Abstract
c-Myc is the most frequently overexpressed oncogene in tumors, including breast cancer, colon cancer and lung cancer. Post-translational modifications comprising phosphorylation, acetylation and ubiquitylation regulate the activity of c-Myc. Recently, it was shown that c-Myc-driven tumors are strongly dependent on the SUMO pathway. Currently, the relevant SUMO target proteins in this pathway are unknown. Here we show that c-Myc is a target protein for SUMOylation, and that SUMOylated c-Myc is subsequently ubiquitylated and degraded by the proteasome. SUMO chains appeared to be dispensable for this process, polymerization-deficient SUMO mutants supported proteolysis of SUMOylated c-Myc. These results indicate that multiple SUMO monomers conjugated to c-Myc could be sufficient to direct SUMOylated c-Myc to the ubiquitin-proteasome pathway. Knocking down the SUMO-targeted ubiquitin ligase RNF4 enhanced the levels of SUMOylated c-Myc, indicating that RNF4 could recognize a multi-SUMOylated protein as a substrate in addition to poly-SUMOylated proteins. Knocking down the SUMO E3 ligase PIAS1 resulted in reduced c-Myc SUMOylation and increased c-Myc transcriptional activity, indicating that PIAS1 mediates c-Myc SUMOylation. Increased SUMOylation of c-Myc was noted upon knockdown of the SUMO protease SENP7, indicating that it also could regulate a multi-SUMOylated protein in addition to poly-SUMOylated proteins. C-Myc lacks KxE-type SUMOylation consensus motifs. We used mass spectrometry to identify 10 SUMO acceptor lysines: K52, K148, K157, K317, K323, K326, K389, K392, K398 and K430. Intriguingly, mutating all 10 SUMO acceptor lysines did not reduce c-Myc SUMOylation, suggesting that SUMO acceptor lysines in c-Myc act promiscuously. Our results provide novel insight into the complexity of c-Myc post-translational regulation.
Introduction
Burkitt's lymphoma patients carry a chromosomal translocation involving a breakpoint in chromosome 8 which was shown to harbor a gene with similarity to the v-Myc myelocytomatosis viral oncogene and therefore was named c-Myc for cellular form of Myc.Citation1-4 This translocation caused overexpression of c-Myc, leading to the disease. Cancer cells are frequently dependent on oncogenes like c-Myc for growth and survival, a process that has been named oncogene addiction.Citation5 c-Myc is the most frequently overexpressed oncogene in tumors including breast cancer, colon cancer and lung cancer.Citation6
The c-Myc protein is a basic Helix-Loop-Helix style transcription factor, driving the expression of a very extensive set of genes via E-boxes for Enhancer box sequences in promoter regions of these genes.Citation7 c-Myc drives cell cycle progression, particularly through a key role in S-phase to control replication.Citation8
Small Ubiquitin-like Modifiers (SUMOs) are ubiquitin family members that are conjugated to extensive sets of target proteins as post-translational modifications.Citation9-11 The SUMO conjugation cascade consists of the SUMO E1 heterodimer (SAE1/2), the unique SUMO E2 Ubc9, and a restricted set of E3 enzymes comprising PIAS family members. SUMOylation is a reversible process; at least 6 SUMO proteases deSUMOylate target proteins and participate in the maturation of SUMO precursor proteins.Citation12 SUMOylation predominantly regulates nuclear processes.
Pharmacological inhibition of c-Myc is notoriously challenging and inhibitors of c-Myc are still not available, despite 3 decades of c-Myc research. To explore indirect manners to block c-Myc, a search for Non-Oncogene Addiction (NOA) of c-Myc-driven tumourigenesis was recently performed to identify cellular pathways cooperating with upregulated c-Myc.Citation13 In a genome-wide shRNA screen, it was found that c-Myc-driven proliferation is strongly dependent on the SUMOylation pathway. Upon knockdown of the SUMO E1 or E2 components, c-Myc-dependent proliferation was strongly inhibited. However, the relevant SUMOylated proteins in this process remained to be identified. Here, we provide evidence that c-Myc is a direct target for SUMOylation. The SUMOylated form of c-Myc is stabilized by proteasome inhibitors, indicating that SUMOylated c-Myc is subsequently ubiquitylated and degraded by the proteasome.
Results
c-Myc is a target for SUMOylation
Previously, it was shown that c-Myc-driven tumors are strongly dependent on SUMOylation.Citation13 Searching for relevant SUMO target proteins in this context, we tested whether c-Myc is a target for SUMOylation. A subsection of SUMOylated proteins is ubiquitylated and degraded by the proteasome,Citation14 therefore we included the proteasome inhibitors MG132 and Epoxomicin in these experiments. U2OS cells stably expressing low levels of His-tagged SUMO2 were treated with one of the proteasome inhibitors or DMSO as a control. Subsequently, cells were lysed in a denaturing buffer and SUMO2 conjugates were enriched using Immobilized Metal Affinity Chromatography (IMAC). SUMO-enriched fractions and input samples were analyzed by immunoblotting. Using this approach, we were able to detect a considerable amount of SUMOylated endogenous c-Myc, indicating that c-Myc is a target for SUMOylation ().
Figure 1. SUMOylation of endogenous c-Myc in a proteasome inhibitor-dependent manner. U2OS cells stably expressing His-tagged SUMO2 were treated for 6 hours with DMSO or a proteasome inhibitor, either MG132 or Epoxomicin. The parental U2OS cell line was included as a negative control for the enrichment of His-SUMO2 conjugates. Input and His pulldown samples were separated by SDS-PAGE, transferred onto a membrane and incubated with antibodies to detect c-Myc, SUMO2/3 or the His-tag. In addition, the membrane was stained with Ponceau S as a loading control. The asterisk represents a non-specific band.
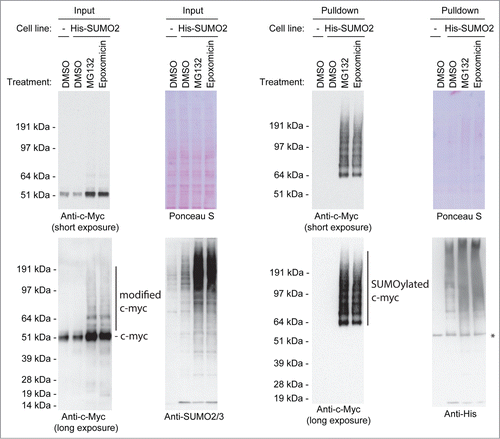
In , the SUMOylated forms of c-Myc are visible in the 65 kDa – 200 kDa range, an exceptionally large region for SUMOylation, suggesting that c-Myc might be conjugated to SUMO polymers. To investigate the potential conjugation of c-Myc to SUMO polymers, we compared the conjugation of c-Myc to wild-type SUMO2 or to a lysine-deficient mutant of SUMO2 (K0) expressed at low levels in U2OS cells. Purified SUMO-conjugates from both cell lines were analyzed by immunoblotting for c-Myc SUMOylation (), but no clear differences in SUMOylation levels could be observed, indicating that SUMO polymers play a limited role. However, we cannot fully rule out a role for SUMO polymers since the observed pattern of c-Myc conjugated to wild-type SUMO2 differs slightly from the pattern of c-Myc conjugated to the lysine-deficient SUMO2 mutant.
Figure 2. For figure legend, see page 1874. Figure 2 (See previous page). SUMO-ubiquitin co-modification of c-Myc. (A) Parental U2OS cells and U2OS cells expressing His-tagged SUMO2 or His-tagged SUMO2-K0 were incubated with MG132. A His pulldown was performed, followed by SDS-PAGE and immunoblotting using anti-c-Myc and anti-SUMO2/3 antibodies. Ponceau S staining was used as a loading control. (B) Cartoon depicting our strategy to identify co-modification of c-Myc by SUMO and ubiquitin. Lysates of cells expressing His-tagged SUMO2 in combination with Flag-tagged ubiquitin contained all modified and unmodified proteins. First, a His-SUMO2 pulldown was performed to enrich for SUMOylated proteins. Subsequently, the SUMO2 and ubiquitin co-modified proteins were selected from this sample via Flag-ubiquitin immunopurification (IP). (C) U2OS cells stably expressing His-SUMO2 and Flag-ubiquitin were treated for 6 hours with DMSO or the proteasome inhibitor MG132. Input samples were taken and a double purification was performed as described above. Input samples, single and double purified samples were separated by SDS-PAGE, transferred onto a membrane and incubated with antibodies to detect c-Myc or the Flag-tag. Equal amounts of starting material were used for the single purification and for the double purified sample, enabling a comparison of these samples. The asterisk represents a non-specific band. (D) U2OS cells stably expressing His-SUMO2 or His-ubiquitin were treated for 6 hours with the proteasome inhibitor MG132. A His pulldown was performed and samples were in vitro deSUMOylated and/or deubiquitylated, followed by SDS-PAGE and immunoblotting using anti-c-Myc, anti-SUMO2/3 or anti-ubiquitin antibodies.
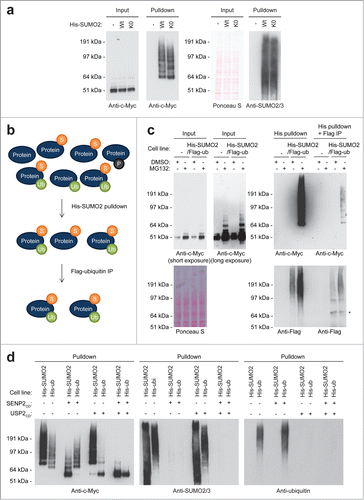
To determine whether c-Myc is co-modified by SUMO and ubiquitin,Citation14 we developed a double purification method to enrich for SUMO2 and ubiquitin (). U2OS cells stably expressing His-SUMO2 and Flag-ubiquitin at low levels were established, and SUMO2 and ubiquitin-conjugates were consecutively enriched and analyzed by immunoblotting (). Additionally, a similar strategy was used to first enrich ubiquitin and then SUMO2-conjugates from U2OS cells stably expressing low levels of His-ubiquitin and Flag-SUMO2 (Fig. S1). The results indicate that high molecular weight c-Myc forms were indeed co-modified by both small protein modifiers as expected, although the amount of co-modified c-Myc appeared to be limited compared to the amount of SUMOylated or ubiquitylated c-Myc.
Additionally, these data were confirmed by performing an in vitro de-SUMOylation and/or de-ubiquitylation assay. His-conjugates were purified from U2OS cells expressing either His-SUMO2 or His-ubiquitin and subsequently incubated with the catalytic domain of SENP2 or/and USP2 (). These results confirmed that a fraction of SUMOylated c-Myc was also ubiquitylated and vice versa.
A limited role for the SUMO-targeted ubiquitin ligase RNF4
Experiments described in the previous section indicated that c-Myc is co-modified by SUMO and ubiquitin to a limited extent. SUMO-targeted ubiquitin ligases (STUbLs) are responsible for ubiquitylating SUMOylated proteins.Citation15 To study whether the STUbL RNF4Citation16-19 is responsible for ubiquitylating SUMOylated c-Myc, we performed RNF4 knockdown experiments (). Knocking down RNF4 resulted in a clear increase in SUMOylated c-Myc, indicating involvement of this STUbL in the co-modification of c-Myc. SUMOylated c-Myc could not be detected upon RNF4 knockdown in the absence of proteasome inhibitors, indicating that RNF4 plays a limited role in c-Myc regulation. One explanation for these findings could be the involvement of another STUbL. The second known human STUbL is RNF111, also known as Arkadia, a component of the DNA damage response.Citation20-22 Similar knockdown experiments were performed as described above (), but no changes in c-Myc SUMOylation could be observed, ruling out a role for RNF111.
Figure 3. SUMO-Targeted Ubiquitin Ligases. U2OS cells stably expressing His-SUMO2 were infected with lentiviruses encoding control, RNF4 (A) or RNF111 (B) shRNAs. Upon knockdown, cells were treated or not with MG132 for 6 hours. A His pulldown was performed, followed by SDS-PAGE and immunoblotting using anti-c-Myc and anti-SUMO2/3 antibodies. Anti-RNF4 and anti-RNF111 antibodies were used to check knockdown efficiency. Ponceau S staining was performed as a loading control. The asterisk indicates a non-specific band. (C) U2OS cells stably expressing His-SUMO2 were infected with lentiviruses encoding control or RNF4 shRNAs. Upon knockdown, cells were treated or not with MG132 for 6 hours. A His pulldown was performed and samples were in vitro deSUMOylated and/or deubiquitylated, followed by SDS-PAGE and immunoblotting using anti-c-Myc and anti-SUMO2/3 antibodies. RNF4 shRNA-mediated knockdown efficiency is shown. The line shows ubiquitylated c-Myc on the SUMOylated c-Myc fraction.
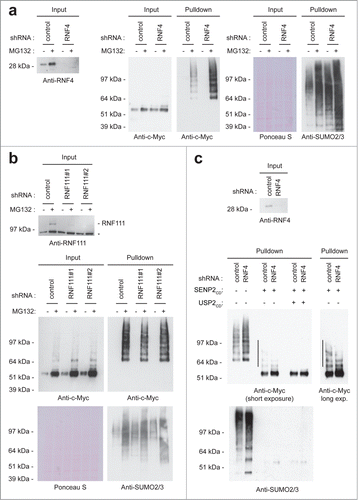
As our results suggest that RNF4 ubiquitylates SUMOylated c-Myc, knocking down RFN4 should increase the ubiquitylated fraction of SUMOylated c-Myc. To address this point, we purified His-SUMO2 conjugates after RNF4 knockdown, in vitro de-SUMOylated and de-ubiquitylated the conjugates and performed immunoblotting to verify c-Myc modification (). In agreement with our hypothesis, knocking down RNF4 resulted in a reduction of the ubiquitylated fraction of SUMOylated c-Myc. We conclude that RNF4 plays a limited role in regulating SUMOylated c-Myc.
PIAS1 and SENP7 regulate reversible SUMOylation ofc-Myc
SUMO conjugation to target proteins is regulated by E3 ligases, including PIAS family members. These E3 ligases contain a so-called SP-RING domain for RING domain identified in Siz and PIAS proteins, with the Siz proteins found in yeast and the PIAS proteins found in mammalian cells.Citation23 Similar to ubiquitin RING E3 ligases, the structure of the SP-RING domain is coordinated by zinc binding.Citation24 In order to identify a c-Myc SUMO ligase, we transfected U2OS cells expressing His-SUMO2 with plasmids to overexpress the different PIAS family members (Fig. S2). Only overexpression of PIAS1 resulted in an increase in SUMOylated c-Myc level. Therefore, we tested knockdown constructs against PIAS1. U2OS cells expressing His-SUMO2 were infected with a lentiviral construct encoding a short hairpin targeting PIAS1. PIAS1 knockdown resulted in a striking reduction of SUMOylated c-Myc, indicating that PIAS1, a major SUMO E3 ligase responsible for the conjugation of many different SUMO substrates, is also responsible for SUMOylating c-Myc ().
Figure 4. For figure legend, see page 1877. Figure 4 (See previous page). PIAS1 and SENP7 mediate the reversible SUMOylation of c-Myc. (A) U2OS cells stably expressing His-SUMO2 were infected with lentiviruses encoding control or PIAS1 shRNAs. Upon knockdown, cells were treated with MG132 and a His pulldown was performed. Samples were analyzed by SDS-PAGE and immunoblotting using anti-c-Myc and anti-SUMO2/3 antibodies. Anti-PIAS1 antibody was used to check knockdown efficiency. Ponceau S staining was used as a loading control. (B) U2OS cells stably expressing His-SUMO2 were infected with lentiviruses encoding control or SENP7 shRNAs. Upon knockdown, cells were treated with MG132 and a His pulldown was performed. Samples were analyzed by SDS-PAGE and immunoblotting using anti-c-Myc and anti-SUMO2/3 antibodies. Anti-SENP7 antibody was used to check knockdown efficiency. Ponceau S staining was used as a loading control. (C) A luciferase assay was performed to check c-Myc activity upon PIAS1 or SENP7 knockdown. The average and SEM of 6 different replicates are shown. The difference between control and PIAS1 knockdown is significant (P < 0.01), indicated by the asterisk.
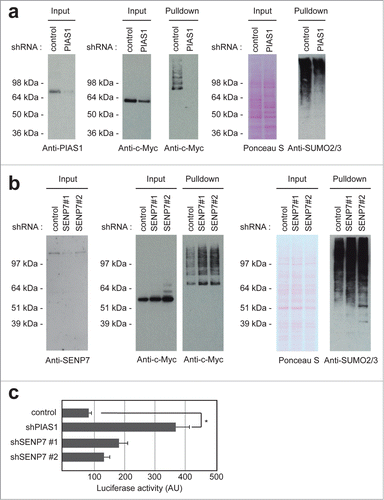
Covalent SUMOylation is a dynamic and reversible process. SENP family members are proteases that remove SUMOs from substrates.Citation12 In order to identify a SENP family member responsible for deconjugating c-Myc, we tested different knockdown constructs. Knockdown of SENP7 resulted in an increase in c-Myc SUMOylation, indicating that SENP7 is responsible for removing SUMOs from c-Myc ().
Subsequently, we tested whether PIAS1 and SENP7 affected c-Myc transcriptional activity, using a c-Myc-driven luciferase reporter. Knocking down PIAS1 enhanced c-Myc-driven transcription about 4-fold (). This result indicates that SUMOylation counteracts c-Myc activity, consistent with the frequently observed repression of transcription factors by SUMOylation.Citation25 Knocking down PIAS1 virtually abolished c-Myc SUMOylation, indicating a direct effect of PIAS1 on c-Myc SUMOylation, although indirect effects cannot be excluded.
SUMOylation and phosphorylation of c-Myc
c-Myc is extensively controlled via post-translational modifications, with a major role for phosphorylation. Two key phosphorylated residues in c-Myc, associated with the stability of the protein, are Thr-58 and Ser-62.Citation26,27 Phosphorylation of Ser-62 is linked to the Ras-ERK pathway and phosphorylation of Thr-58 is carried out by GSK3. Ser-62 phosphorylation of c-Myc leads to subsequent phosphorylation of Thr-58 and degradation via the ubiquitin-proteasome pathway.Citation27
To study whether phosphorylation plays a role in c-Myc SUMOylation via phosphorylation-dependent SUMO modification,Citation28 we tested whether SUMOylated c-Myc is phosphorylated on Ser-62 and Thr-58 using a phosphorylation-specific antibody. In our first attempt, we failed to detect phosphorylated c-Myc in a SUMO-purified fraction, but we noted that the amount of phosphorylated c-Myc that could be detected by the antibody was limited already at total protein level (). To increase the signal, we repeated the experiment, this time cleaving SUMO conjugates after purification using the recombinant catalytic domain of SENP2 (). Using this approach, we visualized the phosphorylated form of c-Myc after SUMO purification, indicating that a fraction of SUMO-conjugated c-Myc is also phosphorylated. Furthermore, the negative results obtained in could potentially be explained by blocking of the epitope for the phosphorylation-specific c-Myc antibody as a result of SUMOylation, or alternatively, the sensitivity of the antibody could be the critical factor.
Figure 5. For figure legend, see page 1879. Figure 5 (See previous page). Co-modification of c-Myc by SUMO and phosphorylation. (A) U2OS cells stably expressing His-SUMO2 were treated with lentiviruses encoding control or PIAS1 shRNA. Upon knockdown, cells were treated with MG132 and a His pulldown was performed. Samples were analyzed by SDS-PAGE and immunoblotting using anti-c-Myc and anti-phospho-c-Myc antibodies. Ponceau S staining was used as a loading control. (B) U2OS cells stably expressing His-SUMO2 were treated with MG132 and a His pulldown was performed. These pulldown samples were treated or not with SENP2 catalytic domain. SDS-PAGE and immunoblotting were performed using anti-c-Myc and anti-phospho-c-Myc antibodies. (C) U2OS cells stably expressing His-SUMO2 were transfected with a control plasmid, HA-tagged c-Myc wild-type or HA-tagged c-Myc T58A/S62A. The parental U2OS cell line was transfected with the c-Myc-HA plasmid and included as a negative control for the enrichment of His-SUMO2 conjugates. Three days after transfection these cells were treated for 6 hours with the proteasome inhibitor MG132, lysed and a His-SUMO2 pulldown was performed. Input and His pulldown samples were separated by SDS-PAGE, transferred onto a membrane and incubated with antibodies to detect the HA-tag or the His-tag. (D) U2OS cells stably expressing His-SUMO2 and Flag-ubiquitin were transfected with a control plasmid, HA-tagged c-Myc wild-type or HA-tagged c-Myc T58A/S62A. Three days after transfection these cells were treated for 6 hours with MG132, lysed and a double His-SUMO2 plus Flag-ubiquitin purification was performed. Input, His pulldown and His plus Flag purified samples were separated by SDS-PAGE, transferred onto a membrane and incubated with antibodies to detect phosphorylated c-Myc, the HA-tag or the Flag-tag.
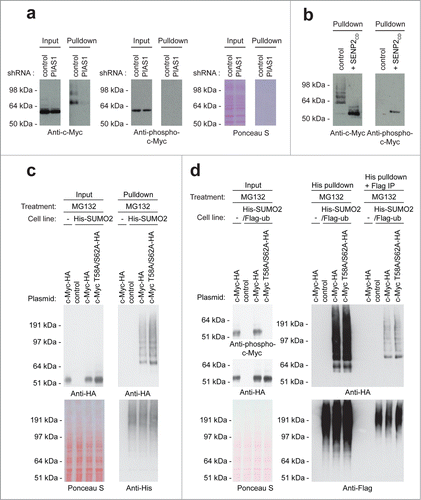
To determine whether phosphorylation is required for c-Myc SUMOylation, we generated a c-Myc phosphorylation-mutant where Thr-58 and Ser-62 were mutated into Ala. U2OS cells were transiently transfected with a control plasmid, HA-tagged c-Myc wild-type or HA-tagged c-Myc T58A/S62A mutant. Subsequently, these cells were treated with the proteasome inhibitor MG132, SUMO-conjugates were enriched and analyzed by immunoblotting (). In agreement with the signaling cascade described above, the c-Myc T58A/S62A mutant was more stable in input samples due to decreased degradation via the ubiquitin-proteasome pathway. Consequently, the SUMOylation level of c-Myc T58A/S62A was modestly increased compared to the SUMOylation level of c-Myc wild-type, corresponding to the increase observed for total levels of c-Myc T58A/S62A. However, no additional changes in SUMOylation level of c-Myc T58A/S62A were observed, indicating that phosphorylation is not required for c-Myc SUMOylation.
Since it is known that phosphorylation at Thr-58 and Ser-62 stimulates c-Myc degradation via the ubiquitin-proteasome system, we studied whether mutating these phosphorylation sites affected the ubiquitylation of SUMOylated c-Myc. U2OS cells stably expressing His-SUMO2 and Flag-ubiquitin at low levels were transiently transfected with a control plasmid, HA-tagged c-Myc wild-type or HA-tagged c-Myc T58A/S62A. Subsequently, SUMO2 and ubiquitin-conjugates were consecutively enriched and analyzed by immunoblotting (). However, no changes in co-modification level by SUMO and ubiquitin were observed between the c-Myc wild-type and c-Myc T58A/S62A mutant. These data indicate that phosphorylation at Thr-58 and Ser-62 does not affect the ubiquitylation levels of SUMOylated c-Myc.
In addition, we investigated whether SUMOylation of c-Myc influences its phosphorylation by knocking down PIAS1 (), based on our finding that PIAS1 is responsible for c-Myc SUMOylation (). Immunoblotting analysis revealed no change in c-Myc phosphorylation on Thr-58 and Ser-62 upon PIAS1 knockdown. Combined, our data indicate that although doubly modified c-Myc can be detected, no functional crosstalk between SUMOylation and phosphorylation of c-Myc could be established.
Mapping SUMO acceptor lysines in c-Myc
During the next phase of the project, we investigated the SUMO acceptor lysines in c-Myc. SUMOs are frequently conjugated to acceptor lysines in target proteins situated in the SUMOylation consensus motif ΨKxE.Citation29-32 Surprisingly, c-Myc lacks lysines situated in this SUMOylation consensus motif. To identify SUMO acceptor lysines, we employed a mass spectrometry approach, similar to the approach described in our previous project.Citation32 HeLa cells stably expressing His-SUMO2 were treated with MG132, lysed and SUMO conjugates were subsequently purified by IMAC, digested by LysC and repurified as described before.Citation32 Site-mapping was enabled by a Q87R mutation, mimicking yeast SUMO, Smt3. In total, we identified 10 SUMO acceptor lysines in c-Myc, including lysines 52, 148, 157, 317, 323, 326, 389, 392, 398, and 430 as summarized in . HeLa cells were particularly useful for these experiments since the methodology for SUMO site identification from HeLa total lysates has been confidently established.
Figure 6. For figure legend, see page 1881. Figure 6 (See previous page). c-Myc SUMO2 acceptor lysines. (A) Cartoon depicting the c-Myc protein including the localization of the SUMO2 acceptor lysines identified by mass spectrometry. The c-Myc protein contains 3 major domains, namely the amino-terminal domain (NTD), the central region (C) and the C-terminal domain (CTD). Within these domains the protein harbours various other regions, including the transactivation domain (TAD), 2 conserved MYC Boxes (MB), a nuclear localization signal (NLS), the basic DNA binding region (BR), a Helix-Loop-Helix domain (HLH) and a leucine zipper motif (LZ). (B) Schematic overview of the experimental strategy used to study the effect of mutating the identified SUMO acceptor lysines on c-Myc SUMOylation. (C) U2OS cells stably expressing His-tagged SUMO2 in combination with either Flag-tagged c-Myc wild-type or 10KR mutant were treated with DMSO or the proteasome inhibitor MG132. Input samples were taken and an His-SUMO2 pulldown was performed on cell lysates. Input and His pulldown samples were separated by SDS-PAGE, transferred onto membranes and incubated with antibodies detecting the Flag-tag, c-Myc or the His-tag. The asterisks represent non-specific bands.
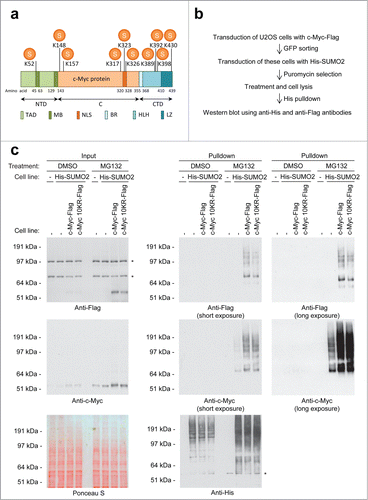
Subsequently, a c-Myc mutant was generated using a synthetic approach to replace SUMO acceptor lysines by arginines (10KR). U2OS cells were infected with lentiviruses encoding GFP and either Flag-tagged c-Myc wild-type or 10KR mutant. Cells were sorted by flow cytometry to obtain cells stable expressing low levels of the transgene (). Subsequently, these cells were transduced with His-tagged SUMO2 and puromycin selected. To determine the effect of mutating the 10 SUMO acceptor lysines in c-Myc on its SUMOylation level, these cells were treated with the proteasome inhibitor MG132 or DMSO as a control. Upon cell lysis, SUMO-conjugates were enriched and analyzed by immunoblotting (). Despite all these mutations, mutant c-Myc was still efficiently conjugated to SUMO. This might indicate that SUMOylation in the ubiquitin-proteasome pathway acts promiscuously, analogous to ubiquitylation.
DISCUSSION
We have uncovered a role for SUMOylation in the regulation of c-Myc. SUMOylated c-Myc was purified and identified from U2OS cells and HeLa cells treated with the proteasome inhibitors MG132 or Epoxomicin, indicating that SUMOylated c-Myc is rapidly degraded by the proteasome. Ten SUMO acceptor lysines were mapped by mass spectrometry, but mutating these 10 sites did not result in a loss of SUMOylation. Conjugation of SUMO to c-Myc could be blocked by PIAS1 knockdown, indicating that PIAS1 is the SUMO E3 ligase responsible for the SUMOylation of c-Myc. Knocking down PIAS1 enhanced c-Myc transcriptional activity 4-fold, consistent with a role for SUMOylation in transcriptional repression of c-Myc (). Deconjugation of SUMO could be reduced by SENP7 knockdown, revealing the SUMO protease responsible for deSUMOylation of c-Myc. Together, these results provide novel insight into the complexity of c-Myc post-translational regulation.
Figure 7. Regulation of c-Myc by SUMOylation: model. Our results indicate that c-Myc is heavily SUMOylated on at least 10 different sites in a promiscuous manner. The SUMO E3 ligase PIAS1 mediates c-Myc SUMOylation. Inhibiting c-Myc SUMOylation by PIAS1 knockdown resulted in increased transcriptional activity, indicating that SUMOylation inhibits c-Myc transcriptional activity. SUMO deconjugation is carried out by the SUMO-specific protease SENP7, possibly recognizing closely-spaced SUMOs on c-Myc. SUMO-Targeted Ubiquitin Ligases (STUbLs), including RNF4, play a role in the ubiquitylation of SUMOylated c-Myc, possibly also by recognizing closely-spaced SUMOs on c-Myc. Subsequently, co-modified c-Myc is targeted to the proteasome for degradation.
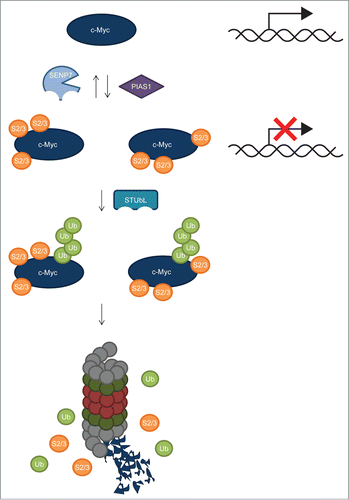
Previously, it was found that the c-Myc oncogenic program is dependent on a properly functioning SUMOylation system.Citation13 Combined with our results, this could indicate that SUMOylation is needed to counteract excessive c-Myc activity by providing cells with a system to limit the activity of overexpressed c-Myc. When the SUMOylation system is defective, changes in the c-Myc transcriptional program caused mitotic catastrophy as observed by Kessler et al. (2012). Nevertheless, it is somewhat surprising to note that Kessler et al. found that inactivation of the SUMO E1 subunit SAE2 switched a transcriptional subprogram of c-Myc from activated to repressed whereas based on our data, one would expect a transcriptional subprogram of c-Myc to be switched from repressed to activated upon impairing SUMOylation. Given the complexity of SUMOylation, targeting hundreds of substrates,Citation11,14,31-34 it is likely that the observations of Kessler et al result from SUMO group modification of many different target proteins.Citation35 Similarly, PIAS1 knockdown affects a broad range of SUMO target proteins in addition to c-Myc.
The promiscuous behavior of SUMO toward c-Myc acceptor lysines is rarely noted in the SUMO field so far. SUMOylation is frequently targeted to lysines situated in SUMOylation consensus motifs.Citation9-11,29-32 By mutating these SUMO acceptor lysines, many different SUMOylation-impaired mutants have been generated. Promiscuous behavior toward acceptor lysines in target proteins is a well-known phenomenon in the ubiquitin field. Weissman and coworkers demonstrated that ubiquitin has little specificity for lysine residues in T cell receptors.Citation36 Even lysines introduced into the protein at aberrant positions acted as ubiquitin acceptor sites. Multiple lysines in cyclin B act as acceptor sites for ubiquitin and no single lysine residue is essential for cyclin B degradation, demonstrating the redundant nature of ubiquitin acceptor lysines in cyclin B.Citation37 In a large scale ubiquitin proteomics study, site promiscuity was deduced from a low level of conservation of ubiquitin acceptor lysines.Citation38 In contrast, mono-ubiquitylation acts in a more site-specific manner.Citation39 Our data would be consistent with site promiscuity of SUMO for proteins that are subsequently ubiquitylated and degraded, if our results can be generalized. We anticipate that site promiscuity is less likely to occur when classical KxE-type SUMOylation consensus motifs are involved.
SENP7 is a SUMO protease with a preference for deconjugating SUMO2 and SUMO3, particularly in a polymeric form.Citation12,40-42 Precursor SUMO1, SUMO2 and SUMO3 and SUMO1 polymers are poorly processed by SENP7.Citation40,41 The catalytic domain of SENP7 consists of 2 separate parts interrupted by a stretch of amino acids.Citation12,43 This feature is thought to contribute to its specificity for SUMO chains.Citation44 We found that SENP7 counteracted c-Myc SUMOylation (). Additionally, little difference was found between the conjugation of c-Myc to wild-type SUMO2 and to a lysine-deficient SUMO2 mutant (), indicating that SUMO polymers play a limited role in the regulation of c-Myc. Three sets of SUMO acceptor lysines in c-Myc are closely spaced, including lysines 148 and 157, lysines 317, 323 and 326 and lysines 389, 392, 398. We hypothesize that these closely spaced SUMOs on c-Myc might be important for the regulation of SUMOylated c-Myc by SENP7 ().
RNF4 is a known regulator of SUMO polymers, enabled by tightly spaced SUMO-interaction motifs.Citation16,42,45,46 Analogous to SENP7, closely spaced SUMOylation events on c-Myc might provide an alternative binding site for RNF4 (). Nevertheless, we cannot fully exclude the presence of SUMO polymers, due to the presence of endogenous SUMO2/3 that can still form polymers together with exogenous SUMO2.
Knocking down RNF4 was not sufficient to detect SUMOylated c-Myc in the absence of proteasome inhibition, indicating that other STUbLs could be involved in the regulation of c-Myc. Since our data did not support a role for RNF111, this could indicate that other STUbLs exist that remain to be identified.
While our work was in progress, a study on the regulation of Myc family members by SUMOylation was published by Amati and coworkers.Citation47 They showed that N-Myc and c-Myc are regulated by SUMOylation with the main focus of their work on N-Myc. They identified K349 in N-Myc as a dominant SUMO acceptor lysine, since mutating this lysine residue reduced N-Myc SUMOylation. However, no difference between N-Myc wild-type and the K349R mutant could be found in functional experiments. Lysine 326 in c-Myc was similarly replaced for an arginine, but this mutation did not reduce SUMOylation of the protein significantly. This is in agreement with our results which show that c-Myc is conjugated to SUMO via at least 10 different SUMO acceptor lysines and that c-Myc SUMOylation is a promiscuous process concerning SUMO acceptor lysines.
In conclusion, SUMOylation adds another level of complexity to the post-translational regulation of c-Myc. Intriguingly, linking c-Myc and SUMOylation might provide novel treatment options for c-Myc-driven tumours.Citation13 Since these tumors are dependent on the SUMOylation system, they are expected to be sensitive to disrupting SUMOylation. Pharmaceutical companies are increasingly interested in disrupting ubiquitin-like signalling,Citation48,49 most notably in the case of Neddylation where the Nedd E1 inhibitor MLN4924 is currently being tested in the clinic.Citation50,51 Analogously, SUMOylation inhibitors might become useful in the future to treat c-Myc-driven tumors.
Materials and Methods
Cell lines and cell culture
Human osteosarcoma (U2OS) cells were cultured at 5% CO2 and 37°C in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal calf serum (FCS, Gibco) and 100 U/ml penicillin plus 100 µg/ml streptomycin (Gibco). Stable cell lines were made by infecting U2OS cells with lentivirus containing the desired cDNA at an MOI of 3. Infected cells were either GFP sorted or puromycin (Calbiochem) selected to obtain low and equal expression levels.
Plasmids, mutagenesis and transfection
The HA-tagged c-Myc plasmid was a kind gift from Prof. R. Bernards (Netherlands Cancer Institute, Amsterdam, The Netherlands). HA-tagged c-Myc 10KR (Genscript) was used to replace the c-Myc wild-type cDNA in this construct, employing restriction enzymes KpnI and BamHI. Subsequently, c-Myc wild-type as well as c-Myc 10KR were cloned in between the PstI and XhoI sites of the plasmid pLV-CMV-IRES-eGFPCitation52 for lentiviral infection to obtain c-Myc encoding virus. The Flag-tag was introduced by PCR.
In addition, a phosphorylation deficient mutant of HA-tagged c-Myc was created by QuickChange site-directed mutagenesis (Stratagene) using oligonucleotides 5'-GAAATTCGAGCTGCTGCCCGCCCCGCCCCTGGCCCCTAGCCGCCGCTCCG-3' and 5'-CGGAGCGGCGGCTAGGGGCCAGGGGCGGGGCGGGCAGCAGCTCGAATTTC-3' to mutate both Threonine 58 and Serine 62 into Alanines. U2OS cells were transfected using 2.5 µl polyethylenimine (PEI, 1 mg/ml; Alfa Aesar) per µg DNA.
Plasmids for Lentivirus mediated knockdown were obtained from the Mission® Library (Sigma-Aldrich); RNF4 (TRCN0000017054), RNF111 (TRCN0000004207, TRCN0000004210), PIAS1 (TRCN0000004145), SENP7 (TRCN0000004544, TRCN0000004547), SHC2 plasmid was used as a control.
Tagged SUMO-2 and ubiquitin constructs have previously been described in.Citation31,32 PIAS expression constructs have previously been described in.Citation53
Proteasome inhibition
To inhibit the proteasome, U2OS cells were treated for 6 hours with 10 µM MG132 (Sigma) or 5 µM Epoxomicin (Sigma). DMSO was used to dissolve both inhibitors and, therefore, added for 6 hours to cells as a negative control.
His-SUMO2 purification
His-tagged SUMO2 conjugates were purified from U2OS cells as described before.Citation31 Briefly, U2OS cells stably expressing His-SUMO2 were washed and collected in PBS. Input samples were taken and the cells were lysed in Guanidinium lysis buffer. After sonication, the lysates were equalized using BCA Protein Assay Reagent (Thermo Scientific). Subsequently, the His-SUMO2 conjugates were enriched using nickel-nitrilotriacetic acid beads (Qiagen). Upon multiple washing steps, the His-SUMO2 conjugates were eluted from the beads and separated by SDS-PAGE.
SUMO – ubiquitin double purification
Co-modified proteins by SUMO2 and ubiquitin were purified using a double purification method. U2OS cells stable expressing either His-SUMO2 and Flag-ubiquitin or His-ubiquitin and Flag-SUMO2 were treated with DMSO or MG132 as described above. Input samples were taken and cells were lysed according to the His-tag pulldown protocol. After enrichment for His-tagged conjugates, a part of the samples was kept separately. Dilution buffer (0.01 M Tris-HCl (pH 7.0), 0.05 M Na2HPO4/NaH2PO4, 0.1 M NaCl and 0.07 M Chloroacetamide) was added to the remaining sample, before it was centrifuged for 45 minutes at 13,200 rpm at 4°C. Flag-M2 beads (Sigma) were added to the supernatant and incubated at 4°C. Subsequently, the samples were washed using wash buffer 1 (0.05 M Tris-HCl (pH 7.5), 0.15 M NaCl and 0.07 M Chloroacetamide) and wash buffer 2 (0.05 M Tris-HCl (pH 7.5) and 0.15 M NaCl). Co-modified proteins were eluted from the beads in 1 × NuPAGE LDS sample buffer (Invitrogen) and separated by SDS-PAGE.
Electrophoresis and immunoblotting
Samples were either separated on 10% SDS-PAGE gels using Tris-Glycine buffer or on Novex 4–12% Bis-Tris gradient gels (Invitrogen) using MOPS buffer. Subsequently, separated proteins were transferred onto a Hybond-C extra membrane (Amersham, BioSciences). Total protein levels were visualized by staining the membrane with Ponceau S (Sigma). After washing in PBS containing 0.05% Tween (PBS/T, MERCK), membranes were blocked in PBS/T containing 8% milk. Membranes were incubated with primary antibodies overnight including rabbit anti-c-Myc (ab32072; Abcam), mouse anti-polyHistidine Clone HIS-1 (H1029; Sigma-Aldrich), mouse anti-SUMO2/3 (ab81375; Abcam), mouse monoclonal anti-Flag M2 (F3165; Sigma), rabbit anti-RNF4,Citation54 mouse anti-RNF111 (H00054778-M05; Abnova), rabbit anti-PIAS1 (GTX62050; Genetex), rabbit anti-SENP7 (A302-995A-1; Bethyl), rabbit anti-phospho c-Myc (ab32029; Abcam), mouse anti-ubiquitin (SC-8017, Santa-Cruz Biotechnology) or mouse monoclonal anti-HA.11 (MMS-101R; Covance). After washing membranes with PBS/T, they were incubated with secondary antibody, either donkey anti-rabbit or goat anti-mouse coupled to HRP (Thermo Scientific). After extensive washing with PBS/T, Pierce ECL 2 western blotting solution (Thermo Scientific) or Amersham ECL Prime (GE Healthcare) were used to visualize signals on FUJI medical X-ray films (FUJIFILM).
shRNA-mediated knockdown
Two million U2OS cells were plated in 15 cm dishes and incubated overnight. Subsequently, cells were infected with either the Mission® Library control shRNA (SHC-2) or shRNAs directed against the indicated targets at an MOI of 2. The following day, medium was replaced for fresh medium. Three days post infection, cells were treated or not with MG132 and harvested for His-SUMO2 pulldown. In the case of RNF4 knockdown, cells were treated and harvested 5 days post infection.
Luciferase activity assay
In order to study endogenous c-Myc activity, a Luciferase assay was employed. 150,000 cells were plated per well in 6 well plates. The following day, cells were transfected in triplicate using 2.5 µl PEI per µg DNA and 0.9 µg of a Luciferase reporter plasmid under the control of the c-Myc promoter with 4 wild-type c-Myc binding sites (wtMBS1-4) or mutated c-Myc binding sites (mutMBS1-4),Citation55 0.9 µg of control or shRNA plasmid and 0.2 µg of pCDNA3-LacZ plasmid.Citation56 The medium was replaced the next day and 2 days after transfection, Luciferase activity was measured using the Luciferase Assay System (Promega). Values were normalized for β-Galactosidase LacZ activity as a transfection efficiency control.
In vitro protease assay
U2OS cells stably expressing His-SUMO2 were treated with MG132 and a His pulldown was performed as described above. In a total volume of 30 µL, 0.5 µL of 50 µM SENP2 and/or USP2 catalytic domain stock solutions (Boston Biochem) were added to the purified samples in deSUMOylation buffer (1.75 M UREA, 0.1 M Na2HPO4/NaH2PO4 and 10 mM Tris-HCl (pH 7.0)). Samples were incubated for 3 hours at room temperature, before the SENP2 or USP2 catalytic domains were added again and incubated for another 3 hours. Samples were analyzed by SDS-PAGE and immunoblotted using anti-c-Myc, anti-phospho-c-Myc, anti-SUMO2/3 or anti-ubiquitin antibodies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1040965_supplemental_files.pdf
Download PDF (3.2 MB)Acknowledgments
We would like to thank Prof. S.J. Elledge (Harvard Medical School, Boston, MA), Dr. T.F. Westbrook (Bayler College of Medicine, Houston, TX) and Prof. R. Bernards (Netherlands Cancer Institute, Amsterdam, NL) for reagents and discussion.
Funding
This work was supported by the Netherlands Organization for Scientific Research (NWO) and the European Research Council (ACOV).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- Colby WW, Chen EY, Smith DH, Levinson AD. Identification and nucleotide sequence of a human locus homologous to the v-myc oncogene of avian myelocytomatosis virus MC29. Nature 1983; 301:722-5; PMID:6298632; http://dx.doi.org/10.1038/301722a0
- Collins S, Groudine M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature 1982; 298:679-81; PMID:6285209; http://dx.doi.org/10.1038/298679a0
- Alitalo K, Ramsay G, Bishop JM, Pfeifer SO, Colby WW, Levinson AD. Identification of nuclear proteins encoded by viral and cellular myc oncogenes. Nature 1983; 306:274-7; PMID:6316149; http://dx.doi.org/10.1038/306274a0
- Ott G, Rosenwald A, Campo E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood 2013; 122:3884-91; PMID:24009228; http://dx.doi.org/10.1182/blood-2013-05-498329
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 2009; 136:823-37; PMID:19269363; http://dx.doi.org/10.1016/j.cell.2009.02.024
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010; 463:899-905; PMID:20164920; http://dx.doi.org/10.1038/nature08822
- Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science 1991; 251:186-9; PMID:1987636; http://dx.doi.org/10.1126/science.1987636
- Heikkila R, Schwab G, Wickstrom E, Loke SL, Pluznik DH, Watt R, Neckers LM. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. Nature 1987; 328:445-9; PMID:3302722; http://dx.doi.org/10.1038/328445a0
- Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 2013; 82:357-85; PMID:23746258; http://dx.doi.org/10.1146/annurev-biochem-061909-093311
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 2010; 11:861-71; PMID:21102611; http://dx.doi.org/10.1038/nrm3011
- Vertegaal AC. Uncovering ubiquitin and ubiquitin-like signaling networks. Chem Rev 2011; 111:7923-40; PMID:22004258; http://dx.doi.org/10.1021/cr200187e
- Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 2012; 13:755-66; PMID:23175280; http://dx.doi.org/10.1038/nrm3478
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 2012; 335:348-53; PMID:22157079; http://dx.doi.org/10.1126/science.1212728
- Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M, Andersen JS, and Vertegaal AC. The ubiquitinproteasome system is a key component of the SUMO-2/3 cycle. Mol Cell Proteomics 2008; 7:2107-22; PMID:18565875; http://dx.doi.org/10.1074/mcp.M800025-MCP200
- Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 2008; 33:201-8; PMID:18403209; http://dx.doi.org/10.1016/j.tibs.2008.02.001
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 2008; 10:538-46; PMID:18408734; http://dx.doi.org/10.1038/ncb1716
- Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 2007; 26:4102-12; PMID:17762864; http://dx.doi.org/10.1038/sj.emboj.7601839
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J 2007; 26:4089-101; PMID:17762865; http://dx.doi.org/10.1038/sj.emboj.7601838
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 2008; 10:547-55; PMID:18408733; http://dx.doi.org/10.1038/ncb1717
- Sun H, Hunter T. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J Biol Chem 2012; 287:42071-83; PMID:23086935; http://dx.doi.org/10.1074/jbc.M112.410985
- Erker Y, Neyret-Kahn H, Seeler JS, Dejean A, Atfi A, Levy L. Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol Cell Biol 2013; 33:2163-77; PMID:23530056; http://dx.doi.org/10.1128/MCB.01019-12
- Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, Wikström M, Choudhary C, Houtsmuller AB, Marteijn JA, et al. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 2013; 201:797-807; PMID:23751493; http://dx.doi.org/10.1083/jcb.201212075
- Hochstrasser M. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 2001; 107:5-8; PMID:11595179; http://dx.doi.org/10.1016/S0092-8674(01)00519-0
- Yunus AA, Lima CD. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell 2009; 35:669-82; PMID:19748360; http://dx.doi.org/10.1016/j.molcel.2009.07.013
- Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev 2005; 15:536-41; PMID:16095902; http://dx.doi.org/10.1016/j.gde.2005.07.004
- Lutterbach B, Hann SR. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol 1994; 14:5510-22; PMID:8035827
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 2000; 14:2501-14; PMID:11018017; http://dx.doi.org/10.1101/gad.836800
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A 2006; 103:45-50; PMID:16371476; http://dx.doi.org/10.1073/pnas.0503698102
- Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 2001; 276:12654-9; PMID:11124955; http://dx.doi.org/10.1074/jbc.M009476200
- Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, Gnad F, Mann M, Vertegaal and AC. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell 2010; 39:641-52; PMID:20797634; http://dx.doi.org/10.1016/j.molcel.2010.07.026
- Schimmel J, Eifler K, Sigurðsson JO, Cuijpers SA, Hendriks IA, Verlaan-de Vries M, Kelstrup CD, Francavilla C, Medema RH, Olsen JV, and Vertegaal AC. Uncovering SUMOylation Dynamics during Cell-Cycle Progression Reveals FoxM1 as a Key Mitotic SUMO Target Protein. Mol Cell 2014; 53:1053-66; PMID:24582501; http://dx.doi.org/10.1016/j.molcel.2014.02.001
- Hendriks IA, D'Souza RC, Yang B, Verlaan-de Vries M, Mann M, and Vertegaal AC. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol 2014; 21:927-36; PMID:25218447; http://dx.doi.org/10.1038/nsmb.2890
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2009; 2: ra24; PMID:19471022; http://dx.doi.org/10.1126/scisignal.2000282
- Becker J, Barysch SV, Karaca S, Dittner C, Hsiao HH, Berriel DM, Herzig S, Urlaub H, Melchior F. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol 2013; 20:525-31; PMID:23503365; http://dx.doi.org/10.1038/nsmb.2526
- Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 2012; 151:807-20; PMID:23122649; http://dx.doi.org/10.1016/j.cell.2012.10.021
- Hou D, Cenciarelli C, Jensen JP, Nguygen HB, Weissman AM. Activation-dependent ubiquitination of a T cell antigen receptor subunit on multiple intracellular lysines. J Biol Chem 1994; 269:14244-7; PMID:8188707
- King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell 1996; 7:1343-57; PMID:8885231; http://dx.doi.org/10.1091/mbc.7.9.1343
- Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, Jensen LJ, Mailand N, Nielsen ML. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics 2011; 10: M110; PMID:21139048; http://dx.doi.org/10.1074/mcp.M110.003590
- Mattiroli F, Sixma TK. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat Struct Mol Biol 2014; 21:308-16; PMID:24699079; http://dx.doi.org/10.1038/nsmb.2792
- Lima CD, Reverter D. Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J Biol Chem 2008; 283:32045-55; PMID:18799455; http://dx.doi.org/10.1074/jbc.M805655200
- Shen LN, Geoffroy MC, Jaffray EG, Hay RT. Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem J 2009; 421:223-30; PMID:19392659; http://dx.doi.org/10.1042/BJ20090246
- Vertegaal AC. SUMO chains: polymeric signals. Biochem Soc Trans 2010; 38:46-9; PMID:20074033; http://dx.doi.org/10.1042/BST0380046
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci 2007; 32:286-95; PMID:17499995; http://dx.doi.org/10.1016/j.tibs.2007.05.002
- Alegre KO, Reverter D. Structural insights into the SENP6 Loop1 structure in complex with SUMO2. Protein Sci 2014; 23:433-41; PMID:24424631; http://dx.doi.org/10.1002/pro.2425
- Rojas-Fernandez A, Plechanovova A, Hattersley N, Jaffray E, Tatham MH, Hay RT. SUMO chain-induced dimerization activates RNF4. Mol Cell 2014; 53:880-92; PMID:24656128; http://dx.doi.org/10.1016/j.molcel.2014.02.031
- Ulrich HD. The fast-growing business of SUMO chains. Mol Cell 2008; 32:301-5; PMID:18995828; http://dx.doi.org/10.1016/j.molcel.2008.10.010
- Sabo A, Doni M, Amati B. SUMOylation of Myc-family proteins. PLoS One 2014; 9:e91072; PMID:24608896; http://dx.doi.org/10.1371/journal.pone.0091072
- Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell 2010; 143:686-93; PMID:21111230; http://dx.doi.org/10.1016/j.cell.2010.11.016
- Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov 2011; 10:29-46; PMID:21151032; http://dx.doi.org/10.1038/nrd3321
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009; 458:732-6; PMID:19360080; http://dx.doi.org/10.1038/nature07884
- Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin Invest Drugs 2012; 21:1563-73; PMID:22799561; http://dx.doi.org/10.1517/13543784.2012.707192
- Vellinga J, Uil TG, de VJ, Rabelink MJ, Lindholm L, Hoeben RC. A system for efficient generation of adenovirus protein IX-producing helper cell lines. J Gene Med 2006; 8:147-54; PMID:16288495; http://dx.doi.org/10.1002/jgm.844
- Roukens MG, Alloul-Ramdhani M, Vertegaal AC, Anvarian Z, Balog CI, Deelder AM, Hensbergen PJ, Baker DA. Identification of a new site of sumoylation on Tel (ETV6) uncovers a PIAS-dependent mode of regulating Tel function. Mol Cell Biol 2008; 28:2342-57; PMID:18212042; http://dx.doi.org/10.1128/MCB.01159-07
- Vyas R, Kumar R, Clermont F, Helfricht A, Kalev P, Sotiropoulou P, Hendriks IA, Radaelli E, Hochepied T, Blanpain C, et al. RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ 2013; 20:490-502; PMID:23197296; http://dx.doi.org/10.1038/cdd.2012.145
- Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O'Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci U S A 2000; 97:2229-34; PMID:10688915; http://dx.doi.org/10.1073/pnas.050586197
- Meulmeester E, Frenk R, Stad R, de GP, Marine JC, Vousden KH, Jochemsen AG. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol 2003; 23:4929-38; PMID:12832478; http://dx.doi.org/10.1128/MCB.23.14.4929-4938.2003
