Abstract
Fibroblast growth factor ligands and receptors (FGF and FGFR) play critical roles in tumorigenesis, and several drugs have been developed to target them. We report the biologic correlates of FGF/FGFR abnormalities in diverse malignancies. The medical records of patients with cancers that underwent targeted next generation sequencing (182 or 236 cancer-related genes) were reviewed. The following FGF/FGFR genes were tested: FGF3, 4, 6, 7, 10, 12, 14, 19, 23 and FGFR1, 2, 3, and 4. Of 391 patients, 56 (14.3%) had aberrant FGF (N = 38, all amplifications) and/or FGFR (N = 22 including 5 mutations and one FGFR3-TACC3 fusion). FGF/FGFR aberrations were most frequent in breast cancers (26/81, 32.1%, p = 0.0003). In multivariate analysis, FGF/FGFR abnormalities were independently associated with CCND1/2, RICTOR, ZNF703, RPTOR, AKT2, and CDK8 alterations (all P < 0.02), as well as with an increased median number of alterations (P < 0.0001). FGF3, FGF4, FGF19 and CCND1 were co-amplified in 22 of 391 patients (5.6%, P < 0.0001), most likely because they co-localize on the same chromosomal region (11q13). There was no significant difference in time to metastasis or overall survival when comparing patients harboring FGF/FGFR alterations versus those not. Overall, FGF/FGFR was one of the most frequently aberrant pathways in our population comprising patients with diverse malignancies. These aberrations frequently co-exist with anomalies in a variety of other genes, suggesting that tailored combination therapy may be necessary in these patients.
Keywords:
Introduction
Fibroblast growth factor (FGF) ligands and receptors (FGFR) are important pathway components of cell proliferation and differentiation, and are vital in embryonic development, wound healing and angiogenesis.Citation1,2 Over the past several decades, there has been extensive progress in understanding the diverse roles FGF/FGFR signaling plays in developmental disorders and oncogenesis.Citation1-4 FGF receptors 1–4 are transmembrane tyrosine kinase receptors that bind 18 different FGF ligands with different affinities, .Citation5 This binding ultimately activates a variety of downstream pathways, including the Ras-dependent MAPK and Ras-independent PI3K-Akt pathways.Citation1 FGF ligands and FGFRs also play a role in angiogenesis, with activation of FGFR1 or FGFR2 leading to endothelial cell proliferation.Citation6 Aberrant activation of these pathways underlies a wide number of cancer types where FGF/FGFR family genes are mutated or amplified.Citation1
Figure 1. Schematic of FGF ligands and their cognate receptors. Relationship between FGF ligands and FGF receptors. A binding relationship is indicated as a red line between ligand and receptor. Various isoforms of the receptors (1c, 1b; 2c, 2b; 3c, 3b; 4D) are included together. FGF14 is not known to bind to any of the FGF receptors. Based on information obtained from (5).
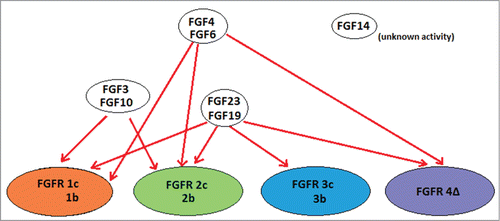
Approximately 50 to 60% of invasive bladder cancers harbor FGFR3 mutations (more common in low-grade papillary tumors and less common in muscle invasive disease), leading to constitutive activation.Citation1 FGFR1 aberrations are also present in about 10% of hormone receptor-positive breast cancers as well as 10–20% of squamous non-small cell lung carcinomas.Citation2 FGFR2 mutations occur in approximately 10% of human endometrial carcinomas, and 15–20% of patients with multiple myeloma overexpress FGFR3 (due to a t(4;14) translocation).Citation7 FGF ligand overexpression is also commonly observed. Indeed, amplification of FGF3 occurs in 15–20% of human breast cancers and is associated with increased invasiveness, and approximately 54% of prostate cancers were found to aberrantly express FGF6.Citation7,8 FGFR aberrations have been observed in glioblastomas, head and neck tumors, gastrointestinal cancers and melanoma.Citation1,2,7,8 Of note, FGF3, FGF4 and FGF19 co-localize on the same amplicon of 11q13 and are frequently co-amplified. Additionally, FGF6 and FGF23 co-localize on 12p13 and are similarly co-amplified, .Citation9
The possibility of using FGF/FGFR aberrations to predict tumor behavior and design targeted therapies is appealing. As an example, FGF3/FGF4 amplification was found to predict increased response to the tyrosine kinase inhibitor sorafenib in patients with hepatocellular carcinoma.Citation10 Additionally, a variety of FGFR specific inhibitors are in early clinical development.Citation1 For instance, an inhibitor of FGFR1–3 (SSR128129E) was found to delay tumor growth and metastasis in human xenograft tumor models, by allosterically inhibiting FGF2 binding to FGFR.Citation11 SSR128129E was also found to improve tumor response to anti-VEGF therapy.Citation11 Indeed, overexpression of FGF ligands and receptors has been shown to increase resistance to anti-VEGF therapy in several tumor types.Citation12 In this regard, some FGFR inhibitors such as lenvatinib and lucitanib also suppress VEGFR.Citation13,14 Both of the latter molecules show considerable activity in the clinic.Citation15,16
The early demonstrated therapeutic effects of FGFR inhibitors in development, in addition to the roles that FGF/FGFR aberrations may play in resistance to other therapies, indicate the clinical relevance of fully characterizing and understanding these aberrations.Citation10,17,18 This is especially relevant for FGF ligand amplifications, which have not been widely studied. We used next generation sequencing results in a population of 391 patients with advanced cancer to study the association between FGF/FGFR aberrations and clinical characteristics, outcomes and co-existent molecular alterations.
Results
Patient characteristics
Three hundred and ninety-one patients were analyzed, 221 (56.5%) of whom were women (). Median age was 54.3 y (range 1.0 to 86.4 years). The most common diagnoses were breast (81/391, 20.7%), glioblastoma (28/391, 7.2%), lung (26/391, 6.6%), and melanoma (25/391, 6.4%). Sixty two patients (15.9%) had metastatic disease at time of diagnosis. Two hundred and thirty-six patients (60.4%) had metastatic disease at the time of biopsy. One hundred and forty-nine biopsies (40.7%) were from a metastatic site, while 217 biopsies (59.3%) were from a primary site.
Table 1. Clinical characteristics of 391 patients with FGF/FGFR aberrations (univariate analysis)
FGF/FGFR abnormalities
Of the 391 patients, 56 (14.3%) had aberrant FGF receptor (FGFR) or FGF ligand (FGF) genes (). FGF/FGFR aberrations were most frequently found in patients with breast cancer (26/81 patients, 32.1%, p = 0.0003) ().
Figure 2. Specific FGF/FGFR aberrations found in patient population. Of 391 patients, 56 had FGF or FGFR aberrations (14%). Of these 56 patients, 22 had aberrations in FGFR1, 2, 3 or 4; and 38 had amplification in FGF3, 4, 6, 10, 14, 19 or 23. The figure shows the frequencies of FGF receptor aberrations and FGF ligand aberrations. Note that some patients had both FGF and FGFR aberrations
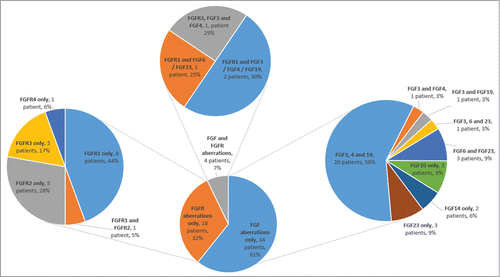
Figure 3. Breakdown of FGF/FGFR aberrations in 391 patients. Frequency chart for 6 main diagnostic categories, showing% of patients in each diagnostic category with FGF or FGFR aberrations and the relative frequencies of amplifications, mutations and fusions. For example, of 81 breast cancer patients, 12 had FGFR aberrations (14.8%) and 16 had FGF aberrations (19.8%).
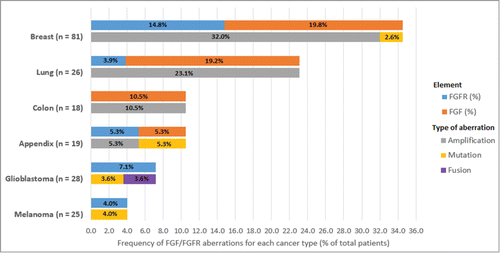
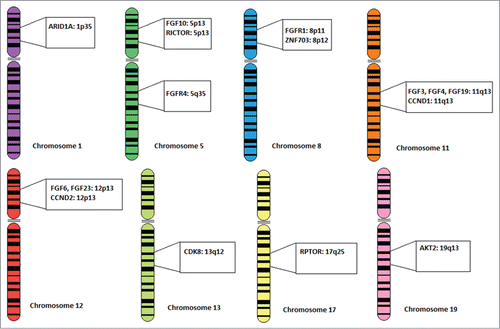
Figure 4. Chromosomal localizations of FGF/FGFR and other correlated genes. Chromosomal localizations of all FGF ligand and FGF receptor genes found in the patient population, along with the 7 genes (CCND1, CCND2, RICTOR, ZNF703, RPTOR, AKT2, and CDK8) that were found to have abnormalities which were independently associated with FGF/FGFR aberrations in multivariate analysis. Several of these genes, such as CCND1, FGF3, FGF4, and FGF19 co-localize on the same amplicon.
Of the 56 patients with FGF/FGFR aberrations, there were 5 mutations, 54 amplifications, and one FGFR3-TACC3 fusion. The five mutations were all in FGFR genes (one in FGFR1, 3 in FGFR2, and one in FGFR3). Among FGF ligand aberrations, only amplifications were seen in 38 patients (with more than one FGF amplified in the tumors of 26 patients), ranging from 2 patients with FGF14 amplifications to 27 patients with FGF3 amplifications (). Included among these were 22 patients that had FGF3, FGF4 and FGF19 aberrations concurrently (39.3%). There were only 2 patients that had both FGF3 and FGF4 aberrations without also having an FGF19 aberration (2/56, 3.6%).
There were 12 patients with FGFR1 aberrations; 6 with FGFR2; 4 with FGFR3 aberrations; and one with an FGFR4 aberration. One patient had both an FGFR1 and FGFR2 aberration (1/56, 1.8%).
In breast cancer patients (N = 81), of 26 patients with an FGF or FGFR abnormality, 13 (16%) also had FGF3, FGF4 and FGF19 amplifications. The other 11 patients showed the following aberrations: FGFR1, N = 7; FGFR2, N = 4; FGF6, N = 1; FGF23, N = 1. The FGF6 and FGF23 amplifications occurred in the same patient.
Characteristics associated with FGF/FGFR aberrations
Women more commonly had FGF/FGFR aberrations than men (17.6% of women vs. 10.0% of men, p = 0.041) (). In univariate analysis, aberrations in FGF/FGFR were also correlated with a diagnosis of breast cancer (N = 26/81; 32.1%, P < 0.0001). There was also a significant association between aberrant FGF/FGFR and liver metastases in univariate analysis (27.6% of FGF/FGFR aberrant patients had liver metastases vs. 11.7% of wild type patients, p = 0.0009) ().
Additionally, there were a significantly higher median number of aberrations in FGF/FGFR-aberrant tumors than in FGF/FGFR wild-type tumors (8 vs. 3, P < 0.0001). The median number of alterations in FGF/FGFR aberrant tumors was significantly higher than for tumors with alterations in P53, KRAS or MYC genes (all P < 0.0001).
Finally, 46/56 patients with FGF/FGFR aberrant tumors (82.1%) had metastases at the time of biopsy, compared to 190/335 patients with FGF/FGFR wild-type tumors (56.7%) (p = 0.0003) (). In a multivariate analysis that included metastasis status at the time of biopsy and the presence of FGF/FGFR abnormalities, only the latter remained independently associated with a higher number of aberrations (P < 0.0001) (Table S3).
Regarding molecular anomalies, we observed a statistically significant association (univariate analysis) between FGF/FGFR aberrations and abnormalities in CCND1, CCND2, ARID1A, MDM2, ERBB2, AURKA, ATM, RICTOR, ZNF703, ZNF217, MYST3, RPTOR, FLT3, NKX2–1, EMSY and AKT2 (all P < 0.05) (Table S1). There was a trend toward a positive association between FGF/FGFR and anomalies in CDK6/8 (p = 0.051) as well as NFKBIA (p = 0.058).
Multivariate analysis showed that CCND1/2, RICTOR, ZNF703, RPTOR, AKT2 and CDK8 aberrations were significantly (all P < 0.02) and independently associated with FGF/FGFR aberrations ().
Table 2. Multivariate analysis of patient characteristics (N = 391) associated with FGF/FGFR aberrations
Clinical outcomes and FGF/FGFR status
The median time from diagnosis to metastasis was not significantly different in patients with and without FGF/FGFR aberrations (p = 0.402) (Table S2). Similarly, there was no statistically significant difference in overall survival between patients with FGF/FGFR aberrant versus wild-type (p = 0.324) (Table S2).
There was no significant difference in overall median PFS for first-line therapy between patients with FGF/FGFR-aberrant vs. wild-type cancers (p = 0.330). When the PFS analysis was performed by treatment type, there was no significant difference between FGF/FGFR aberrant versus wild-type tumors though there was a trend (p = 0.068) toward improved PFS for FGF/FGFR aberrant malignancies treated with taxane-based regimens (Table S2).
Discussion
Aberrations of the fibroblast growth factor family, both ligands and receptors, are frequently found in a wide variety of cancers. Most work so far has focused on FGF receptor aberrations, but FGF ligand amplifications may also be an important molecular feature of certain cancers.Citation1 In the population considered in this study, 56 of 391 patients (14.3%) had either an FGF or FGFR aberration; 22 of 391 (5.6%) had at least one FGFR aberration and 38 of 391 (9.7%) had at least one FGF ligand aberration. FGF or FGFR aberrations were most frequent in breast cancer (p = 0.0003), with 32.1% (of 81 breast cancer patients) harboring an alteration in either FGFR (14.8%) or FGF (19.8%). The most common FGF/FGFR abnormality in breast cancer was FGF3/FGF4/FGF19 co-amplification, which occurred in 15 patients (18.5%), followed by FGFR1 amplification seen in 8 patients (9.9%).
There was no difference in median overall survival, median time from diagnosis to metastasis, or median PFS for first line therapy between FGF/FGFR-aberrant and FGF/FGFR-wild-type patients (Table S2). Similarly, we did not observe any association between FGF or FGFR status and overall survival or time to metastasis in any histology subtype, including breast cancer. Previous studies have shown that FGFR amplifications were associated with increased tumor invasiveness and worsened patient outcomes in several cancer types.Citation19-24 It is possible that no similar associations were seen in our study due to any one or more of several factors: (i) the low number of death events in our population so far (among 56 FGF/FGFR=-aberrant patients, there were 10 death events at the time of analysis); (ii) the fact that our study examined not only FGFR amplifications but also mutations and rearrangements, as well as ligand amplifications; and (iii) the small number of patients in each subgroup.
FGFR1 has previously been found to be aberrant in a variety of cancer types; 8–10% of breast cancers show FGFR1 amplification.Citation25 As mentioned, in our study, 8 of 81 (9.9%) of breast cancers displayed FGFR1 amplification. Additionally, FGFR1 amplification was significantly associated with estrogen receptor (ER) status: 8 of 52 ER-positive tumors were FGFR1 amplified vs. 0 of 29 ER-negative tumors (p = 0.046). Previous studies found FGFR2 to be associated with ER-positive breast cancer, however we found no correlation between estrogen receptor status and FGFR2, FGFR3 or FGFR4 alterations.Citation25 FGFR2 aberrations were found in only 3 of 52 ER-positive (6%) breast cancers.
We found 4 FGFR3 aberrations (out of 391 patients, 1.0%), including 2 amplifications, one R399C mutation, and one FGFR3-TACC3 fusion in a glioblastoma sample. Transforming fusions of FGFR3 and TACC3 have previously been reported in the literature, with the fusion protein inducing chromosomal segregation defects and aneuploidy in astrocytes.Citation27 Of interest, FGFR3 aberrations are reported in about 29% of primary urothelial tumors, and in the germline form are associated with dwarfism.Citation28,29 Our study cohort, however, had only one patient with a urothelial tumor.
A large number of co-existing aberrations were observed for FGF/FGFR-aberrant tumors. In univariate analysis, aberrations in 15 genes were significantly associated with FGF/FGFR aberration (Table S1). A multiple logistic regression method employing a forward selection model identified 6 aberrations that were independently associated with FGF/FGFR status: CCND1/2, RICTOR, ZNF703, RPTOR, AKT2 and CDK8 (all P < 0.02). RPTOR, AKT2 and CDK8 do not co-localize with any FGF or FGFR genes. CCND1 co-localizes with FGF3, FGF4 and FGF19 at chromosome location 11q13. Twenty-two patients (5.6%) had co-amplification of FGF3/FGF4/FGF19 and CCND1 (P < 0.0001). Similarly, CCND2 co-localizes with FGF6 and FGF23 at location 12p13. CCND2 was co-amplified with FGF6 and FGF23 in 5 patients (1.3%, P < 0.0001). These findings are consistent with a recently published study demonstrating association of FGF/FGFR alterations with cyclin gene amplifications.Citation30 RICTOR co-localizes with FGF10 at chromosome location 5p13. Of three patients with FGF10 amplifications, all also had RICTOR amplifications. ZNF703 co-localizes with FGFR1 at chromosome location 8p12. Of 9 patients with ZNF703 amplifications in our population, 7 had FGFR1 amplifications (P < 0.0001).
A significantly higher median number of aberrations were found in FGF/FGFR-aberrant tumors vs. wild-type (8 vs. 3, P < 0.0001). A multivariate analysis showed that FGF/FGFR aberrations were independently associated with an increased median number of total alterations (P < 0.0001), while metastasis at time of diagnosis or biopsy was not. We performed another multivariate analysis and found that co-alterations of FGF3/4/19 and CCND1 also associated with a higher median number of alterations (p = 0.0004). This indicates that the co-localization of FGF3/4/19 and CCND1 explains, at least in part, the association between FGF/FGFR alterations and a higher number of alterations in total (Tables S3 and S4). These results may be of importance, as a larger total number of aberrations is of prognostic value in several tumor types, with more aberrations predicting a shorter progression-free survival.Citation31,32
Our study has several limitations. For many histologies, there were only a small number of patients available for analysis. Several alterations occurred in only a few tumors. Furthermore the study evaluated patients with diverse cancers, though it is possible that this aspect could also suggest generalizability of the observations across tumor types. Finally, the study was retrospective and associations with outcome of specific therapy may require a more robust prospective analysis.
In conclusion, abnormalities in both FGF and FGFR are frequent across diverse cancers, with 14% of our patients harboring these aberrations, making aberrations in FGF/FGFR the third most common to occur in our patient population (behind TP53 and KRAS anomalies). Most abnormalities in FGF or FGFR are due to amplifications, although mutations and rearrangements can also be discerned in FGFR (but not in FGF genes), at least in our population. FGF and FGFR co-amplified with multiple other genes that affect key oncogenic pathways, often because they reside on the same amplicon. The latter suggests that patients that harbor FGF or FGFR abnormalities may require tailored combination therapy, which also targets the protein products of those genes that are frequently co-amplified. Common aberrations that coexisted with FGFR/FGF alterations included components of the cyclin and PI3K/Akt/mTOR axis. Therefore combinations that could be explored include FGFR inhibitors such as lenvatinib, pazopanib, or ponatinib with cyclin dependent kinase (CDK) inhibitors such as palbociclib, or with mTOR inhibitors such as temsirolimus or everolimus (all approved drugs). Alternatively, combinations of experimental drugs that target these pathways could also be explored.
Materials and Methods
Patients
Using next generation sequencing, we analyzed the molecular profile of patients with diverse malignancies (all advanced cancers) that were evaluated at the UC San Diego Moores Cancer Center between October 2012 and May 2014. We retrospectively analyzed patient's molecular profile results, demographic data, as well as clinical characteristics such as response to therapy, time to metastasis, overall survival and progression-free survival. The study was carried out in accordance with the guidelines of the UCSD Institutional Review Board.
Tissue samples and genetic analysis
Tissue samples obtained during diagnostic and therapeutic procedures were used to characterize molecular aberrations. Histologies were confirmed at UC San Diego Moores Cancer Center. Formalin-fixed paraffin-embedded samples were used for next generation sequencing at Foundation Medicine (Cambridge, Massachusetts, http://foundationone.com/). The Foundation One test sequences the full coding regions of 236 cancer-related genes, with 47 introns from 19 genes that are frequently altered in cancers. (Nine patients were tested with a prior version of the test comprising 182 genes). The average depth of coverage is greater than 250×.
The Foundation One test detects substitutions, insertions and deletions, copy number alterations (amplifications) and rearrangements. Amplifications were considered present if there was a >8-fold change in copy number. The gene panel used tested the following FGF/FGFR genes: 3, 4, 6, 7, 10, 12, 14, 19, 23, and FGFR1, 2, 3, and 4 (http://www.foundationone.com).
Endpoints and statistical methods
Patient demographics and characteristics were summarized using descriptive statistics. To analyze the association between categorical variables, Fisher's exact test was used. Time to metastasis was measured from the date of diagnosis to the first reported date of metastasis or last follow up date. Overall survival (OS) was defined as time from diagnosis to last follow up date or death. Progression-free survival (PFS) was defined as the time interval between the initiation of therapy and the first date of disease progression (or the end of therapy, whichever occurred first), or last follow up date. The Kaplan-Meier method was used to estimate the time to metastasis, PFS and OS. The log-rank test and Cox regression methods were used to compare subgroups of patients; all P-values were 2-sided. Any variables with P-value <0.1 in univariate analyses were included for multivariate analysis. As appropriate, a forward selection model using a stepwise probability of 0.05 for entry and removal was used to further narrow characteristics included for multivariate analysis (). P-values less than 0.05 were considered significant. Statistical analyses were carried out by MS using SPSS version 22.0 (Chicago, IL, USA) and packages written in the R programming language (http://www.R-project.org/). Gene chromosome localization information was found using the GeneCards database (http://www.genecards.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Funded in part by the Joan and Irwin Jacobs Fund.
References
- Brooks AN, Kilgour E, Smith PD. Molecular Pathways. Fibroblast growth factor signaling. a new therapeutic opportunity in cancer. Clin Cancer Res 2012; 18(7):1855-62; PMID:22388515; http://dx.doi.org/10.1158/1078-0432.CCR-11-0699
- Dienstmann R, Rodon J, Prat A, Perez-Garcia J, Adamo B, Felip E, Cortes J, Iafrate AJ, Nuciforo P, Tabernero J. Genomic aberrations in the FGFR pathway. opportunities for targeted therapies in solid tumors. Ann Oncol 2014; 25(3):552-63; PMID:24265351; http://dx.doi.org/10.1093/annonc/mdt419
- Turner N, Grose R. Fibroblast growth factor signaling. from development to cancer. Nat Rev Cancer 2010; 10:116-29; PMID:20094046; http://dx.doi.org/10.1038/nrc2780
- Nie X, Luukko K, Kettunen P. FGF signaling in craniofacial development and developmental disorders. Oral Dis 2006; 12(2):102-11; PMID:16476029; http://dx.doi.org/10.1111/j.1601-0825.2005.01176.x
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. the complete mammalian FGF family. J Biol Chem 2006; 281:15694-700; PMID:16597617; http://dx.doi.org/10.1074/jbc.M601252200
- Presta M, Dell'Era P, Mitola S, Moroni E, Ronca R, Rusnati M. Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev 2005; 16:159-78; PMID:15863032; http://dx.doi.org/10.1016/j.cytogfr.2005.01.004
- Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J 2011; 437:199-213; PMID:21711248; http://dx.doi.org/10.1042/BJ20101603
- Ropiquet F, Giri D, Kwabi-Addo B, Mansukhani A, Ittmann M. Increased expression of fibroblast growth factor 6 in human prostatic intraepithelial neoplasia and prostate cancer. J Canc Res 2000; 60(15):4245-50
- Itoh N, Ornitz D. Evolution of the FGF and FGFR gene families. Trends Genet 2004; 20:563-9;; PMID:15475116; http://dx.doi.org/10.1016/j.tig.2004.08.007
- Arao T, Ueshima K, Matsumoto K, Nagai T, Kimura H, Hagiwara S, Sakurai T, Haji S, Kanazawa A, Hidaka H et al. FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology 2013; 57(4):1407-15; PMID:22890726; http://dx.doi.org/10.1002/hep.25956
- Bono F, De Smet F, Herbert C, De Bock K, Georgiadou M, Fons P, Tjwa M, Alcouffe C, Ny A, Bianciotto M et al. Inhibition of tumor angiogenesis and growth by a small-molecule multi-FGF receptor blocker with allosteric properties. Cancer Cell 2013; 23(4):477-88; PMID:23597562; http://dx.doi.org/10.1016/j.ccr.2013.02.019
- Lieu C, Heymach J, Overman M, Tran H, Kopetz S. Beyond VEGF. inhibition of the fibroblast growth factor pathway and antiangiogenesis. Clin Cancer Res 2011; 17(19):6130-9; PMID:21953501; http://dx.doi.org/10.1158/1078-0432.CCR-11-0659
- Okamoto K, Kodama K, Takase K, Sugi NH, Yamamoto Y, Iwata M, Tsuruoka A. Antitumor activities of the targeted multi-tyrosine kinase inhibitor lenvatinib (E7080) against RET gene fusion-driven tumor models. Cancer Lett 2013; 340:97-103; PMID:23856031; http://dx.doi.org/10.1016/j.canlet.2013.07.007
- Soria J, DeBraud F, Bahleda R, Adamo B, Andre F, Dientsmann R, Delmonte A, Cereda R, Isaacson J, Litten J et al. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Ann Oncol 2014; 25:2244-51; PMID:25193991; http://dx.doi.org/10.1093/annonc/mdu390
- Molina AM, Hutson TE, Larkin J, Gold AM, Wood K, Carter D, Motzer R, Michaelson MD. A phase 1b clinical trial of the multi-targeted tyrosine kinase inhibitor lenvatinib (E7080) in combination with everolimus for treatment of metastatic renal cell carcinoma (RCC). Cancer Chemother Pharmacol 2013; 73:181-9; PMID:24190702; http://dx.doi.org/10.1007/s00280-013-2339-y
- Capdevila J, Dienstmann R, Adamo B, Cereda R, Litten J, Collin J, Legrand F, Robert R, Saba C, De Braud FGM et al. Prolonged anti-tumor activity of lucitanib in advanced thyroid cancer. Ann Oncol 2014; 25(suppl 4):iv340-56
- Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, Rooney C, Coleman T, Baker D, Mellor MJ et al. AZD4547; an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res 2012; 72(8):2045-56; PMID:22369928; http://dx.doi.org/10.1158/0008-5472.CAN-11-3034
- Dey JH, Bianchi F, Voshol J, Bonenfant D, Oakeley EJ, Hynes NE. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res 2010; 70(10):4151-62; PMID:20460524; http://dx.doi.org/10.1158/0008-5472.CAN-09-4479
- Elsheikh ES, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, Ellis IO, Reis-Filho JS. FGFR1 amplification in breast carcinomas. a chromogenic in situ hybridisation analysis. Breast Cancer Res 2007; 9(2):R23; PMID:17397528; http://dx.doi.org/10.1186/bcr1665
- Suyama K, Shapiro I, Guttman M, Hazan RB. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell 2002; 2:301-14; PMID:12398894; http://dx.doi.org/10.1016/S1535-6108(02)00150-2
- Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res 2010; 70:2085-94; PMID:20179196; http://dx.doi.org/10.1158/0008-5472.CAN-09-3746
- Wang J, Yu W, Cai Y, Ren C, Ittmann MM. Altered fibroblast growth factor receptor 4 stability promotes prostate cancer progression. Neoplasia 2008; 10:847-56; PMID:18670643; http://dx.doi.org/10.1593/neo.08450
- Weiss J, Sos M, Seidel D, Peifer M, Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010; 2(62):62ra93; PMID:21160078; http://dx.doi.org/10.1126/scitranslmed.3001451
- Koziczak M, Holbro T, Hynes N. Blocking of FGFR signaling inhibits breast cancer cell proliferation through downregulation of D-type cyclins. Oncogene 2004; 23:3501-8; PMID:15116089; http://dx.doi.org/10.1038/sj.onc.1207331
- Hynes N, Dey J. Potential for targeting the fibroblast growth factor receptors in breast cancer. Cancer Res 2010; 70:5199-202; PMID:20570901; http://dx.doi.org/10.1158/0008-5472.CAN-10-0918
- Garcia-Closas M, Chanock S. Genetic susceptibility loci for breast cancer by estrogen receptor status. Clin Cancer Res 2008; 14:8000-9; PMID:19088016; http://dx.doi.org/10.1158/1078-0432.CCR-08-0975
- Singh D, Chan J, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012; 337:1231-5; PMID:22837387; http://dx.doi.org/10.1126/science.1220834
- Guancial E, Werner L, Bellmunt J, Bamias A, Choueiri TK, Ross R, Schutz F, Park R, O'Brein R, Hirsch M et al. FGFR3 expression in primary and metastatic urothelial carcinoma of the bladder. Cancer Med 2014; 3:835-44; PMID:24846059; http://dx.doi.org/10.1002/cam4.262
- Foldynova-Trantirkova S, Wilcox W, Krejci P. Sixteen years and counting. The current understanding of fibroblast growth factor receptor 3 (FGFR3) signaling in skeletal dysplasias. Hum Mutat 2011; 33:29-41; PMID:22045636; http://dx.doi.org/10.1002/humu.21636
- Schwaederle M, Daniels G, Piccioni D, Fanta P, Schwab R, Shimabukuro K, Parker B, Kurzrock R. Cyclin alterations in diverse cancers: Outcome and co-amplification network. Oncotarget 2015; 6(5):3033-42; PMID:25596748
- Simon R, Bürger H, Brinkschmidt C, Bocker W, Hertle L, Terpe HJ. Chromosomal aberrations associated with invasion in papillary superficial bladder cancer. J Pathol 1998; 185:345-51; PMID:9828832; http://dx.doi.org/10.1002/(SICI)1096-9896(199808)185:4%3c345::AID-PATH109%3e3.0.CO;2-0
- Dellas A, Torhorst J, Jiang F, Proffitt J, Schultheiss E, Holzgreve W, Sauter G, Mihatsch MJ, Moch H. Prognostic value of genomic alterations in invasive cervical squamous cell carcinoma of clinical stage IB detected by comparative genomic hybridization. Cancer Res 1999; 59:3475-9; PMID:10416613
