutant Drosophila line (BDSC 18316) also exhibited markedly enhanced latency to respond to noxious heat (). To evaluate the developmental impact of TRHDE perturbations on nociceptor neurons, we finally expressed the fluorescent membrane label UAS-CD8 GFP in PPK1-positive nociceptor neurons, and analyzed the impact of TRDHE knockdown on neuronal morphology. Larvae with TRDHE knockdown showed a marked reduction in branching and dendritic field coverage (). Moreover, RNAi-mediated knockdown of the fly TRDHE homolog markedly reduced dendritic nerve terminals in type IV nociceptive neurons (). These data show that the Drosophila TRHDE homolog controls dendritic morphology of nociceptive neurons, ultimately resulting in altered pain sensitivity.
Discussion
Patients with HSAN present early in life, suggesting a likely developmental component to this condition.Citation15 There are at least 11 known mutations that cause HSAN, however, none of the identified HSAN genes are transcriptional regulators.Citation15 Moreover, little is known of the transcriptional regulation required for the establishment of peripheral sensory neurons. In the manuscript submitted elsewhere, mutations in PRDM12 have been linked to impaired pain perception in human patients.14b Our results extend these data by showing that PRDM12 controls the induction of sensory neuronal markers in Xenopus. These data are in agreement with the recent observation that Prdm12 is important for preplacodal developmentCitation32 and neuronal specification in the spinal cord.Citation7,8 Furthermore, we show that the fly PRDM12 homolog Hamlet controls nociceptive behavior in Drosophila larvae. In support of our results that Hamlet is the relevant evolutionary conserved PRDM-family member involved in nociception, it has been previously shown that Hamlet is involved in the specification of peripheral sensory neurons in the fly embryo and regulates the differentiation of neuroblasts in the central nervous system.Citation33,34 Whether PRDM12 also controls the proliferation of neuronal progenitors and specifies defined neuronal lineages in trigeminal and dorsal root ganglia in mammals needs to be tested in future experiments.
The conservation of PRDM12/Hamlet in nociception and sensory neuronal specification allowed us to model the relevance of the identified human PRDM12 mutations, also because all human mutant residues are conserved between Xenopus and human PRDM12 and 2/3 of the mutant residues in the conserved domains are identical between human PRDM12 and fly Hamlet. We indeed found that the PRDM12 mutations impact the structure stability of the protein and its subcellular localization. Functionally the human mutations affect induction of sensory neuronal markers in Xenopus and change nocifensive behavior in Drosophila. As PRDM12 is expressed in multiple human tissues, might act as a tumor suppressor gene in human chronic myeloid leukemiaCitation35 and in fly was identified as a transcriptional regulator essential for the proliferation of neuroblasts in the central nervous systemCitation34 the effect of the human PRDM12 mutation in pain perception could be secondary to consequences in other organ systems. Importantly, our experiments in Xenopus and Drosophila allowed us to directly assess the effect of PRDM12/Hamlet in sensory pain neurons in vivo, avoiding potential confounding effects of the PRDM12 mutations on other tissues. All data taken together clearly support the notion that PRDM12 and its functional fly homolog Hamlet are evolutionary conserved master regulators of sensory neuronal specification and play a critical role in pain perception.
Using RNA-seq in patient fibroblasts harboring 2 distinct PRDM12 mutations, we found a small number of genes deregulated. Some of the targets have been previously linked to pain or neuro-development. Moreover, we found an overlap of these targets in RNA-seq and Prdm12 ChIP-seq assays in our Xenopus experiments, e.g. S1PR1, CGNL1, IL1R1, or TRH, the target for TRHDE.Citation8 Fly genetics allowed us to test the role these candidates in nociception in vivo using sensory neuron specific knock-down of the respective fly homologues. We therefore functionally linked multiple PRDM12 target genes to impaired nocifensive behavior in Drosophila. One of the genes functionally validated was the Drosophila TRHDE homolog. Cell fate mapping showed that the Drosophila TRHDE homolog controls dendritic morphology of nociceptive neurons, ultimately resulting in altered pain sensitivity. Whether other identified PRDM12 targets also control the development and morphology of sensory neurons remains to be tested. Importantly, our data in multiple species extend beyond the description of human PRDM12 mutations to uncover key pathways that regulate evolutionary conserved pain perception.
Methods
Functional assays in Xenopus
Prdm12 wild type (NM021619) and mutants (480G>C to generate Trp160Cys, 91G>T to generate Asp31Tyr, 502C>T to generate Arg168Cys and 516G>C to generate Glu172Asp mutations) were made (GenScript) and cloned into pCMV6 Empty vector with a DDK-tag at C-terminus (OriGene). The Prdm12 expression vectors used in frog microinjection experiments were obtained by PCR from these pCMV6 constructs. The PCR products were cloned between the EcoRI and XhoI sites of the pCS2-Flag vector. Xenopus embryos were obtained from adult frogs by hormone induced egg-laying, in vitro fertilized using standard methodsCitation36 and staged.Citation37 Synthetic mRNAs were prepared using SP6 mMESSAGE mMACHINE (Ambion). Expression vectors were linearized with NotI and transcribed with SP6 RNA polymerase. The Prdm12 antisense morpholino oligonucleotide (MO) and the control Prdm12 antisense mismatch have been described.Citation8 For in situ analyses, embryos were injected in one-cell of 2 to 4-cell stage embryos (200pg mRNA or 20 ng of morpholino/blastomere), and fixed at tailbud or early tadpole stages. In all experiments, embryos were coinjected with LacZ mRNA (50 pg/blastomere) to mark the manipulated side. For animal cap assays, synthesized mRNA was microinjected into the animal region of each blastomere of 4-cell stage embryos (200 pg mRNA/blastomere). Animal caps were dissected at the blastula stage (stage 9) and cultured in 1× Steinberg medium, 0.1% BSA with or without RA (µ10 M) until sibling embryos reached the desired stage. All microinjection experiments have been repeated independently at least 2 times.
For RT-qPCR analysis of Xenopus animal caps, total RNA was extracted using the RNAspin Mini RNA isolation kit (GE Healthcare), cDNAs were synthesized with iScript cDNA synthesis kit (Biorad), RT-PCR reactions were carried out following the Gene Amp PCR kit protocol (Perkin Elmer) and real time RT-PCR was performed using the Step One Plus Real Time PCR system (Applied Biosystems) with Q-PCR core kits for SYBR Green I (Eurogentec). Samples were normalized with Xenopus GAPDH. Primers used are available upon request. All PCR-based assays were carried out in duplicates or triplicates. Error bars represent SEM. For in situ analysis, Xenopus embryos were fixed in 4% paraformaldyde for 1–2 hours. Whole mount in situ hybridizations were performed as describedCitation36 using digoxigenin- or fluorescein-labeled antisense probes revealed with NBT/BCIP. The Sncg, Tlx3 and Islet1 probes were generated as described.Citation13,38,50
Structural modeling of PRDM12 mutant proteins and alignments
For structural modeling, mutations were mapped onto the 3D X-ray structure of the PRDM12 SET domain (PDB: 3EP0, amino-acid residues 60–229). The mutations were modeled into the structure by PyMol (http://www.pymol.org/)Citation39 which was also used for visualization. Salt bridge analysis was carried out using VMD.Citation40 Sequence alignments alignment was performed using Mafft (PMID: 23329690, MAFFT multiple sequence alignment software version 7: improvements in performance and usability) and rendered with ESPript (PMID: 24753421 Deciphering key features in protein structures with the new ENDscript server). Homology searches were performed using Ensembl Compara (http://www.ensembl.org/info/genome/compara/index.html).
Subcellular localization of human PRDM12 mutants in HEK293 cells
For immunofluorescence, wild type and mutated versions of human PRDM12 in pCMV6 vector with a DDK-tag at C-terminus (OriGene) were used. HEK293 cells were plated on coverslips and transiently transfected the next day using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. 24 hours after transfection, cells were washed with phosphate buffered saline (PBS) and fixed for 10 min with 4% Roti-Histofix (Roth). Following a wash with PBS, cells were blocked with 1% bovine serum albumin (BSA, Sigma) and 0.01% Triton-X 100 in PBS, washed again with PBS and incubated for 3 hours at room temperature with anti-Flag antibody (Sigma) at 1:500 dilution in blocking buffer. Cells were then washed with PBS and incubated for 1 hour at room temperature with Alexa Fluor 568 goat anti-mouse antibodies (Molecular Probes) at 1:400 dilution in PBS. Following a wash with PBS, coverslips were mounted onto the slides using Vectashield Mounting Medium with DAPI (Vector Laboratories) to visualize the nuclei. Images were taken on Axio Imager Z2 Stativ and taken at 100× magnification.
Drosophila stocks, thermal nociception assay, immunohistochemistry and confocal analysis
UAS-dicer-2,Citation41,42 pickpocket1.9-GAL4,Citation43 UAS-mCD8-GFP, FRT40A, and UAS-RNAi-hamlet (26728) were obtained from the Bloomington Drosophila Stock Center. Hamlet mutants and UAS-hamlet were gifts from H.Bellen.Citation44,45 All other RNAi lines to knockdown gene expression in sensory neurons were obtained from the Vienna Drosophila RNAi Center. For neuronal morphology analysis, driver lines carrying a fluorescent reporter (UAS-DCR2;ppk-GAL4;UAS-mCD8-GFP) were crossed to include a single copy of the UAS-RNAi-hamlet or UAS-hamlet construct. Flies were reared on cornmeal-molasses-yeast agar at 25°C, 70% humidity over a 12 hr light/dark cycle.
Larval nociceptive behavior was analyzed using previously described methods.Citation46,47,20 Briefly, third instar larvae were transferred to a 100 mm petri dish containing a thin film of distilled water and allowed a 10 minute incubation period. After this time, a heat probe (46°C) was applied at abdominal segments A4-A6. The response time was recorded as the time elapsed between application of the heat probe and the elicitation of the characteristic nocifensive response, a 360° rolling motion about the lateral axis to escape the noxious heat. For immunohistochemistry, third instar larvae were dissected in HL-3 saline solution on a Sylgard dissecting plate. Specimens were then fixed in 4% paraformaldehyde for 20 minutes at room temperature. Specimens underwent 4 15 minute washes with PBT (PBS with 0.4% triton X-100) and were incubated in 500 μl of 0.25% BSA (bovine serum albumin diluted in PBT) for 45 minutes under gentle shaking at room temperature. Primary antibodies were diluted in 500 μl of 0.25% BSA at a dilution of 1/1000 for rabbit anti-GFP (Invitrogen) and 1/200 for mouse Nc82 (Developmental Studies Hybridoma Bank, Iowa) and the larvae were then incubated with the antibodies overnight at 4°C. After four 10 minute washes in PBT, the conjugated secondary antibodies (goat anti-rabbit Alexa488 and goat anti-mouse Alexa555 (Invitrogen) were diluted to 1/500 in 500 μl of 0.25% BSA and added to the larvae for 1.5 hours at room temperature under gentle shaking. Prior to mounting for microscopy, the larvae were washed in PBT 6 times, each for 10 minutes.
For confocal microscopy, Drosophila specimens were mounted in water and visualized using a Leica DMI-6000 inverted microscope and an SP8 Scanning Laser Confocal system using a 20× (dry, 0.7 numerical aperture) objective (Leica, Wetzlar, Germany). Excitation was achieved using 488 nm and 552 nm lasers with respective intensities of 3.55% and 4.33% and gain of 550V. The class IV ddaC neuron found at abdominal segment A5 from the left and right aspects of each larva was visualized and captured. Z-stack images were collected at 1024 × 1024 resolution in 0.8 μm steps, speed 400 Hz with a zoom factor of 0.8. Z-stacks were condensed into a maximum projection using the Leica Application Suite 4.2 program and exported as .jpg files. Images of class IV sensory neurons were processed to 8-bit black and white images using ImageJ.Citation48 These neurons were analyzed for total number of branch terminals and dendritic branch order quantification, using the centrifugal counting method as described previously.Citation49
Western blot analysis
Small biopsy of the gingiva during a reconstructive surgical procedure obtained from a female patient carrying the D31Y PRDM12 mutation, and skin biopsy from her mother as a control (from Dr. Auer-Grumbach, AKH, Vienna), were diced with a surgical blade and incubated for several weeks in Cnt.BM.2 media with appropriate supplements (CELLnTEC) and 1 unit/ml of Pen/Strep (Invitrogen) in a humidified 37°C, 5% CO2 chamber. Once the fibroblasts from the biopsy created a confluent monolayer the cells were then transferred to a 10 cm tissue culture dish or frozen at −80°C for further analysis. Fibroblasts from a male patient carrying the E172D PRDM12 mutation and those of his father as a sex matched control were a kind gift from Dr. Senderek (Ludwig-Maximilians-University Munich). Confluent cells on a 10 cm culture dish were harvested and lysed using RIPA buffer (Sigma). Protein concentrations were determined using BioRad Protein Assay (BioRad), and 20 μg of each lysate were ran on a precast 4–20% Bis Tris Gel (Novex, Life Technologies), transferred to a nitrocellulose membrane and subjected to Western blot analysis using standard protocols. Briefly, anti-PRDM12 rabbit polyclonal antibodies (Sigma) at 1:1000 dilution were incubated with membranes overnight at 4°C, washed 4 times with 0.1% Tween-20 in PBS, and finally incubated with anti-rabbit horseradish peroxidase (HRP)-linked whole antibodies (donkey; GE Healthcare) for 1 hour at room temperature. Chemiluminescence was generated using the ECL kit (GE Healthcare) according to manufacturer's protocol, and membranes were exposed to film (Amersham Hyperfilm ECL, GE Healthcare) for several exposure times. For transiently transfected HEK cells, the procedure was identical except for the primary antibody step, where we used anti-flag (Sigma) antibodies at 1:500. For detection of TRHDE and S1PR1 proteins in fibroblasts, anti-TRHDE (N-18, Santa Cruz Biotechnology) and anti-S1PR1 antibodies (Millipore) were used at 1:200 dilution followed by appropriate secondary antibodies. To confirm equal loading, membranes were blotted with anti-GAPDH HRP linked antibodies (Cell Signaling) for 1 hour at room temperature.
Gene expression profiling
Total RNA was prepared from confluent fibroblasts grown in 10 cm tissue culture using RNeasy Mini Kit (Qiagen) according to manufacturer's instructions. Quality of RNA samples were confirmed by Agilent Bioanalyzer 2100. Libraries of PolyA-mRNA of parental control and patient fibroblasts were sequenced by 50-bp single-end Illumina mRNA sequencing (http://www.csf.sc.st/facilities/ngs/). Reads were aligned using Tophat v2.0.10 and bowtie v0.12.9, FPKM estimation was performed with Cufflinks v2.1.1, aligned reads were counted with HTSeq, and differential expression analysis was performed with DESeq2. Differentially expressed genes were selected using a cut-off at a P value of less than 0.05 (FDR adjusted for multiple testing). The data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE61600 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE61600).
Statistical analyses
For analysis of larval pain behavior, we have performed the Kruskal-wallis non-parametric test for median comparison followed by the Dunn's post-hoc test. For analysis of dendritic branching and dendritic branch terminals we used a 2-way ANOVA test and an unpaired Student's t-test, respectively. Unless otherwise stated, data are represented as mean values ± SEM.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
V.N. designed the experiments in human fibroblasts, cloned the mutant proteins, expressed proteins in HEK cells and coordinated the project. DC helped to clone the mutant proteins. M.N. performed the sequence alignments. D.W. and A.M. helped to establish human fibroblast lines. A.A.P. performed structural 3D modeling. I.K. assisted with Western blotting. T.C., T.M.K., and C.L., under the supervision and coordination of G.G.N. performed all fly experiments. C.V.C., S.V., and E.B. designed and performed all experiments in Xenopus. J.M.P. coordinated the project, and together with V.N. designed experiments and wrote the manuscript.
1036209_Supplemental_Material.pdf
Download PDF (1.3 MB)Acknowledgements
We thank all members of our laboratories for helpful discussions and technical support. We thank all members of the IMP-IMBA Biooptics service facility for assistance in imaging and Dr. Bojan Zagrovic for discussion of the 3D models. We thank Drs. Senderek and Auer-Grumbach for providing access to PRDM12 mutations in patients and sharing human fibroblasts.
Funding
This work was supported by grants from the Belgian Fonds de la Recherche Scientifique (FRS-FNRS) (FRFC), the Walloon region (First International Epigene) and the Van Buuren Foundation to E.B. J.M.P. is supported by grants from IMBA, the Austrian National Foundation, the Austrian Academy of Sciences, and an EU ERC Advanced Grant.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- Kang K., Pulver SR, Panzano VC, Chang EC, Griffith LC, Theobald DL, Garrity PA. Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature 2010; 464:597-600; PMID:20237474; http://dx.doi.org/10.1038/nature08848
- Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, Griffin RS, Belfer I, Dai F, Smith SB, et al. A genome-wide Drosophila screen for heat nociception identifies alpha2delta3 as an evolutionarily conserved pain gene. Cell 2010; 143:628-38; PMID:21074052; http://dx.doi.org/10.1016/j.cell.2010.09.047
- Khuong TM, Neely GG. Conserved systems and functional genomic assessment of nociception. FEBS J 2013; 280:5298-306; PMID:23910505; http://dx.doi.org/10.1111/febs.12464
- Lumpkin EA, Caterina MJ. Mechanisms of sensory transduction in the skin. Nature 2007; 445:858-65; PMID:17314972; http://dx.doi.org/10.1038/nature05662
- Tracey I, Mantyh PW. The cerebral signature for pain perception and its modulation. Neuron 2007; 55:377-91; PMID:17678852; http://dx.doi.org/10.1016/j.neuron.2007.07.012
- Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell 2009; 139:267-84; PMID:19837031; http://dx.doi.org/10.1016/j.cell.2009.09.028
- Zannino DA, Downes GB, Sagerstrom CG. prdm12b specifies the p1 progenitor domain and reveals a role for V1 interneurons in swim movements. Dev Biol 2014; 390:247-60; PMID:24631215; http://dx.doi.org/10.1016/j.ydbio.2014.02.025
- Thélie A, Desiderio S, Hanotel J, Kricha S, Quigley I, Borromeo MD, Lahaye F, Croce J, Cerda-Moya G, Bolle B, et al. Prdm12 ispecifies V1 interneurons downstream of Pax6 and through cross-repressive interactions with Dbx1 and Nkx6.1. Submitted.
- McEvilly RJ, Erkman L, Luo L, Sawchenko PE, Ryan AF, Rosenfeld MG. Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature 1996; 384:574-7; PMID:8955272; http://dx.doi.org/10.1038/384574a0
- Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci 2008; 11:1283-93; PMID:18849985; http://dx.doi.org/10.1038/nn.2209
- Chen ZF, Rebelo S, White F, Malmberg AB, Baba H, Lima D, Woolf CJ, Basbaum AI, Anderson DJ. The paired homeodomain protein DRG11 is required for the projection of cutaneous sensory afferent fibers to the dorsal spinal cord. Neuron 2001; 31:59-73; PMID:11498051; http://dx.doi.org/10.1016/S0896-6273(01)00341-5
- Feng Y, Xu Q. Pivotal role of hmx2 and hmx3 in zebrafish inner ear and lateral line development. Dev Biol 2010; 339:507-18; PMID:20043901; http://dx.doi.org/10.1016/j.ydbio.2009.12.028
- Wang C, Liu Y, Chan WY, Chan SO, Grunz H, Zhao H. Characterization of three synuclein genes in Xenopus laevis. Dev Dyn 2011; 240:2028-33; PMID:21761485; http://dx.doi.org/10.1002/dvdy.22693
- a. Lopes C, Liu Z, Xu Y, Ma Q. Tlx3 and Runx1 act in combination to coordinate the development of a cohort of nociceptors, thermoceptors, and pruriceptors. J Neurosci 2012; 32:9706-15; PMID:22787056; http://dx.doi.org/10.1523/JNEUROSCI.1109-12.2012; b. Chen YC, Auer-Grumbach M, Matsukawa S, Zitzelsberger M, Themistocleous AC, Strom TM, Samara C, Moore AW, Cho LT, Young GT, et al. Transcriptional regulator PRDM12 is essential for human pain perception. Nat Genet 2015; http://dx.doi.org/10.1038/ng.3308
- Auer-Grumbach M, Mauko B, Auer-Grumbach P, Pieber, TR. Molecular genetics of hereditary sensory neuropathies. Neuromolecular Med 2006; 8:147-58; PMID:16775373; http://dx.doi.org/10.1385/NMM:8:1-2:147
- Neely GG, Keene AC, Duchek P, Chang EC, Wang QP, Aksoy YA, Rosenzweig M, Costigan M, Woolf CJ, Garrity PA, et al. TrpA1 regulates thermal nociception in Drosophila. PLoS One 2011; 6:e24343; PMID:21909389; http://dx.doi.org/10.1371/journal.pone.0024343
- Simonetti M, Hagenston AM, Vardeh D, Freitag HE, Mauceri D, Lu J, Satagopam VP, Schneider R, Costigan M, Bading H, et al. Nuclear calcium signaling in spinal neurons drives a genomic program required for persistent inflammatory pain. Neuron 2013; 77:43-57; PMID:23312515; http://dx.doi.org/10.1016/j.neuron.2012.10.037
- Xu SY, Cang CL, Liu XF, Peng YQ, Ye YZ, Zhao ZQ, Guo AK. Thermal nociception in adult Drosophila: behavioral characterization and the role of the painless gene. Genes Brain Behav 2006; 5:602-13; PMID:17081265; http://dx.doi.org/10.1111/j.1601-183X.2006.00213.x
- Zhong L, Hwang RY, Tracey WD. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol 2010; 20:429-34; PMID:20171104; http://dx.doi.org/10.1016/j.cub.2009.12.057
- Tracey WD Jr, Wilson RI, Laurent G, Benzer S. painless, a Drosophila gene essential for nociception. Cell 2003; 113:261-73; PMID:12705873; http://dx.doi.org/10.1016/S0092-8674(03)00272-1
- Manev H, Dimitrijevic N. Drosophila model for in vivo pharmacological analgesia research. Eur J Pharmacol 2004; 491:207-8; PMID:15140638; http://dx.doi.org/10.1016/j.ejphar.2004.03.030
- Wu Z., Guo H, Chow N, Sallstrom J, Bell RD, Deane R, Brooks AI, Kanagala S, Rubio A, Sagare A, et al. Role of the MEOX2 homeobox gene in neurovascular dysfunction in Alzheimer disease. Nat Med 2005; 11:959-65; PMID:16116430
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, Kranzler HR, Farrer L. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet 2006; 15:3498-507; PMID:17085484; http://dx.doi.org/10.1093/hmg/ddl426
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Association of haplotypic variants in DRD2, ANKK1, TTC12 and NCAM1 to alcohol dependence in independent case control and family samples. Hum Mol Genet 2007; 16:2844-53; PMID:17761687; http://dx.doi.org/10.1093/hmg/ddm240
- Kim SJ, Qu Z, Milbrandt J, Zhuo M. A transcription factor for cold sensation! Mol Pain 2005; 1:11; PMID:15813958; http://dx.doi.org/10.1186/1744-8069-1-11
- Ernsberger U. Role of neurotrophin signalling in the differentiation of neurons from dorsal root ganglia and sympathetic ganglia. Cell Tissue Res 2009; 336:349-84; PMID:19387688; http://dx.doi.org/10.1007/s00441-009-0784-z
- Mair N, Benetti C, Andratsch M, Leitner MG, Constantin CE, Camprubí-Robles M, Quarta S, Biasio W, Kuner R, Gibbins IL, et al. Genetic evidence for involvement of neuronally expressed S1P(1) receptor in nociceptor sensitization and inflammatory pain. PLoS One 2011; 6:e17268; PMID:21359147; http://dx.doi.org/10.1371/journal.pone.0017268
- Xie W, Strong JA, Kays J, Nicol GD, Zhang JM. Knockdown of the sphingosine-1-phosphate receptor S1PR1 reduces pain behaviors induced by local inflammation of the rat sensory ganglion. Neurosci Lett 2012; 515:61-5; PMID:22445889; http://dx.doi.org/10.1016/j.neulet.2012.03.019
- Boschi G, Desiles M, Reny V, Rips R, Wrigglesworth S. Antinociceptive properties of thyrotropin releasing hormone in mice: comparison with morphine. Br J Pharmacol 1983; 79:85-92; PMID:6409194; http://dx.doi.org/10.1111/j.1476-5381.1983.tb10499.x
- Eto K, Kim SK, Nabekura J, Ishibashi H. Taltirelin, a thyrotropin-releasing hormone analog, alleviates mechanical allodynia through activation of descending monoaminergic neurons in persistent inflammatory pain. Brain Res 2011; 1414:50-7; PMID:21872219
- Tanabe M, Tokuda Y, Takasu K, Ono K, Honda M, Ono H. The synthetic TRH analogue taltirelin exerts modality-specific antinociceptive effects via distinct descending monoaminergic systems. Br J Pharmacol 2007; 150:403-14; PMID:17220907; http://dx.doi.org/10.1038/sj.bjp.0707125
- Matsukawa S, Miwata K, Asashima M, Michiue T. The requirement of histone modification by PRDM12 and Kdm4a for the development of pre-placodal ectoderm and neural crest in Xenopus. Dev Biol 2015; 399:164-76; PMID:25576027
- Moore AW, Jan LY, Jan YN. hamlet, a binary genetic switch between single- and multiple- dendrite neuron morphology. Science 2002; 297:1355-8; PMID:12193790; http://dx.doi.org/10.1126/science.1072387
- Eroglu E, Burkard TR, Jiang Y, Saini N, Homem CC, Reichert H, Knoblich JA. SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell 2014; 156:1259-73; PMID:24630726; http://dx.doi.org/10.1016/j.cell.2014.01.053
- Reid AG, Nacheva EP. A potential role for PRDM12 in the pathogenesis of chronic myeloid leukaemia with derivative chromosome 9 deletion. Leukemia 2004; 18:178-80; PMID:14523459; http://dx.doi.org/10.1038/sj.leu.2403162
- Sive HL, et al. Early Development of Xenopus Laevis: A Laboratory Manual. Cold Spring Harbor: CSHL Press. 2000.
- Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis (Daudin), North Holland, Publishing Co, Amsterdam, The Netherlands. 1967.
- Patterson KD, Krieg PA. Hox11-family genes XHox11 and XHox11L2 in xenopus: XHox11L2 expression is restricted to a subset of the primary sensory neurons. Dev Dyn 1999; 214:34-43; PMID:9915574; http://dx.doi.org/10.1002/(SICI)1097-0177(199901)214:1%3c34::AID-DVDY4%3e3.0.CO;2-R
- The PyMOL Molecular Graphics System, Version 1.3r1. Schrödinger, LLC 2010. http://www.pymol.org/citing.
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph 1996; 14:33-8; 27–8; http://dx.doi.org/10.1016/0263-7855(96)00018-5
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 2007; 448:151-6; PMID:17625558; http://dx.doi.org/10.1038/nature05954
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004; 117:69-81; PMID:15066283; http://dx.doi.org/10.1016/S0092-8674(04)00261-2
- Ainsley JA, Pettus JM, Bosenko D, Gerstein CE, Zinkevich N, Anderson MG, Adams CM, Welsh MJ, Johnson WA. Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr Biol 2003; 13:1557-63; PMID:12956960; http://dx.doi.org/10.1016/S0960-9822(03)00596-7
- Andrews HK, Giagtzoglou N, Yamamoto S, Schulze KL, Bellen HJ. Sequoia regulates cell fate decisions in the external sensory organs of adult Drosophila. EMBO Rep 2009; 10:636-41; PMID:19444309; http://dx.doi.org/10.1038/embor.2009.66
- Moore AW, Roegiers F, Jan LY, Jan YN. Conversion of neurons and glia to external-cell fates in the external sensory organs of Drosophila hamlet mutants by a cousin-cousin cell-type respecification. Genes Dev 2004; 18:623-8; PMID:15075290; http://dx.doi.org/10.1101/gad.1170904
- Aldrich BT, Kasuya J, Faron M, Ishimoto H, Kitamoto T. The amnesiac gene is involved in the regulation of thermal nociception in Drosophila melanogaster. J Neurogenet 2010; 24:33-41; PMID:19995327; http://dx.doi.org/10.3109/01677060903419751
- Chattopadhyay A, Gilstrap AV, Galko MJ. Local and global methods of assessing thermal nociception in Drosophila larvae. J Vis Exp 2012;, e3837; PMID:22643884
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9:671-5; PMID:22930834; http://dx.doi.org/10.1038/nmeth.2089
- Cupp CJ, Uemura E. Age-related changes in prefrontal cortex of Macaca mulatta: quantitative analysis of dendritic branching patterns. Exp Neurol 1980; 69:143-63; PMID:6771151; http://dx.doi.org/10.1016/0014-4886(80)90150-8
- Brade T, Gessert S, Kuhl, M, Pandur, P. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev Biol 2007; 311:297-310; PMID:17900553; http://dx.doi.org/10.1016/j.ydbio.2007.08.004
