Emerging evidence in a growing number of biological systems indicates that the functions played by small non-coding RNAs (sRNAs) impact on an ever-increasing number of cell biological functions. Our laboratory has previously contributed some results to this complex landscape by the identification of a novel class of sRNA that are critical components of the DNA damage response (DDR), the signaling pathway that promptly arrests cell proliferation allowing DNA repair.Citation1,2 These sRNA, that we named DDRNAs (DNA damage response RNAs), are generated in a DICER- and DROSHA-dependent manner at the proximity of DNA double-strand break (DSB) and are crucial for DDR focus formation and DDR signaling at the site of damage.Citation1 Similar observations have been reported in plantsCitation3 and DrosophilaCitation4 and they point to a previously unappreciated engagement of some components of the RNA interference (RNAi) machinery in DDR modulation and genome stability.Citation5
Among the RNAi components, members of Argonaute (Ago) family of proteins play a fundamental role, acting as the effectors of sRNA-mediated gene silencing.Citation6 Ago proteins (Agos in brief) are evolutionary conserved in each of the 3 domains of life; indeed, all eukaryotic genomes, with the exception of S. cerevisiae, and many prokaryotic ones, encode for Agos.Citation6-8 During the last decade, besides their well-established cytoplasmic role in post-transcriptional gene silencing, novel functions for eukaryotic Agos have been described to impact in different ways on gene expression, also by acting in the nucleus.Citation7
In order to exert their functions, Agos require sRNA fragments as guiding molecules for target-RNA recognition. However, both eukaryotic and prokaryotic catalytically-active Agos share similar structural features with RNase H8-10, which makes them potential DNA/RNA hybrid interactors. Indeed, intriguingly, in vitro, prokaryotic Agos display a greater affinity for short DNA molecules than for RNA.Citation6 Most strikingly, bacterial Agos have been lately shown to associate and cleave single-strand DNA in vivo, using both sRNA and DNA fragments as guide molecules.Citation8,11,12 Furthermore, it has been recently reported that AGO2 interacts with RAD51, a DNA repair factor involved in homologous recombination (HR), and mediates its accumulation at resected DNA damage sites in human cells.Citation13
These enticing findings, along with the reported Mn2+-dependent affinity of human AGO2 for short DNAs in in vitro assays,Citation14 led us to conceive the provocative hypothesis that also human Ago (specifically Ago2, the only catalytically active member of Agos in mammalsCitation8,10) could cleave DNA and contribute to resect DSB DNA ends to promote DNA repair by HR. At present, direct evidence of an ability of eukaryotic Agos to bind and process DNA molecules in vivo has not been provided, although invoked and amply discussed in some recent reviews.Citation15
We thus decided to experimentally address this open question and we exposed HeLa cells to ionizing radiations to induce DSB formation and trigger the DDR cascade, including RAD51 recruitment to the genomic lesions, a process proposed to be mediated by AGO2.Citation13 One hour post-irradiation, we purified cell nuclei and immunoprecipitated endogenous AGO2 from this specific cell compartment. The efficiency of AGO2 immunoprecipitation (IP) was confirmed by immunoblotting, thus also proving the presence of AGO2 in the nucleus (). Then, in order to characterize the nature of the nucleic acids associated with nuclear AGO2, we followed the very same experimental approach previously used to demonstrate the in vivo association of bacterial Agos with small DNA.Citation11 Specifically, AGO2-bound nucleic acids were isolated using neutral phenol-chloroform extraction and dephosphorylated, prior to 5′ labeling with [γ-32P]ATP. Labeled nucleic acids were then subjected to RNase A or DNase I treatment and fractionated onto 10% urea PAGE. We observed that AGO2-associated nucleic acids are sensitive to RNase A treatment and resistant to DNase I digestion.
Figure 1. (A) AGO2 IP efficiency was tested by Western blot. Five% of each AGO2 IP (lanes AGO2) or mock IP (lanes IgG), performed in nuclear extracts of irradiated (20 Gy) or not irradiated (NO IR) HeLa cells, was resolved onto a 4–15% SDS-PAGE along with 1% of the input lysates (lanes IN). Proteins were transferred onto nitrocellulose membrane and probed for AGO2. The arrowhead indicates endogenous AGO2, whereas the asterisks mark unspecific bands. No AGO2 is retained in the mock IPs. (B) Human AGO2 binds to small RNAs only and not to small DNAs in the nucleus. AGO2 co-precipitated nucleic acids (lanes AGO2) from irradiated (20 Gy) or not irradiated (NO IR) HeLa nuclei were 5′-radioactively labeled, treated with RNase A or DNase I and fractionated onto a 10% urea PAGE along with a size marker (lane M). As controls, both untreated samples (lanes -) and nucleic acids from mock IP (lane IgG) were also fractionated.
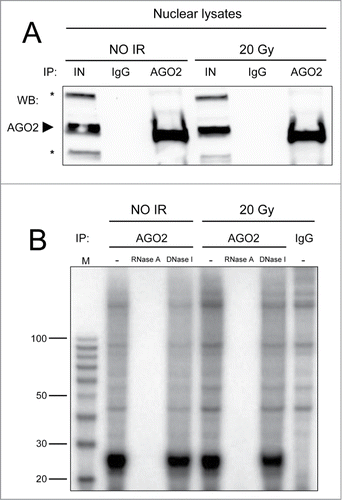
This result therefore demonstrates that human AGO2, differently from its prokaryotic orthologues,Citation11 interacts only with sRNAs (20–30 nt in length), and not with DNA fragments. Furthermore, DNA damage generation does not alter this selective affinity ().
Although AGO2 has been proposed to participate to the HR repair in a process that necessitates both its ability to interact with sRNA as well as its catalytic activity,Citation13 our data suggest that the underlying mechanism does not require a direct processing of DNA ends by this enzyme as suggested by some.Citation15 Rather, our results point to an involvement of AGO2 in DNA repair related to its ability to bind and process RNA molecules, although their identity and their functions remain unclear. It is possible that RNA association of eukaryotic Agos favors their functions at the chromatin level in guiding DNA repair events similarly to their already described roles in transcriptional regulationCitation16,17 and heterochromatin formation.Citation18
In summary, our results show that throughout evolution the Ago clade of proteins has lost the ability to directly associate with DNA molecules and catalyze their hydrolysis, and that these features are exclusive of prokaryotic Agos and absent in human AGO2.
References
- Francia S, et al. Nature 2012; 488:231–5; PMID:22722852; http://dx.doi.org/10.1038/nature11179.
- Jackson SP, et al. Nature 2009; 461:1071-8; PMID:19847258; http://dx.doi.org/10.1038/nature08467.
- Wei W, et al. Cell 2012; 149:101-12; PMID:22445173; http://dx.doi.org/10.1016/j.cell.2012.03.002.
- Michalik KM, et al. Nucleic Acids Res 2012; 40:9596-603; PMID:22848104; http://dx.doi.org/10.1093/nar/gks711.
- d'Adda di Fagagna F. Trends Cell Biol 2014; 24:171-8; PMID:24156824; http://dx.doi.org/10.1016/j.tcb.2013.09.008.
- Swarts DC, et al. Nat Struct Mol Biol 2014; 21:743-53; PMID:25192263; http://dx.doi.org/10.1038/nsmb.2879.
- Meister G. Nat Rev Genet 2013; 14:447-59; PMID:23732335; http://dx.doi.org/10.1038/nrg3462.
- Willkomm S, et al. Life (Basel) 2015; 5:538-53; PMID:25692904.
- Song JJ, et al. Science 2004; 305:1434-7; PMID:15284453; http://dx.doi.org/10.1126/science.1102514.
- Liu J, et al. Science 2004; 305:1437-41; PMID:15284456; http://dx.doi.org/10.1126/science.1102513.
- Olovnikov I, et al. Molecular Cell 2013; 51:594-605; PMID:24034694; http://dx.doi.org/10.1016/j.molcel.2013.08.014.
- Swarts DC, et al. Nature 2014; 507:258-61; PMID:24531762; http://dx.doi.org/10.1038/nature12971
- Gao M, et al. Cell Res 2014:1-10; 24:532-41; http://dx.doi.org/10.1093/nar/gku1387
- Lima WF, et al. J Biol Chem 2009; 284:26017-28; PMID:19625255; http://dx.doi.org/10.1074/jbc.M109.010835.
- Smalheiser NR, et al. Biol Direct 2014; 10:27; PMID:25472905; http://dx.doi.org/10.1186/preaccept-1466302485137399.
- Huang V, et al. PLoS Genet 2013; 9:e1003821; PMID:24086155; http://dx.doi.org/10.1371/journal.pgen.1003821.
- Carissimi C, et al. Nucleic Acids Res 2015; 43:1498-512; PMID:25605800; http://dx.doi.org/10.1093/nar/gku1387
- Irvine DV, et al. Science 2006; 313:1134-7; PMID:16931764; http://dx.doi.org/10.1126/science.1128813.
