Abstract
Nuclear DNA duplication in the absence of cell division (i.e. endoreplication) leads to somatic polyploidy in eukaryotic cells. In contrast to some invertebrate neurons, whose nuclei may contain up to 200,000-fold the normal haploid DNA amount (C), polyploid neurons in higher vertebrates show only 4C DNA content. To explore the mechanism that prevents extra rounds of DNA synthesis in these latter cells we focused on the chick retina, where a population of tetraploid retinal ganglion cells (RGCs) has been described. We show that differentiating chick RGCs that express the neurotrophic receptors p75 and TrkB while lacking retinoblastoma protein, a feature of tetraploid RGCs, also express p27Kip1. Two different short hairpin RNAs (shRNA) that significantly downregulate p27Kip1 expression facilitated DNA synthesis and increased ploidy in isolated chick RGCs. Moreover, this forced DNA synthesis could not be prevented by Cdk4/6 inhibition, thus suggesting that it is triggered by a mechanism similar to endoreplication. In contrast, p27Kip1 deficiency in mouse RGCs does not lead to increased ploidy despite previous observations have shown ectopic DNA synthesis in RGCs from p27Kip1−/− mice. This suggests that a differential mechanism is used for the regulation of neuronal endoreplication in mammalian versus avian RGCs.
Introduction
In eukaryotes, the cell cycle is regulated by Cyclins and their specific cyclin-dependent kinases (Cdks). The complex formed by Cyclin D and Cdk4/6 phosphorylates Retinoblastoma protein (Rb), thus leading to the release of the transcription factor E2F1,Citation1 which in turn induces the synthesis of the proteins necessary for DNA replication.Citation2 G1/S progression is then regulated by the activation of Cdk2 by Cyclin E, which phosphorylates additional residues of Rb,Citation3 thus facilitating cell cycle progression. DNA synthesis is driven by the association of Cdk2 with Cyclin A, and the Cdk1/Cyclin A complex regulate late G2 phase progression.Citation4-6 Finally, G2/M transition is regulated by the formation of the Cdk1/Cyclin B complex.Citation7 The transitions between the different cell cycle phases are further regulated by 2 families of Cdk inhibitors (CKI): the Ink and Cip/Kip protein families. The Ink family, composed by p16Ink4a, p15Ink4b, p18Ink4c and p19Ink4d, regulates the quiescent state by specifically binding to Cdk4/6 thus preventing its union with Cyclin D.Citation8 The members of the Cip/Kip family, constituted by p21Cip1, p27Kip1 and p57Kip2, preferentially inactivate Cyclin/Cdk complexes but they can also modulate their activity.Citation3,9 Through all these mechanisms, cell cycle is strictly regulated to obtain 2 daughter cells with the same chromosomal number as that of the original cell.
Ploidy can be defined as the number of chromosome sets that are present in the cell nucleus and it is expressed as the N value (i.e., the set of chromosomes present in a gamete, or haploid cell). In contrast, DNA content (expressed as the C value) represents the amount of DNA present in a gamete or haploid cell. While the C value increases during S-phase, remaining duplicated during G2, the N value remains unaltered as the cell progresses through the cell cycle.Citation10-13 Polyploidy, defined as the presence in the cell nucleus of more than 2 sets of chromosomes, can affect all the cells of the organism. In this case, it is referred to as germinal polyploidy. A strategy to increase the body size, based on germinal polyploidy, has been widely used by plants,Citation14,15 and also by protozoa (Tetrahymena),Citation16 fish,Citation17 as well as by amphibians including Salamandra salamandra and Xenopus laevis.Citation18,19 Although germinal polyploidy cannot be observed in higher vertebrates because it compromises their viability, polyploidy affecting to specific cell types or tissues (i.e. somatic tetraploidy) can be widely observed in these organisms. In this case, the term polyploidy is used in a broad sense as a synonym of the C value, since the actual number of chromosomes in the affected cells remains usually unknown. Somatic polyploidy has several advantages for the affected cells including a better response to metabolic and/or genotoxic stress and a lower sensibility to apoptotic stimuli.Citation10,11,20,21 In mammals and higher vertebrates different cell types have been shown to be polyploid, including trophoblast giant cells,Citation10,11,,22 hepatocytes,Citation23,24 megakaryocytes,Citation25 keratinocytes,Citation26 and vascular smooth cells.Citation27 Endoreplication represents a major mechanism leading to developmental programmed somatic polyploidy.Citation10-12 This process consists of successive rounds of DNA synthesis in the absence of mitosis (endocycles), thus leading to cells with a DNA content higher than 2C. Although the mechanism of endoreplication is far from being completely understood, it seems to be dependent on the levels and activity of the Cdk/Cyclin complex after the completion of every S phase, the oscillations of Cyclin E and Geminin levels, and the localization of the minichromosome maintenance protein complexes.Citation10-12,28 Endoreplication can be mimicked by inhibition of Cdk1 activity,Citation11 and the subsequent prevention of the mitotic process. Moreover, cyclin E/Cdk2 is the G1 Cdk complex crucial for DNA synthesis initiation during the endoreplicative process.Citation29-32
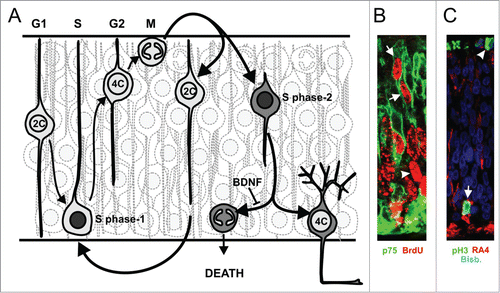
Somatic polyploid neurons generated by endoreplication are very common in the nervous system of lower vertebrates and invertebrates.Citation33,34 For instance, the nucleus of giant neurons from Aplysia californica has been shown to contain 200,000-fold the normal amount of haploid DNA (i.e., 200,000C).Citation33 These neurons have routinely been subjected to electrophysiological analyses,Citation35 proving that they are fully functional. In humans, around 10% of the cortical neurons show DNA contents higher than 2C, being tetraploid around 1% of these neurons.Citation36 Tetraploid neurons have also been found in the murine retina and cerebral cortex,Citation37,38 as well as in the retina, optic tectum, dorsal root ganglia, cerebellum, telencephalon and spinal cord of the chick.Citation37,38 In the chick retina, tetraploid ganglion cells are generated through cell cycle reactivation as they migrate to the ganglion cell layer, soon after their final mitosisCitation37 (see ). Cell cycle reactivation in neurons fated to become tetraploid occurs in response to the interaction of nerve growth factor (NGF) with the neurotrophin receptor p75 (p75NTR).Citation37-40 Tetraploid RGCs remain in a G2-like state in the presence of brain-derived neurotrophic factor (BDNF), which activates the TrkB receptor to decrease Cdk1 expression and activity in these neurons, thus blocking G2/M transition.Citation41 In contrast, in the absence of BDNF these neurons undergo mitosis followed by apoptosisCitation37 ().
Figure 1. Scheme of the mechanism inducing tetraploid RGCs in the chick retina. (A) Retinal precursors undergo S-phase (dark gray nucleus) at the basal neuroepithelium (S-phase-1), and they displace their nuclei to the apical neuroepithelium during G2, showing 4C DNA content. Then, they undergo mitosis at the apical portion of the neuroepithelium. This division gives rise to precursors with 2C DNA content that undergo a new round of interkinetic nuclear movement (see ref.Citation78). Alternatively, daughter cells may undergo neuronal differentiation (gray cytoplasm). Some of the differentiating RGCs can reactivate the cell cycle (S-phase-2) in response to NGF as they migrate to the basal neuroepithelium, where the GCL will be located. In the presence of BDNF, RGCs remain with 4C DNA content (i.e. tetraploid neurons), whereas in its absence they undergo ectopic mitosis at the basal neuroepithelium and die. (B) An illustrative image showing p75NTR-positive differentiating RGCs undergoing S-phase-2 at the apical neuroepithelium (arrows). In contrast, precursors undergoing S-phase-1 (arrowhead) are located basally. (C) An illustrative image showing an RA4-positive differentiating RGC undergoing ectopic mitosis, revealed with phosphoHistone H3 immunolabeling (pH3), at the basal neuroepithelium (arrow). In contrast, precursors undergo mitosis at the apical neuroepithelium (arrowhead). Bisb.: bisbenzimide.
So far, no polyploid neurons with DNA levels above 4C have been found in the normal brain of higher vertebrates.Citation37,42 Furthermore, Rb-deficient neurons have been shown to undergo cell cycle re-entry in vivo, remaining alive with a 4C DNA content but not higher.Citation43 Therefore, the question that arises is why tetraploid neurons cannot undergo successive rounds of endoreplication similar to those performed by giant neurons from lower vertebrates. As there should be a mechanism that prevents tetraploid neurons from undergoing successive rounds of endoreplication we have hypothesized p27Kip1 as being a potential candidate for this function. In this study we provide evidence that this CKI is expressed by all RGCs in the developing chick retina, and that the downregulation of this CKI in cultured RGCs results in DNA synthesis and increased DNA content in these neurons. This mechanism seems to be specific for chick RGCs as the retina of p27Kip1−/− miceCitation44 lack RGCs with a DNA content higher than 4C.
Results
p27Kip1 is expressed by layered RGCs in the developing chick neural retina, including those that become tetraploid
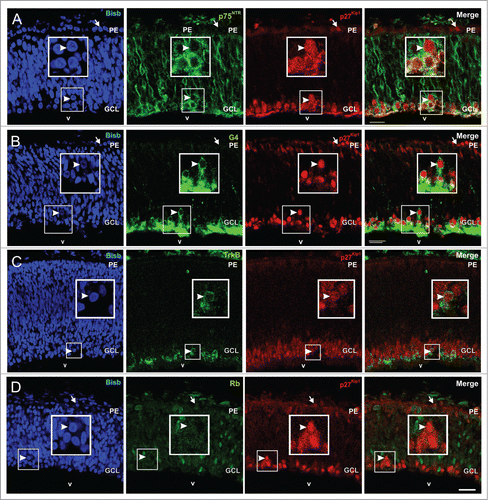
To determine whether p27Kip1 may be involved in preventing successive rounds of S phase in tetraploid neurons, the expression pattern of p27Kip1 was characterized in the E5 chick retina, a stage at which tetraploid RGCs are being generated in this tissue.Citation37 This analysis demonstrated that p27Kip1 presents a conspicuous expression pattern. On the one hand, p27Kip1-specific immunoreactivity can be clearly observed in the retinal pigmented epithelium (arrows in ), as previously shown by ref.Citation45. On the other hand, p27Kip1 colocalizes with the RGC differentiating marker p75NTR in the basal region of the neural retina [99,4 ± 0,6% (n = 2) of the p27Kip1-positive cells express p75NTR] (), thus suggesting that p27Kip1 is expressed at the end of the migration of all differentiating RGCs. This assumption is consistent with the co-localization of p27Kip1 with G4 (), a marker that preferentially detects differentiating layered RGCs.Citation46 Therefore, the majority of the p27Kip1-positive cells present in the retina at this stage are differentiating neurons localized in the ganglion cell layer (GCL), as previously described by ref.Citation47. All cells that express TrkB receptor, known to prevent G2/M transition in tetraploid RGCs,Citation41 show p27Kip1 immunoreactivity (), suggesting that tetraploid RGCs also express p27Kip1. In this regard, most laminated RGCs that express p27Kip1 lack Rb [99,2 ± 0,4% (n = 3)] (), a feature of RGCs that re-enter cell cycle and become tetraploid.Citation37 In sum, we conclude that p27Kip1 is expressed by all layered RGCs, including those that become tetraploid.
Figure 2. p27Kip1 expression in the E5 chick retina. Confocal sections from retinas of E5 chick embryos double labeled with an anti-p27Kip1 specific antibody (red) and antibodies (green) specific for p75NTR (A), G4 (B), TrkB (C), or Rb (D) are shown. Nuclear staining with bisbenzimide in blue (Bisb.). p27Kip1 expression can be detected in the pigment epithelium (arrows) and layered differentiating RGCs in the GCL (arrowheads). (A) p75NTR-positive cells co-localize with p27Kip1. (B) p27Kip1 is expressed in differentiated CGRs expressing G4. (C) TrkB expressing cells also express p27kip1. (D) p27kip1 is expressed by RGCs lacking Rb. Boxes: high magnification of the framed areas. GCL: ganglion cell layer; PE: pigment epithelium; v: vitreous body. Bar: 20 μm.
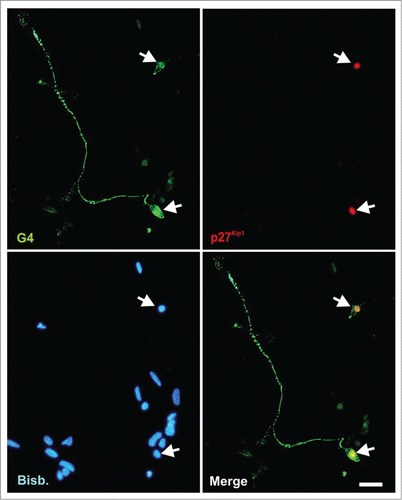
p27Kip1 expression was also analyzed by immunocytochemistry in E6 chick retina cultures maintained under neurogenic conditions, known to mimick RGC differentiation in vivo.Citation37,39,41,48-50 As expected from its expression pattern in vivo, p27Kip1-specific immunoreactivity was detected only in the nucleus of G4-positive neurons (i.e. differentiating RGCs) ().
Figure 3. p27Kip1 is expressed by RGCs in vitro. E6 chick retinal cells were cultured under neurogenic conditions for 20 h, fixed and immunostained with antibodies against G4 (green) and p27Kip1 (red). Only differentiated RGCs neurons (G4+ cells) express p27Kip1. Nuclear staining with bisbenzimide (Bisb.) is shown in blue. Bar: 20 μm.
Knock-down of p27Kip1 expression with specific interfering RNA constructs
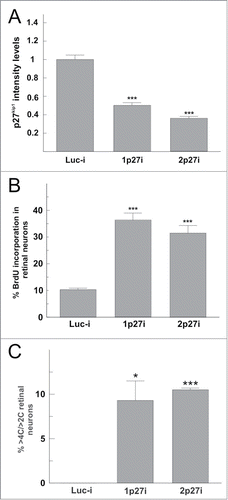
To verify the participation of p27Kip1 in preventing tetraploid neurons from undergoing successive rounds of endoreplication, specific shRNA vectors against p27Kip1 were constructed following the procedure described by ref.Citation51. Two independent vectors were designed, one targeting the 5′ region (1p27i) and another one targeting the 3′ region (2p27i) of the Cdkn1b mRNA. A shRNA vector known to interfere with luciferaseCitation51 was used as a control (Luc-i). All these vectors co-express RFP. To evaluate the efficiency of the interfering RNAs, E6 chick retinal cells were electroporated with the shRNA vectors and then cultured under neurogenic conditions to give rise to differentiating chick retinal neurons (DCRNs).Citation48,49 After 20 h, these neurons were subjected to double immunostaining with G4- and p27Kip1-specific antibodies. p27Kip1 expression levels were quantified by image analysis in G4/RFP positive cells. Both interference vectors (1p27i and 2p27i) decreased the expression levels of p27Kip1 by 50% and 67%, respectively, in comparison with the expression levels observed in RGCs transfected with the control vector (Luc-i) ().
Figure 4. p27Kip1 downregulation increases BrdU incorporation and DNA amount in cultures enriched in RGCs. (A) E5 chick retinal cells were transfected with shRNA vectors designed against different regions of the Cdkn1b gene (1p27i and 2p27i) or a control shRNA vector against luciferase (Luc-i), and then cultured for 20 h under neurogenic conditions. Then, p27Kip1 expression levels were quantified by image analysis in differentiated chick retinal neurons transfected with the shRNA or control vectors. Both Cdkn1b-specific constructs were able to significantly decrease the expression levels of p27Kip1 as compared with the control construct. (B) RGC-enriched retinal cultures were obtained from E7 chick embryos as described by ref.Citation52. These cells were then lipofected with Luc-i, 1p27i, or 2p27i vectors and then treated with 0.5 mg/ml BrdU for 20 h. The percentaje of neurons (NeuN+ cells) incorporating BrdU was significantly increased in the presence of p27Kip1-specific shRNA compared to the control (RFPi). (C) The frequency of non-diploid neurons (>2C) with a DNA content higher than 4C (>4C) was significantly increased in the presence of p27Kip1-specific shRNA compared to the control (RFPi). *P < 0.01; ***P < 0.001 (Student´s t test, n = 3).
p27Kip1 knock-down facilitates DNA synthesis and increased ploidy in differentiating RGCs
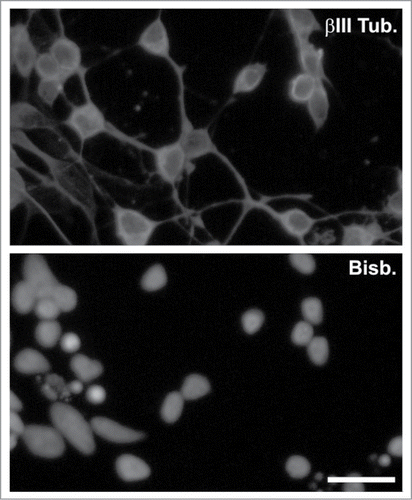
The interfering RNAs described above were used to test whether p27Kip1 knock-down could induce BrdU incorporation in differentiated RGCs. To increase the proportion of these latter neurons in our cultures we employed a procedure previously described by ref.Citation52, based on the centrifugation of E7 chick retinal cells through a Percoll gradient. shows an example of a neurogenic culture enriched in RGCs obtained with this protocol and immunostained with βIII tubulin, a marker that is expressed at high levels by the RGCs.Citation53 After 20 h in culture, RGC-enriched cultures were lipofected with the 1p27i, the 2p27i, or the Luc-i vectors, and treated with BrdU during an additional 20 h period. BrdU incorporation was then quantified in lipofected cells (i.e., RFP-positive cells) expressing the neuronal marker NeuN. This analysis demonstrated a statistically significant increase of BrdU incorporation in neurons transfected with any of the p27Kip1 shRNA vectors ().
Figure 5. Enrichment of RGCs through a Percoll gradient. βIII tubulin staining (βIII Tub.) performed in a culture enriched in RGCs. Nuclei were stained with bisbenzimide (Bisb.). Bar: 20 μm.
According to our hypothesis, the increase of DNA synthesis in RGCs triggered by p27Kip1 knock-down should result in a concomitant increase of DNA content in these neurons through an endoreplicative mechanism. In accordance with this assumption, none of the neurons lipofected with any of the shRNA vectors (1p27i, n = 266; 2p27i, n = 188) as well as with the control vector (n = 188) did show mitotic figures at the end of the culture period, thus suggesting that the increase of DNA content in RGCs is not reduced as a result of cell division. To directly prove that the p27Kip1 knock-down in RGCs actually results in an increase of the C value, DNA intensity level, evidenced by DAPI staining, was quantified by image analysis in the lipofected NeuN-positive neurons. In all these cultures 8.73 ± 1.12% (mean ± s.e.m.; n = 6) of the lipofected NeuN-positive cells were observed to be tetraploid, a proportion in the range to that previously observed in E6 chick retinal cells cultured under neurogenic conditions as well as in the posthatch chick retina in vivo.Citation37 This analysis also indicated that around 10% of the more-than-diploid (>2C), p27Kip1-knocked-down neurons showed DNA content higher than 4C and below 8C irrespective of the shRNA vector used. In contrast, >2C neurons transfected with the Luc-i control vector did not show DNA levels above 4C ().
In the differentiating chick retina, tetraploid neurons that undergo mitosis finally die by apoptosis.Citation37,41 Therefore, the lack of mitotic figures in the neurons lipofected with the shRNA vectors is consistent with the absence of apoptosis in these cells. In this regard, none of the neurons transfected with either the 1p27i (n = 266) or the 2p27i (n = 188) vectors showed pyknotic nuclei. A similar situation was observed in those neurons transfected with the Luc-i vector (n = 188).
DNA synthesis in RGCs expressing p27Kip1-specific shRNAs is independent from Cdk4/6 activity
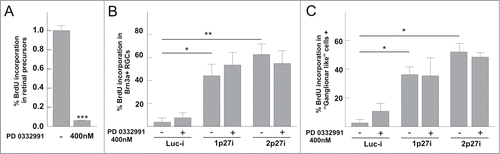
To test whether the increase of BrdU incorporation in neurons expressing p27Kip1-specific shRNAs was due to endoreplication or, alternatively, to conventional cell cycle progression, a potent and specific inhibitor against Cdk4/6 (PD0332991) was used. As previously shown,Citation39 PD0332991 was able to prevent cell cycle progression in retinal precursor cells (P < 0.05) (), thus indicating that Cdk4/6 is required for normal cell cycle progression in these proliferating cells. In contrast, PD0332991 was unable to inhibit DNA synthesis in cultured RGCs lipofected with the p27Kip1-specific shRNA vectors and treated for 20 h with BrdU (P > 0.05, n = 3; ANOVA test). For this latter analysis, RGCs were identified by using 2 different criteria: expression of Brn3a (), a known marker of a subpopulation of RGCs,Citation54 or just by morphology (monoaxonal cells with rounded cytoplasm were identified as RGCs) (). In both cases, the increase of BrdU incorporation in response to the p27Kip1-specific shRNA vectors was similar to that obtained in the experiments illustrated in . Overall, these data indicate that the mechanism triggering DNA synthesis in p27Kip1-knocked-down RGCs is different from the classical Cdk4/6-dependent mechanism leading to cell cycle progression in retinal precursor cells.
Figure 6. BrdU incorporation in RGCs, but not in retinal precursors, is independent of Cdk4/6 activity. (A) The selective Cdk4/6 inhibitor PD 0332991, used at 400 nM, prevents the capacity of E6 retinal precursor cells to incorporate BrdU after a short pulse (30 min). The analysis was performed after 20 h of inhibitor treatment. (B) RGC-enriched retinal cultures were lipofected with Luc-i, 1p27i, or 2p27i vectors and treated with 0.5 mg/ml BrdU for 20 h in the presence (+) or absence (−) of 400 nM PD 0332991. BrdU incorporation was evaluated in Brn3a-positive cells (a subpopulation of RGCs). (C) RGC-enriched retinal cultures treated as described above. BrdU incorporation was evaluated in cells that display RGC morphology (monoaxonic cells with round cytoplasm). *P < 0.01; **P < 0.005; ***P < 0.001 (Student´s t test, n = 3).
No increase of ploidy in RGCs from p27Kip1 knock-out mice
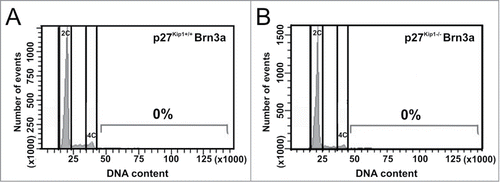
A previous study has demonstrated that differentiated RGCs from p27Kip1 knockout mice are able to undergo DNA synthesis.Citation44 Moreover, p27Kip1 interacts with MCM7, a DNA replication licensing factor, to inhibit initiation of DNA replication.Citation55 These findings make p27Kip1 a candidate to block further rounds of endoreplication in mouse tetraploid RGCs. To directly test this hypothesis, the DNA content of Brn3a-positive cell nuclei isolated from the retina of wild-type and p27Kip1−/− mice at E20-P2 was analyzed by flow cytometry. At this developmental stage, RGCs have already been born,Citation56 and most of them express Brn3a.Citation57 This analysis confirmed previous results indicating that a proportion of mouse RGCs are tetraploid.Citation37 In contrast, no Brn3a-positive neurons were observed showing DNA contents higher than 4C in both wild-type and p27Kip1−/− retinas (). Similarly, the proportion of Brn3a-negative nuclei with a DNA content above 4C did not significantly differ in p27Kip1−/− vs. wild-type retinas [wild-type: 0.47 ± 0,12% (n = 3); p27Kip1−/−: 0.23 ± 0.07% (n = 3)], thus indicating that the absence of p27Kip1 is not translated into hyperploidy in any retinal cell type.
Figure 7. RGCs with a DNA content higher than 4C are not present in the p27Kip1−/− mouse retina. Cell nuclei isolated from the retina of either p27Kip1+/+ (A) or p27Kip1−/− (B) P2 mice were immunostained for the RGC marker Brn3a, treated with PI (DNA content), and then analyzed by flow cytometry (n = 3 for each genotype). Representative DNA content histograms are shown for Brn3a+ nuclei. No net increase of RGCs with a DNA content above 4C can be seen in p27Kip1−/− retinas.
Discussion
In this study we have demonstrated that p27Kip1 is expressed by layered, differentiating chick RGCs, including those previously shown to be tetraploid. In addition, we provide evidence that p27Kip1 downregulation induces DNA synthesis and hyperploidy in chick RGCs. We propose that p27Kip1 participates in the prevention of extra rounds of genome duplication in tetraploid avian RGCs. This mechanism may also keep diploid chick RGCs with normal DNA levels.
Some populations of vertebrate projection neurons replicate its DNA content and become tetraploid in the context of a normal developmental program.Citation37,38,42 This process results in neurons with increased size and extensive dendritic arbors.Citation37 In contrast, neurons with an elevated degree of ploidy have been observed both in invertebrates and in lower vertebrates.Citation33,34 Polyploidization in these latter neurons takes place through successive endoreplicative cycles that seem to be abolished in higher vertebrates, in which neurons are maintained with 4C DNA content. Our results suggest that p27Kip1, a CKI known to prevent Cdk2 activity and block G1/S transition,Citation3,9,58 is likely involved in the mechanism that prevents tetraploid neurons to undergo successive rounds of endoreplication in the chick retina. This conclusion is consistent with a previous study showing that p27Kip1 can modulate endoreplication in Solanum lycopersicum L.Citation59
We have demonstrated that p27Kip1 is expressed by differentiating chick RGCs in the E5 chick neural retina, an observation consistent with previous published work.Citation47 This conclusion is supported by the co-localization of p27Kip1 with both G4, a neuronal marker mostly expressed by layered, differentiating chick RGCs,Citation46 and p75NTR, known to be an early marker of RGC differentiation in the chick retina.Citation37 In the neural chick retina, a subset of p27Kip1-expressing neurons lack Rb, known to be absent in tetraploid neurons,Citation37 and many of the p27Kip1-positive neurons co-express TrkB, the BDNF receptor known to prevent ectopic mitosis in tetraploid chick RGCs.Citation41 Overall, these observations suggest that p27Kip1 is expressed by all chick RGCs, including those that become tetraploid. Importantly, p27Kip1 initiates its expression once that DNA duplication is already finished in migrating, tetraploid chick RGCs (see ), as it would be expected for a blocker of extra rounds of endoreplication. In older stages of chick retinal development such as E12, p27Kip1 is also expressed by horizontal and amacrine cells,Citation47 following a pattern coincident with the onset of differentiation of the different retinal neuronal types, from which RGCs are the first ones in coming out from the cell cycle.Citation60 This observation suggests that p27Kip1 can maintain the tetraploid state of horizontal cells previously shown to remain with duplicated DNA levels in the chick retina.Citation61,62
We have shown that p27Kip1 downregulation causes an increase of BrdU incorporation in chick RGCs that does not lead to cell division, thus suggesting that p27Kip1 prevents DNA synthesis and the subsequent increase of DNA amount in these neurons. This observation is consistent with the capacity of p27Kip1 to inhibit a MCM7, necessary for the duplication license.Citation55 For this analysis, we used chick RGC-enriched cultures containing tetraploid neurons and maintained under conditions that mimic the in vivo situation.Citation37,39,41,48-50 Different criteria for neuron identification, including the expression of the neuronal marker NeuN in chick RGC-enriched cultures, the presence of the RGC marker Brn3a, or just by identifying RGCs in base to their morphology, yielded similar results, thus stressing the robustness of our analysis. Our results provide direct evidence that p27Kip1 downregulation actually leads to an increase of DNA amount in at least 10% of the tetraploid chick RGCs. As this effect is not blocked by the Cdk4/6 selective inhibitor PD0332991, which prevents cell cycle progression in chick retinal precursors, we conclude that the canonic mechanism leading to S-phase in the chick retina is not involved in this process. This suggests that this new DNA synthesis upon p27Kip1 downregulation could be due to a mechanism similar to endoreplication, where what is crucial is the activity of Cdk2.Citation63
BrdU incorporation in chick RGCs triggered by p27Kip1 downregulation is reminiscent of the phenotype observed in the retina of adult p27Kip1 knock-out mice, where differentiated RGCs also undergo DNA synthesis.Citation44,64 Nevertheless, we could not find evidence for an increase of ploidy in mouse RGCs depleted of p27Kip1. In mice, the effect of p27Kip1 is synergic with that of p19Ink4d since the proportion of mouse RGCs that incorporate BrdU is enhanced in the retina of double p27Kip1/p19Ink4d knockout mice.Citation44 In contrast, there is no evidence for the existence of a p19Ink4d homolog in the chick genome, thus suggesting that this synergic mechanism does not take place in avian RGCs. Nevertheless, we cannot rule out that in the chick retina, a similar cooperative effect between p27Kip1 and other members of the Ink family could actually take place. Indeed, homologs of mammalian p15Ink4b, p16Ink4a, and p18Ink4c have been identified in the chick (accession numbers: NP_989764, NP_989765, and XP_004936839, respectively). This may explain why only a relatively modest proportion of chick RGCs actually increases their DNA amount in response to p27Kip1 downregulation. In contrast with the expression pattern of p27Kip1 in the E5 chick retina, which is restricted to the differentiated RGCs (this study; ref.Citation47), in the murine retina p27Kip1 is also expressed by precursor cells during late G2/early G1.Citation65 The expression of p27Kip1 in the mouse retinal precursors may explain both the increase of RGC axons in the optic nerveCitation66 and the retinal dysplasiaCitation67-69 observed in p27Kip1−/− mice. Nevertheless, the increase of RGC number in the retina of p27Kip1−/− mice that would explain this phenotype is not dramatic,Citation66 and seems to be compensated by apoptosis.Citation44,65 In this context, interfering RNA against p27Kip1 has been shown to induce cell death in cultured cortical neurons.Citation70 In contrast, no signs of apoptosis were evident in RGCs incorporating BrdU in our experiments, thus indicating that the mechanisms regulating endoreplication in the chick retina may differ from those in the mouse. This differential apoptotic response that allows the survival of avian RGCs with a DNA content above 4C may explain the presence of hyperploid RGCs in the chick but not in the mouse retina.
Our results suggest that p27Kip1 is involved in blocking successive rounds of endoreplication in tetraploid chick neurons. Nevertheless, p27Kip1 can also participate in the blockage of classical cell cycle progressionCitation9 as well as neuronal differentiation and migration.Citation71 Therefore, p27Kip1 is probably playing multiple roles in RGCs, including the blockade of tetraploid neurons to undergo extra rounds of endoreplication and the inhibition of cell cycle reactivation in diploid neurons that have not been programmed to become tetraploid.
Materials and Methods
Chick embryos
Fertilized eggs from White Leghorn hens (Gallus gallus L.) were obtained from a local supplier (Granja Santa Isabel, Spain) and incubated at 38.5°C in an atmosphere of 70% humidity. The embryos were staged as described previously.Citation72 All experiments were performed in accordance with the European Union and Spanish guidelines.
Mice
Mice of the p27Kip1 mutant strainCitation67 were crossed and maintained at the University of Valencia animal core facility in accordance with Spanish regulations. Heterozygous mice were mated to produce all 3 genotypes. Embryo littermates were staged as described by ref.Citation73. All experiments were performed in accordance with the European Union and Spanish guidelines.
Antibodies
The mouse monoclonal antibody (mAb) against neuron-specific βIII tubulin (clone 5G8; Millipore), which specifically recognizes RGCs from early stages of differentiation,Citation53 was diluted 1/2,000 for immunohistochemistry and 1/1,000 for immunocytochemistry. The rabbit polyclonal antibody (pAb) against G4, a chicken glycoprotein mostly expressed by differentiated RGCs,Citation46 was kindly provided by E.J. de la Rosa (Centro de Investigaciones Biológicas, CSIC). This antibody was used at 1/1,000 for immunohistochemistry and 1/500 for immunocytochemistry. The mouse mAb against Brn3a MAB1585 (Millipore), which specifically recognizes a subpopulation of RGCs in both chickCitation54 and mouseCitation57 retinas, was diluted 1/500 for immunohistochemistry and 1/300 for flow cytometry. The rabbit poylclonal antibody against NeuN ABN78 (Millipore) was used at 1/1,000 dilution for immunohistochemistry. The rabbit polyclonal antiserum [9650] against the extracellular domain of human p75NTR was a generous gift of Moses V. Chao (New York University), and it was diluted 1/1,000 for immunohistochemistry. The rabbit polyclonal antiserum against the extracellular domain of chick TrkB was a kind gift from Louis F. Reichardt (University of California, San Francisco), and it was used at 1/2,000 dilution for immunohistochemistry. The mouse monoclonal antiserum against BrdU (Developmental Studies Hybridoma Bank) was used at 1/4,000 dilution for immunohistochemistry. The rat monoclonal antibody against BrdU (AbD Serotec, MCA2060) was used at 1/200 dilution for immunocytochemistry. The anti-Rb pAb Ab39690 (Abcam) was used at 1/50 dilution for immunohistochemistry. The mouse monoclonal antibody against p27Kip1 (BD Transduction Laboratories) was used at 1/500 for immunohistochemistry. The anti-phosphoHistone H3 rabbit polyclonal antiserum (Upstate Biotechnology) is a well characterized marker of mitosis and it was diluted 1/400 for immunohistochemistry. The mAb anti-RA4, a generous gift from Stephen C. McLoon (University of Minnesota), recognizes an epitope expressed by RGCs soon after their final mitosis,Citation74,75 and it was diluted 1/400 for immunohistochemistry. All secondary antibodies were purchased from Invitrogen. The Alexa Fluor 594 goat anti-rabbit antibody was used at a 1/1,000 dilution for immunohistochemistry. The Alexa Fluor 647 donkey anti-mouse and the Alexa Fluor 488 goat anti-rat antibodies were used at 1/1,000 for immunohistochemistry. The Alexa Fluor 488 goat anti-mouse antibody was used at a 1/1,000 dilution for immunocytochemistry and 1/500 for cytometric studies
Cell culture
Retinal cultures were performed under neurogenic conditions as described previously.Citation76 To this aim, neuronal precursors isolated from the E6 chick retina were cultured in the presence of laminin-1 and insulin to give rise to DCRNs.Citation48,49 Briefly, E6 retinas were dissected out of the pigmented epithelium and treated for 5 min at 37°C with 0.5 mg/ml trypsin prepared in PBS containing 3 mg/ml bovine serum albumin (BSA). The reaction was stopped with 1 mg/ml soybean trypsin inhibitor (Sigma-Aldrich) and cells were mechanically disaggregated by passing them 5 times through a Pasteur pipette whose tip has previously been polished with a flame. Dissociated retinal cells were suspended in 50% Dulbecco's-modified Eagle's Medium (DMEM)/50% F12 (Sigma-Aldrich) plus N2 supplement (DMEM/F12/N2), and plated at a density of 10,000-200,000 cells/cm2 on 10-mm round glass coverslips previously coated with 500 μg/ml poly (D-L) ornithine (PLO; Sigma-Aldrich) and 10 μg/ml laminin-1 (natural mouse laminin; Invitrogen). Cultures were maintained overnight (ON) in 4-well dishes (Greiner Bio-One, Germany). Cells were cultured in DMEM/F12/N2 for 20 h at 37°C in a water-saturated atmosphere containing 5% CO2. BrdU (Roche) was used at 0.5 μg/ml. In some instances, cells were cultured for 20 h in the presence or absence of the Cdk4/6 inhibitor PD0339921 (Axon MedChem), used at 400 nM.
RGCs enrichment
RGCs enrichment was carried out following the protocol described by ref.Citation52. For each experiment, 30 retinas from E7 chick embryos were dissected away from the pigment epithelium, and then treated for 5 min at 37°C with 0.5 mg/ml trypsin prepared in PBS containing 3 mg/ml BSA. The reaction was stopped with 1 mg/ml soybean trypsin inhibitor and then, cells were mechanically disaggregated as described above. Cell suspensions were diluted using DMEM/F12/N2 to reach 19.8 ml and distributed in 3 6.60 ml aliquots. Each aliquot was loaded as described below on a discontinuous Percoll gradient prepared in a 15 ml falcon tube. The bottom layer (d = 1.09) was obtained by mixing 1.12 ml DMEM/F12/N2 with 1.70 ml Percoll (Sigma-Aldrich); the second layer (d = 1.06) is formed by the mix of 1.80 ml of the cell suspension with 1.44 ml Percoll; the third layer (d = 1.04) was obtained by mixing 2.40 ml of the cell suspension with 0.52 ml Percoll; the forth layer (d = 1.02) was generated by mixing 2.40 ml of the cell suspension with 0.28 ml Percoll, and the top layer was composed by 2.70 ml DMEM/F12/N2. Then, discontinuous Percoll gradients were centrifuged at 351 × g for 30 min at 4°C. As a result of the centrifugation, 2 bands were generated. The top layer, enriched with RGCs, was isolated and washed with DMEM/F12/N2. After centrifuging the cells at 300 × g for 1 min, pellets were resuspended in DMEM/F12/N2 and plated on 10-mm round coverslips previously coated with PLO and laminin-1 (20,000–200,000 cells/cm2). Cultures, performed in 4-well dishes, were maintained in DMEM/F12/N2 for 20–22 h at 37°C in a water-saturated atmosphere containing 5% CO2.
shRNA
Two shRNA constructs targeting different regions of the gene Cdkn1b (accession number: NM_204256.2) were generated, based on the protocol described by ref.Citation51. The 1p27i construct (1p27i) was generated by targeting the sequence found between 93 and 112 base pairs (bp) (GCCATGGAGGATTACACGAA). The 2p27i construct (2p27i) was generated by targeting the sequence found between 1043 and 1062 bp (GAGGAGGTTTCAGAAGACT). Once generated, these constructs were cloned into the pRFPRNAiC vector.Citation51 pRFPRNAi luciferase (Luc-i)Citation51 was used as negative control. All these plasmids co-express red fluorescent protein (RFP).
Lipofection
RGC-enriched cultures were lipofected with 3 μg of the desired plasmid using Lipofectamin 2000 (Invitrogen) following the manufacturer's instructions. Cultures were immediately treated with 0.5 μg/ml BrdU. In some cases, cultures were also treated with the Cdk4/6 inhibitor PD0339921, used at 400 nM, or vehicle. After 4–6 h, the culture medium was exchanged with fresh DMEM/F12/N2 containing 0.5 μg/ml BrdU and cells were cultured for 24 h at 37°C. 400 nM PD0339921 (or vehicle) was included in the replaced culture medium when necessary.
Explant electroporation
Electroporation of retinal explants was performed as described previously.Citation37,41 E6 chick retinas were dissected away from the pigment epithelium and fragmented into small pieces of around 10 mm2. These retinal fragments were laid onto a glass coverslips and subsequently immersed in 4 μl PBS containing different plasmid combinations (0,75−1 μg/μl). Electroporation was performed with four 50 milliseconds pulses of 25–27 V, at a 200 milliseconds frequency. After electroporation, the explants were grown in suspension for 3–4 h in 50% DMEM/F12/N2. Explants were then collected, dissociated, and cultured as previously described.
In vivo BrdU treatment
E5-6 embryos were treated for 15 min with 40 μl of a solution containing 10 μg/μl BrdU, which was applied to the chorioallantoic membrane.
Immunocytochemistry
Immunocytochemistry was performed in cultures fixed with 4% paraformaldehyde for 15 min at room temperature (RT), and permeabilized for 30 min with phosphate buffered saline (PBS) containing 0.05% Triton X-100 (Sigma-Aldrich) (PBTx) and 10% Fetal Calf Serum (FCS; Invitrogen). Cultures were then incubated for 2 h (at RT) or ON (at 4°C) with PBTx containing 1% FCS and the appropriate primary antibodies. After five washes in PBTx, cultures were incubated for 1 h in PBTx containing 1% FCS and a 1/1,000 dilution of Alexa Fluor 488 goat anti-mouse antibody, Alexa Fluor 594 goat anti-mouse antibody, Alexa Fluor 647 donkey anti-mouse antibody, Alexa Fluor 647 goat anti-rabbit antibody, or Alexa Fluor 488 goat anti-rat antibody. All these secondary antibodies were purchased from Life Technologies.
For BrdU immunolabeling, cultures were previously subjected to DNA denaturation by incubation for 30 min at RT with 2N HCl/0.33× PBS, following by a neutralization step consisting of 3 15-min washes with 0.1 M sodium borate, pH 8.9, and 2 washes of 5 min each with PBS/0.05% Triton X-100. G4 immunostaining was performed in living cells as described previously.Citation50 To this aim, cultures were washed 4 times with Krebs-Ringer-Hepes buffer (KRH), and incubated for 20 min at RT with a 1/400 dilution of the anti-G4 antibody prepared in KRH. Cells were then washed 4 times with KRH and incubated for 20 min at RT with a 1/1,000 dilution of the Cy2-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) prepared in KRH. After 4 washes with KRH, cells were fixed with 4% PFA for 15 min. At this stage, cells can be double immunolabeled using the standard protocol described above. Nuclear labeling was performed with PBS containing either 1 μg/ml bisbenzimide (Sigma-Aldrich) or 100 ng/mL DAPI (Sigma-Aldrich), and the preparations were then mounted in glycerol (Panreac)/PBS (1:1).
Immunohistochemistry
For immunohistochemistry, E5-6 chick embryos were fixed for 6–8 h at 4°C with 4% paraformaldehyde (Merck), cryopreserved ON at 4°C in PBS containing 30% sucrose (Merck), embedded in the OCT compound Tissue-Tek (Sakura), and cryosectioned (Cryostat 1950; Leica). Cryosections (12 μm) were permeabilized and blocked for 30 minutes at RT in 0.5% PBTx and 10% FCS, and they were then incubated ON at 4°C with the primary antibodies in PBTx plus 1% FCS. After 5 washes with PBTx, the sections were incubated for 1 h at RT with Alexa 594-coupled anti-mouse IgG (Invitrogen) and Cy2-conjugated Affinipure Goat Anti-Rabbit IgG (H+L), each diluted 1/1,000. The sections were finally washed 5 times in PBTx, once in PBS, and they were then incubated with 1 μg/ml bisbenzimide (Sigma-Aldrich) in PBS before mounting in glycerol/PBS (1:1). Negative controls performed using only secondary antibodies resulted in lack of specific immunostaining (data not shown). For BrdU immunostaining, cryosections were incubated with the mouse anti-BrdU antibody as previously described.Citation38 Briefly, cryosections were permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature, rinsed twice with PBS, and incubated for 1 h at 37 °C in 100 mM Tris-HCl buffer (pH 8.5) containing 2.5 mM MgCl2, 0.5 mM CaCl2, and 10 μg/ml DNAse I (Roche). Then, cryosections were rinsed twice with PBS and subjected to immunohistochemistry as described above.
DNA quantification
DNA quantification was performed by measuring DAPI levels in lipofected cells, after being immunostained as described above. DAPI labeling was recorded with a Nikon E80i microscope equipped with a DXM 1200 digital camera (Nikon, Melville, NY), using a 40× magnification objective. The integral density of DAPI, obtained as arbitrary units, was quantified using the freely available ImageJ software (National Institute of Health, Bethesda, Maryland). Intensity values were normalized to control conditions using the mean value of the integral density of DAPI in the RNAi control. Cells with DNA amounts around the modal value were considered as diploid. The rest of the cells were classified as follows: cells with DNA amount above 1.5-fold the diploid level (>2C cells), cells showing 1.8-/2.4-fold the diploid level (4C cells), and cells containing more than 2.5-fold the diploid level (>4C cells). At least 88 cells were analyzed per treatment.
Flow cytometry
Flow cytometry of unfixed frozen tissue was performed as described previously.Citation42,77 E20-P2 mouse retinas, dissected out of the pigmented epithelium, were homogeneized with a Dounce tissue grinder in DNase-free PBS containing 0.1% Triton X-100 and 1× protease inhibitor cocktail (Roche) (2 retinas/600 µl, 3 strokes with the loose pestle and 3 strokes with the tight pestle). After addition of 5% FCS and 1.25 mg/ml BSA (final concentrations), nuclei were immunostained with anti-Brn3a (1/300) and Goat anti mouse alexa 488 secondary antibody (1/500), added both together. In the control sample (generated from small aliquots of each sample), the primary antibody was excluded. Finally, nuclei were incubated ON at 4°C in the dark, and then sieved through a 40 µm nylon filter to remove nuclear aggregates. Finally, propidium iodide (PI) (Sigma-Aldrich) and DNase-free RNase I (Sigma-Aldrich) were added at a final concentration of 25 µg/ml and 25 µg/ml, respectively, and incubated for 30 min at RT. In some instances, nuclei were sieved through a 100 µm nylon filter, with similar results. The quality of the nuclei and specificity of immunostaining signal was checked by fluorescence microscopy after labeling with 100 ng/ml DAPI. Flow cytometry was performed with a FACSAria cytometer (BD Biosciences) equipped with a double argon (488 nm) and helium-neon laser (633 nm). Data were collected by using a linear digital signal process. The emission filters used were BP 530/30 for Alexa 488 and BP 616/23 for PtdIns. Data were analyzed with FACSDiva (BD Biosciences) software.
Image analysis
p27Kip1 levels were quantified using the equatorial confocal plane from cells transfected with either 1p27i, 2p27i, or Luc-i vectors. For each experimental point, p27Kip1 levels were quantified in at least 200 electroporated cells.
Statistical analyses
Quantitative data are represented as the mean ± s.e.m. Statistical differences were analyzed using either Student's t-test or ANOVA.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Aknowledgments
We are deeply in debt with I. Fariñas for her help in developing this work as well as her valuable comments on the manuscript. We thank M. J. Román for technical assistance and M. J. Palop and F. Durupt for help with the mouse colony. We are grateful to M. V. Chao for the [9992] antiserum, L. F. Reichardt for the chick TrkB antiserum, E. J. de la Rosa for the G4 antiserum, S. C. McLoon for the anti-RA4 antibody, and S. Wilson for the pRFPRNAi vectors. The monoclonal antibody G3G4 (S. Kaufman) was obtained from the Developmental Studies Hybridoma Bank (University of Iowa).
Funding
This work was supported by grants from “Ministerio de Economía y Competitividad” (SAF2012-38316) and “Fundación Ramón Areces” (CIVP16A1815).
References
- Kitagawa M, Higashi H, Jung HK, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, et al. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J 1996; 15:7060-9; PMID:9003781
- Liu N, Lucibello FC, Engeland K, Müller R. A new model of cell cycle-regulated transcription: repression of the cyclin A promoter by CDF-1 and anti-repression by E2F. Oncogene 1998; 16:2957-63; PMID:9662327
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999; 13:1501-12; PMID:10385618
- Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci USA 2009; 106:3184-9; PMID:19221029; http://dx.doi.org/10.1073/pnas.0809350106
- Gong D, Ferrell JE Jr. The roles of cyclin A2, B1, and B2 in early and late mitotic events. Mol Biol Cell 2010; 21:3149-61; PMID:20660152; http://dx.doi.org/10.1091/mbc.E10-05-0393
- Merrick KA, Fisher RP. Why minimal is not optimal: driving the mammalian cell cycle–and drug discovery–with a physiologic CDK control network. Cell Cycle 2012; 11:2600-5; PMID:22732498; http://dx.doi.org/10.4161/cc.20758
- Yasutis KM, Kozminski KG. Cell cycle checkpoint regulators reach a zillion. Cell Cycle 2013; 12:1501-9; PMID:23598718; http://dx.doi.org/10.4161/cc.24637
- Cánepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, Ogara MF. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life 2007; 59:419-26; PMID:17654117
- Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell 2008; 14:159-69; PMID:18267085; http://dx.doi.org/10.1016/j.devcel.2008.01.013
- Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell 2001; 105:297-306; PMID:11348589
- Ullah Z, Lee CY, Lilly MA, DePamphilis ML. Developmentally programmed endoreduplication in animals. Cell Cycle 2009; 8:1501-9; PMID:19372757
- Nordman J, Orr-Weaver TL. Regulation of DNA replication during development. Development 2012; 139:455-64; PMID:22223677; http://dx.doi.org/10.1242/dev.061838
- Edgar BA, Zielke N, Gutierrez C. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat Rev Mol Cell Biol 2014; 15:197-210; PMID:24556841; http://dx.doi.org/10.1038/nrm3756
- Sugimoto-Shirasu K, Roberts K. “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 2003; 6:544-53; PMID:14611952
- Madlung A, Wendel JF. Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet Genome Res 2013; 140:270-85; PMID:23751292; http://dx.doi.org/10.1159/000351430
- Li S, Yin L, Cole ES, Udani RA, Karrer KM. Progeny of germ line knockouts of ASI2, a gene encoding a putative signal transduction receptor in Tetrahymena thermophila, fail to make the transition from sexual reproduction to vegetative growth. Dev Biol 2006; 295:633-46; PMID:16712831
- Leggatt RA, Iwama GK. Occurrence of polyploidy in the fishes. Rev Fish Biol Fish 2003; 13:237-46
- Vernon JA, Butsch J. Effect of tetraploidy on learning and retention in the salamander. Science 1957; 125:1033-4; PMID:13432750
- Szaro BG, Tompkins R. Effect of tetraploidy on dendritic branching in neurons and glial cells of the frog, Xenopus laevis. J Comp Neurol 1987; 258:304-16; PMID:3584543
- Biesterfeld S, Gerres K, Fischer-Wein G, Böcking A. Polyploidy in non-neoplastic tissues. J Clin Pathol 1994; 47:38-42; PMID:8132807
- Ullah Z, Lee CY, Depamphilis ML. Cip/Kip cyclin-dependent protein kinase inhibitors and the road to polyploidy. Cell Div 2009; 4:10; PMID:19490616; http://dx.doi.org/10.1186/1747-1028-4-10
- Ullah Z, Kohn MJ, Yagi R, Vassilev LT, DePamphilis ML. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev 2008; 22:3024-36; PMID:18981479; http://dx.doi.org/10.1101/gad.1718108
- Gentric G, Celton-Morizur S, Desdouets C. Polyploidy and liver proliferation. Clin Res Hepatol Gastroenterol 2012; 36:29-34; PMID:21778131; http://dx.doi.org/10.1016/j.clinre.2011.05.011
- Duncan AW. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol 2013; 24:347-56; PMID:23333793; http://dx.doi.org/10.1016/j.semcdb.2013.01.003
- Vitrat N, Cohen-Solal K, Pique C, Le Couedic JP, Norol F, Larsen AK, Katz A, Vainchenker W, Debili N. Endomitosis of human megakaryocytes are due to abortive mitosis. Blood 1998; 91:3711-23; PMID:9573008
- Zanet J, Freije A, Ruiz M, Coulon V, Sanz JR, Chiesa J, Gandarillas A. A mitosis block links active cell cycle with human epidermal differentiation and results in endoreplication. PLoS One 2010; 5:e15701; PMID:21187932 http://dx.doi.org/10.1371/journal.pone.0015701
- Nagata Y, Jones MR, Nguyen HG, McCrann DJ, St Hilaire C, Schreiber BM, Hashimoto A, Inagaki M, Earnshaw WC, Todokoro K, et al. Vascular smooth muscle cell polyploidization involves changes in chromosome passenger proteins and an endomitotic cell cycle. Exp Cell Res 2005; 305:277-91; PMID:15817153
- Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development 2013; 140:3-12; PMID:23222436; http://dx.doi.org/10.1242/dev.080531
- Lilly MA, Spradling AC. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev 1996; 10:2514-26; PMID:8843202
- Follette PJ, Duronio RJ, O'Farrell PH. Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr Biol 1998; 8:235-8; PMID:9501987
- Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cyclin E ablation in the mouse. Cell 2003; 114:431-43; PMID:12941272
- Parisi T, Beck AR, Rougier N, McNeil T, Lucian L, Werb Z, Amati B. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J 2003; 22: 4794-803; PMID:12970191
- Lasek RJ, Dower WJ. Aplysia californica: analysis of nuclear DNA in individual nuclei of giant neurons. Science 1971; 172: 278-80; PMID:5548710
- Yamagishi M, Ito E, Matsuo R. DNA endoreplication in the brain neurons during body growth of an adult slug. J Neurosci 2011; 31:5596-604; PMID:21490200; http://dx.doi.org/10.1523/JNEUROSCI.0179-11.2011
- Smith TG Jr, Futamachi K, Ehrenstein G. Site of action potential generation in a giant neuron of Aplysia californica. Brain Res 1982; 242:184-9; PMID:6286048
- Mosch B, Morawski M, Mittag A, Lenz D, Tarnok A, Arendt T. Aneuploidy and DNA replication in the normal human brain and Alzheimer's disease. J Neurosci 2007; 27:6859-67; PMID:17596434
- Morillo SM, Escoll P, de la Hera A, Frade JM. Somatic tetraploidy in specific chick retinal ganglion cells induced by nerve growth factor. Proc Natl Acad Sci USA 2010; 107:109-14; PMID:20018664; http://dx.doi.org/10.1073/pnas.0906121107
- López-Sánchez N, Ovejero-Benito MC, Borreguero L, Frade JM. Control of neuronal ploidy during vertebrate development. Results Probl Cell Differ 2011; 53:547-63; PMID:21630159; http://dx.doi.org/10.1007/978-3-642-19065-0_22
- Morillo SM, Abanto EP, Román MJ, Frade JM. Nerve growth factor-induced cell cycle reentry in newborn neurons is triggered by p38MAPK-dependent E2F4 phosphorylation. Mol Cell Biol 2012; 32:2722-37; PMID:22586272; http://dx.doi.org/10.1128/MCB.00239-12
- López-Sánchez N, Ovejero-Benito MC, Rodríguez-Ruiz C, Frade, JM. NGF/p75NTR in cell cycle and neuronal tetraploidy. In: Kostrzewa R, editor. Handbook of Neurotoxicity. Heidelberg: Springer Verlag 2014:1877-97
- Ovejero-Benito MC, Frade JM. Brain-derived neurotrophic factor-dependent cdk1 inhibition prevents G2/M progression in differentiating tetraploid neurons. PLoS One 2013; 8:e64890; PMID:23741412; http://dx.doi.org/10.1371/journal.pone.0064890
- López-Sánchez N, Frade JM. Genetic evidence for p75NTR-dependent tetraploidy in cortical projection neurons from adult mice. J Neurosci 2013; 33:7488-500; PMID:23616554; http://dx.doi.org/10.1523/JNEUROSCI.3849-12.2013
- Lipinski MM, Macleod KF, Williams BO, Mullaney TL, Crowley D, Jacks T. Cell-autonomous and non-cell-autonomous functions of the Rb tumor suppressor in developing central nervous system. EMBO J 2001; 20:3402-13; PMID:11432828
- Cunningham JJ, Levine EM, Zindy F, Goloubeva O, Roussel MF, Smeyne RJ. The cyclin-dependent kinase inhibitors p19(Ink4d) and p27(Kip1) are coexpressed in select retinal cells and act cooperatively to control cell cycle exit. Mol Cell Neurosci 2002; 19:359-74; PMID:11906209
- Tsukiji N, Nishihara D, Yajima I, Takeda K, Shibahara S, Yamamoto H. Mitf functions as an in ovo regulator for cell differentiation and proliferation during development of the chick RPE. Dev Biol 2009; 326:335-46; PMID:19100253; http://dx.doi.org/10.1016/j.ydbio.2008.11.029
- Rathjen FG, Wolff JM, Frank R, Bonhoeffer F, Rutishauser U. Membrane glycoproteins involved in neurite fasciculation. J Cell Biol 1987; 104:343-53; PMID:3805123
- Portugal LC, Ventura AL. Localization of p27kip1 in the developing avian retina: sustained expression in the mature tissue. Neurosci Lett 2009; 466:35-40; PMID:19786071; http://dx.doi.org/10.1016/j.neulet.2009.09.046
- Frade JM, Martí E, Bovolenta P, Rodríguez-Peña MA, Pérez-García D, Rohrer H, Edgar D, Rodríguez-Tébar A. Insulin-like growth factor-I stimulates neurogenesis in chick retina by regulating expression of the alpha 6 integrin subunit. Development 1996; 122:2497-506; PMID:8756294
- Frade JM, Martínez-Morales JR, Rodríguez-Tébar A. Laminin-1 selectively stimulates neuron generation from cultured retinal neuroepithelial cells. Exp Cell Res 1996; 222:140-9; PMID:8549656
- Frade JM. Unscheduled re-entry into the cell cycle induced by NGF precedes cell death in nascent retinal neurones. J Cell Sci 2000; 113:1139-48; PMID:10704365
- Das RM, Van Hateren NJ, Howell GR, Farrell ER, Bangs FK, Porteous VC, Manning EM, McGrew MJ, Ohyama K, Sacco MA, et al. A robust system for RNA interference in the chicken using a modified microRNA operon. Dev Biol 2006; 294:554-63; PMID:16574096
- de Curtis I, Quaranta V, Tamura RN, Reichardt LF. Laminin receptors in the retina: sequence analysis of the chick integrin alpha 6 subunit. Evidence for transcriptional and posttranslational regulation. J Cell Biol 1991; 113:405-16; PMID:1826298
- Watanabe M, Rutishauser U, Silver J. Formation of the retinal ganglion cell and optic fiber layers. J Neurobiol 1991; 22: 85-96; PMID:2010752
- Liu W, Khare SL, Liang X, Peters MA, Liu X, Cepko CL, Xiang M. All Brn3 genes can promote retinal ganglion cell differentiation in the chick. Development 2000; 127: 3237-47; PMID:10887080
- Nallamshetty S, Crook M, Boehm M, Yoshimoto T, Olive M, Nabel EG. The cell cycle regulator p27Kip1 interacts with MCM7, a DNA replication licensing factor, to inhibit initiation of DNA replication. FEBS Lett 2005; 579:6529-36; PMID:16289477
- Dräger UC. Birth dates of retinal ganglion cells giving rise to the crossed and uncrossed optic projections in the mouse. Proc R Soc Lond B Biol Sci 1985; 224:57-77; PMID:2581263
- Quina LA, Pak W, Lanier J, Banwait P, Gratwick K, Liu Y, Velasquez T, O'Leary DD, Goulding M, Turner EE. Brn3a-expressing retinal ganglion cells project specifically to thalamocortical and collicular visual pathways. J Neurosci 2005; 25:11595-604; PMID:16354917
- Foster DA, Yellen P, Xu L, Saqcena M. Regulation of G1 cell cycle progression: distinguishing the pestriction point from a nutrient-sensing cell growth checkpoint(s). Genes Cancer 2010; 1:1124-31; PMID:21779436; http://dx.doi.org/10.1177/1947601910392989
- Bisbis B, Delmas F, Joubès J, Sicard A, Hernould M, Inzé D, Mouras A, Chevalier C. Cyclin-dependent kinase (CDK) inhibitors regulate the CDK-cyclin complex activities in endoreduplicating cells of developing tomato fruit. J Biol Chem 2006; 281:7374-83; PMID:16407228
- Prada C, Puga J, Pérez-Méndez L, López R, Ramírez G. Spatial and temporal patterns of neurogenesis in the chick retina. Eur J Neurosci 1991; 3:559-569; PMID:12106488
- Donovan SL, Corbo JC. Retinal horizontal cells lacking Rb1 sustain persistent DNA damage and survive as polyploid giant cells. Mol Biol Cell 2012; 23:4362-72; PMID:23015754; http://dx.doi.org/10.1091/mbc.E12-04-0293
- Shirazi Fard S, Jarrin M, Boije H, Fillon V, All-Eriksson C, Hallböök F. Heterogenic final cell cycle by chicken retinal Lim1 horizontal progenitor cells leads to heteroploid cells with a remaining replicated genome. PLoS One 2013; 8(3):e59133; PMID:23527113; http://dx.doi.org/10.1371/journal.pone.0059133
- Kim JA, Lee J, Margolis RL, Fotedar R. SP600125 suppresses Cdk1 and induces endoreplication directly from G2 phase, independent of JNK inhibition. Oncogene 2010; 29:1702-16; PMID:20062077; http://dx.doi.org/10.1038/onc.2009.464
- Levine EM, Close J, Fero M, Ostrovsky A, Reh TA. p27(Kip1) regulates cell cycle withdrawal of late multipotent progenitor cells in the mammalian retina. Dev Biol 2000; 219:299-314; PMID:10694424
- Dyer MA, Cepko CL. p27Kip1 and p57Kip2 regulate proliferation in distinct retinal progenitor cell populations. J Neurosci 2001; 21:4259-71; PMID:11404411
- Lopez-Sanchez E, Frances-Muñoz E, Chaques V, Sanchez-Benavent ML, Menezo JL. Optic nerve alterations in P27(Kip1) knockout mice. Eur J Ophthalmol 2007; 17:377-82; PMID:17534820
- Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell 1996; 85:733-44; PMID:8646781
- Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh DY, Nakayama K. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 1996; 85:707-20; PMID:8646779
- Kiyokawa H, Kineman RD, Manova-Todorova KO, Soares VC, Hoffman ES, Ono M, Khanam D, Hayday AC, Frohman LA, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1). Cell 1996; 85:721-32; PMID:8646780
- Akashiba H, Matsuki N, Nishiyama N. p27 small interfering RNA induces cell death through elevating cell cycle activity in cultured cortical neurons: a proof-of-concept study. Cell Mol Life Sci 2006; 63:2397-404; PMID:17006629
- Nguyen L, Besson A, Heng JI, Schuurmans C, Teboul L, Parras C, Philpott A, Roberts JM, Guillemot F. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev 2006; 20:1511-24; PMID:16705040
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol 1951; 88:49-92; PMID:24539719
- Kaufman MH. The Atlas of Mouse Development. San Diego: Academic Press, Inc.; 1992
- McLoon SC, Barnes RB. Early differentiation of retinal ganglion cells: an axonal protein expressed by premigratory and migrating retinal ganglion cells. J Neurosci 1989; 9:1424-32; PMID:2703885
- Waid DK, McLoon SC. Immediate differentiation of ganglion cells following mitosis in the developing retina. Neuron 1995; 14:117-24; PMID:7826629
- Frade JM, Rodríquez Tébar A. Neuroepithelial differentiation induced by ECM molecules. Methods Mol Biol 2000; 139:257-64; PMID:10840793
- López-Sánchez N, Frade JM. Flow cytometric analysis of DNA synthesis and apoptosis in central nervous system using fresh cell nuclei. Methods Mol Biol 2015; 1254:33-42; PMID:25431055; http://dx.doi.org/10.1007/978-1-4939-2152-2_3
- Frade JM. Interkinetic nuclear movement in the vertebrate neuroepithelium: encounters with an old acquaintance. Prog Brain Res 2002; 136:67-71; PMID:12143404
