Abstract
The COP9 signalosome subunit 6 (CSN6), which is involved in ubiquitin-mediated protein degradation, is overexpressed in many types of cancer. CSN6 is critical in causing p53 degradation and malignancy, but its target in cell cycle progression is not fully characterized. Constitutive photomorphogenic 1 (COP1) is an E3 ubiquitin ligase associating with COP9 signalosome to regulate important target proteins for cell growth. p27 is a critical G1 CDK inhibitor involved in cell cycle regulation, but its upstream regulators are not fully characterized. Here, we show that the CSN6-COP1 link is regulating p27Kip1 stability, and that COP1 is a negative regulator of p27Kip1. Ectopic expression of CSN6 can decrease the expression of p27Kip1, while CSN6 knockdown leads to p27Kip1 stabilization. Mechanistic studies show that CSN6 interacts with p27Kip1 and facilitates ubiquitin-mediated degradation of p27Kip1. CSN6-mediated p27 degradation depends on the nuclear export of p27Kip1, which is regulated through COP1 nuclear exporting signal. COP1 overexpression leads to the cytoplasmic distribution of p27, thereby accelerating p27 degradation. Importantly, the negative impact of COP1 on p27 stability contributes to elevating expression of genes that are suppressed through p27 mediation. Kaplan-Meier analysis of tumor samples demonstrates that high COP1 expression was associated with poor overall survival. These data suggest that tumors with CSN6/COP1 deregulation may have growth advantage by regulating p27 degradation and subsequent impact on p27 targeted genes.
Introduction
The COP9 signalosome (CSN) is involved in protein ubiquitination, transcriptional activation,Citation1,2 signal transduction,Citation3-6 DNA damage response/genome integrity,Citation7 and tumorigenesis.Citation5,8-11 The role of the CSN's subunits in cancer has not been well characterized. We previously showed that Csn6−/− mice developed until 7.5 d post-coitus.Citation10 We showed that Csn6 haplo-insufficiency mitigated the development of cancer in a Csn6+/− mouse tumor experiment,Citation10 which indicates that CSN6 expression level is pivotal for tumorigenesis. CSN6 expression is elevated in cancers and leads to poor survival,Citation5,12,13 but the level and biological consequence of CSN6 expression in cancer remain unclear. Constitutive photomorphogenic 1 (COP1) is a RING-containing E3 ligase, which is involved in various cellular functions, including cell proliferation and survival. COP1 facilitates the degradation of many substrates via ubiquitin-mediated protein degradation.Citation14,15 COP1 itself is self-ubiquitinated, and this process is under the regulation of COP9 signalosome subunit 6 (CSN6).Citation6
The tumor suppressor p27 inhibits Cyclin-dependent kinase (CDK) and is important for regulating the cell cycle G1 to S transition Citation16 and contact inhibition.Citation17 p27 is regulated by many oncogenic signals for facilitating cell cycle progression.Citation18-26 Protein levels of p27 are tightly controlled for cell cycle progression,Citation16,17,27 and p27 levels are positively regulated by the activity of many tumor suppressors.Citation22,28 As a negative regulator of the cell cycle, p27 is also functioning like a tumor suppressor. p27 levels are mainly regulated by polyubiquitination,Citation29 and p27 are downregulated in many types of cancer. However its downregulation mechanism in cancers is not fully characterized.
In this study, we show that CSN6 has a negative impact on p27 stability and further indicate that CSN6-associated protein, COP1, is involved in negatively regulating p27 stability. We characterized the mechanism of CSN6-COP1 link in destabilizing p27, and demonstrated that COP1-mediated p27 nuclear export is critical for p27 degradation. Significantly, we showed that COP1 overexpression alleviates p27-mediated gene suppression in cancers. Our data provide important insight into the signal of the CSN6-COP1 axis in regulating p27 to promote cancer growth.
Results
CSN6 associates with p27 and increases p27 turnover rate
We previously show that CSN6 +/− MEF has elevated p27 when compared with wt CSN6 +/+ MEFs,Citation11 we thus hypothesized that CSN6 and p27 have an interactive or regulatory relationship. Co-immunoprecipitation experiments showed endogenous interaction of the 2 proteins in cells (). We then showed that CSN6 was able to downregulate steady-state expression of exogenous p27 in a dose-dependent manner in cells (). Similar results were obtained for levels of endogenous p27 ().
Figure 1. CSN6 interacts with p27 and regulates its stability. (A) Indicated expression vectors were transfected into 293T cells. Lysates were analyzed by IP with Flag and IB with anti-HA. CSN6 interacts with endogenous p27. Lysates of HCT116 cells were prepared and equal amounts of cell lysates were analyzed by immunoprecipitation (IP) with either control mouse IgG or CSN6 and analyzed by immunoblotting (IB) with anti-p27. (B) CSN6 reduced the steady-state expression of p27 in a dose-dependent manner. 293T cells were co-transfected with the indicated expression vectors. Equal amounts of protein from cell lysates were analyzed by immunoblotting with the indicated antibodies. Knockdown of CSN6 upregulates the expression of p27 protein level. Lysates of HCT116 cells infected with either CSN6-shRNA or control shRNA were analyzed by IB with the indicated antibodies. (C) CSN6-mediated destabilization of p27 is proteasome-dependent. 293T cells co-transfected with the either Myc-CSN6 or vector control was treated with or without proteasome inhibitor MG132 before collecting lysates. Lysates were immunoblotted with indicated antibodies. (D) CSN6 increases the turnover of p27. 293T cells co-transfected with the indicated expression vectors were treated with cycloheximide (CHX) (100 µg/ml) for the indicated times. Equal amounts of protein from cell lysates were immunoblotted with indicated antibodies.
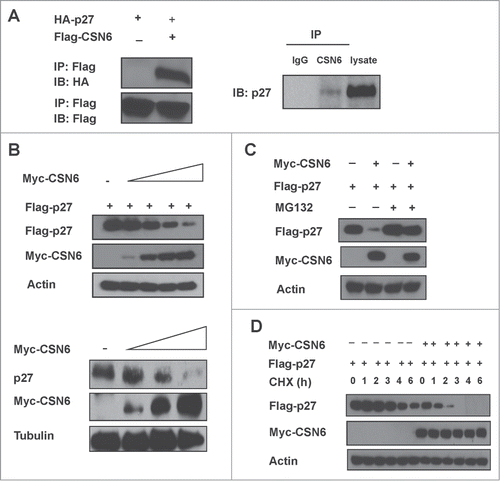
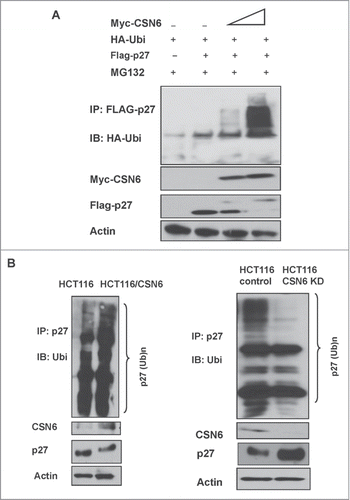
CSN6-mediated p27 downregulation can be rescued by the proteasome inhibitor MG132 (). Also, CSN6 increased the turnover rate of p27 in the presence of the de novo protein synthesis inhibitor cycloheximide (). We then found that overexpression of CSN6 increased the ubiquitination level of p27 in a dose-dependent manner (). Also, CSN6 facilitated the ubiquitination process of endogenous p27, whereas CSN6 knockdown reduced the endogenous ubiquitination level of p27 (). Together, these results suggest that CSN6 downregulates p27 by enhancing ubiquitin-mediated degradation.
Figure 2. CSN6 increases p27 poly-ubiquitination. (A) 293T cells were transfected with indicated expressing plasmids. MG132 was added 6 h before they were harvested. The cell lysates was then immunoprecipitated with anti-Flag and immunoblotted with anti-HA antibody. Equal amount of whole cell lysates were immunoblotted with anti-myc or Actin. (B) HCT116 cells were transfected with CSN6 or knocked down with CSN6 shRNA. The cell lysates from indicated cells were immunoprecipitated with anti-p27 and immunoblotted with anti-ubiquitin antibody. Equal amounts of cell lysates were analyzed by IB with the indicated antibodies.
CSN6 cooperates with COP1 to downregulate p27
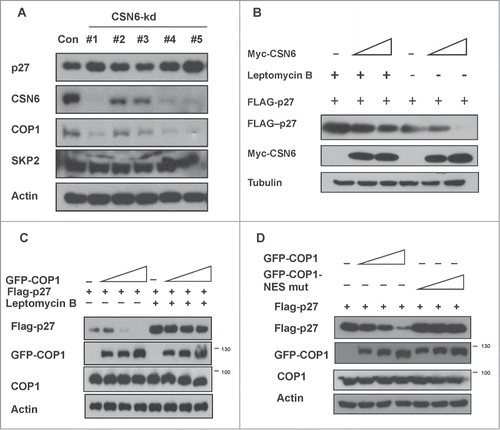
CSN6 usually collaborate with other E3 ligase to regulate target proteins. We then examined whether any E3 ligase is involved in CSN6-mediated 27 degradation. We found that p27 levels were elevated when cells were treated with CSN6-shRNA virus to perform CSN6 knockdown (). As expected, levels of p27 are elevated in cells with CSN6 knockdown. We examined 2 E3 ligases expression level and found that COP1 is downregulated following the CSN6 knockdown, while Skp2, a known E3 ligase for p27, is not changed (). Also, we showed that CSN6-mediated p27 degradation could be antagonized by leptomycin B, an inhibitor of nuclear export, suggesting that CSN6-mediated p27 degradation involves the subcellular localization of p27 (). Given that COP1 is critical in regulating target proteins through nuclear exporting and that COP1 is downregulated following CSN6 knockdown, we then examined whether COP1 is critical in regulating p27 stability and whether this process is depending on nuclear exporting. We showed that COP1 could mediate downregulation of p27 in a dose-dependent manner, and found that COP1-mediated p27 degradation depends on the nuclear export of p27 as blocking p27 nuclear export with leptomycin B diminished COP1-mediated p27 degradation (). Furthermore, the COP1 NES mutant (L242A/L244A) failed to downregulate p27 levels compared with wt COP1 (), suggesting that the COP1 nuclear export signal is coupled with p27 degradation.
Figure 3. COP1-mediated nuclear export of p27 is involved in CSN6-mediated p27 ubiquitination (A) COP1 is downregulated following the CSN6 knockdown. 293T cells were co-transfected with the indicated expression vectors. Lysates were immunoblotted with the indicated antibodies. (B) CSN6-mediated p27 downregulation is diminished by the leptomycin B. 293T cells were co-transfected with increasing myc-CSN6 expression vectors. Cells were treated with or without leptomycin B (20 ng/ml) for 6 hours before lysates were collected. Lysates were immunoblotted with the indicated antibodies. (C) Leptomycin B rescued COP1-mediated p27 downregulation. 293T cells were co-transfected with the indicated expression vectors. Cells were treated with or without leptomycin B (20 ng/ml) for 6 hours before lysates were collected. Lysates were immunoblotted with the indicated antibodies. (D) Mutation in the COP1 nuclear export signal (NES) sequence impaired the ability of COP1 to downregulate p27 expression. 293T cells were co-transfected with the indicated expression vectors. Lysates were immunoblotted with the indicated antibodies.
COP1-mediated p27 nuclear export depends on NES
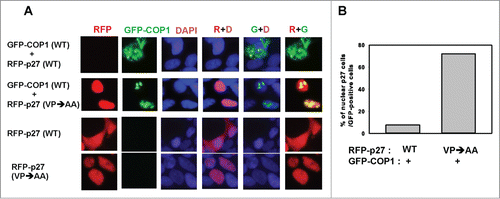
To further investigate the relationship between COP1-mediated nuclear export of p27 and degradation, we performed the immunofluorescence. Immunofluorescence studies showed that leptomycin B reduced cytoplasmic accumulation of COP1 (punctate green staining), leading to p27 accumulation (). The COP1 NES mutant also showed reduced levels in the cytoplasm, again resulting in p27 accumulation (). The percentage of nuclear p27 was quantitated and is presented as a bar graph (). We also showed that the nuclear staining of p27 (VPAA) mutant, which cannot bind COP1 and is more stable,Citation30 was not diminished by COP1, as demonstrated by abundant levels of nuclear p27 (), suggesting that COP1-mediated p27 nuclear export occurs through direct binding. Taken together, these results show that dynamic distribution of COP1 and directing binding of p27 are critical in COP1-mediated p27 degradation.
Figure 4. COP1 induces p27 nuclear export. (A) Leptomycin B increased p27 nuclear accumulation in the presence of COP1. 293T cells co-transfected with either wild-type (wt) or NES mutant GFP-COP1 and RFP-p27, cultured with or without leptomycin B (LMB), were stained with DAPI. (B) Percentages of nuclear p27 signals among GFP-positive cells in A are shown.
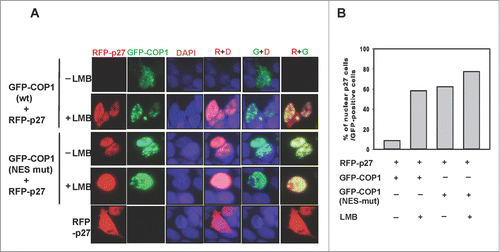
Figure 5. COP1 interacts with p27 to mediate nuclear export. (A) The Flag-p27 (VPAA) mutant nuclear staining is not diminished in the presence of COP1. 293T cells were co-transfected with either wild-type (wt) or VPAA mutant RFP-p27 and GFP-COP1 and stained with DAPI. (B) Percentages of nuclear p27 signals among GFP-COP1-positive cells in A are shown.
COP1-p27 axis deregulation in cancers triggers the expression of several target genes suppressed by p27
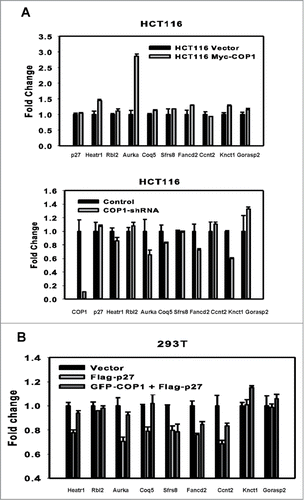
p27 plays a role in suppressing genes involved in mitosis or cell proliferation.Citation31 Transfection studies showed that COP1 can rescue suppression of genes suppressed by p27, such as activating the gene expression of Aurora A (), while COP1 knockdown suppressed the gene expression of Aurora A, among others (). To confirm the antagonizing effect of COP1 on the p27 transcriptional suppression activity, we found that enforced expression of COP1 in p27 expressing cells can rescue the expression of several target genes suppressed by p27 (). Together, COP1 overexpression resulted in p27 downregulation and subsequent elevation of p27-mediated suppression of targeted genes.
Figure 6. COP1 overexpression leads to upregulation of p27-mediated suppressed genes. (A, B) COP1 regulated the genes that are suppressed through p27 mediation. COP1-overexpression or COP1 knockdown on gene expression of p27 target genes are shown. mRNA levels of the indicated p27 target genes were determined by quantitative reverse transcriptase PCR. (C) COP1 antagonized the suppressive impact of p27 on the expression of several targeted genes.
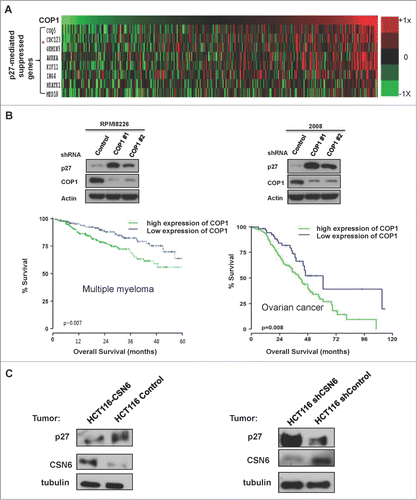
To further understand the COP1–p27 link in influencing gene expression in cancer, we performed transcriptome analysis in cancer samples and demonstrated that COP1 overexpression in human cancer samples was associated with elevated expression of genes that are suppressed through p27 mediation (). Cell line studies showed that COP1 knockdown caused p27 upregulation in multiple myeloma cell line (RPMI8226) or ovarian cancer cell line (2008) (). We then found that high levels of COP1 expression were associated with poor overall survival in a cohort of patients with multiple myeloma and another cohort of patients with ovarian cancer by Kaplan-Meier analysis (). In addition, in mouse xenograft HCT116 cancer model studies, p27 level is regulated according to the status of CSN6 cells. CSN6 expressing tumors have low levels of p27, while CSN6 knockdown tumors have high p27 levels (). These data suggest that tumors with COP1/CSN6 deregulation may have growth advantage by regulating p27 expression and affecting p27 target genes accordingly.
Figure 7. COP1-p27 link correlates with poor survival. (A) COP1 overexpression in cancer led to increased expression of genes suppressed through p27 mediation. Relative expression of COP1 and genes suppressed through p27 mediation (COQ5, CDC123, GEMIN5, AURKA, KIF11, ING4, HEARTR1, and MED18) were analyzed from a data set of multiple myeloma samples. Data are presented as a heat map. (B) High expression of COP1 was associated with poor overall survival. Lysates of indicated multiple myeloma cells (RPMI8226) or ovarian cancer cells (2008) infected with either COP1-shRNA or control shRNA were analyzed by IB with the indicated antibodies. Kaplan-Meier overall survival curves for 414 patients with multiple myeloma (left) or ovarian cancer (right), classified by COP1 expression, are shown. (C) Protein levels of p27 were regulated according to COP1 expression in xenograft mouse models. HCT116 cells with CSN6 overexpression or knockdown were subcutaneously injected into the nude mice. Tumor samples were collected and lysates of indicated tumor samples were analyzed by IB with the indicated antibodies.
Discussion
We demonstrated that CSN6 is involved in regulating p27. Both CSN6 and E3 ligase COP1 are working together to regulate the ubiquitination process of p27. In biochemical studies, depletion of CSN6 expression resulted in reduction of COP1 and concurrent elevation of p27, indicating that CSN6-COP1 link has a critical role in p27 stabilization. CSN6 is a critical ubiquitination regulator involved in tumorigenesis and cell cycle regulation,Citation5,6 but its role in cell cycle remains not fully understood. We showed that CSN6 has uncharacterized biological activity in degrading p27, and that COP1 acts as an E3 ligase to catalyze this process, thereby mitigating p27 targeted gene suppression. This study demonstrates CSN6's role in promoting cancer development by influencing the stability of a major CDK inhibitor––p27 in cell cycle progression.
Since COP1 can suppress p53 activity,Citation32 it is deemed as an oncoprotein. On the other hand, COP1 knockout mouse model study shows that COP1 could act like a tumor suppressor by compromising the oncogenic activity of c-Jun and ETV1 Citation14,33,34 in certain tissues. Thus, its role in cancer is controversial. To define its role in cancer, its many substrates need to be characterized. The fact that COP1 functions as an E3 ligase of p27 to mediate p27 ubiquitination suggests that COP1 has an oncogenic role. On the other hand, the COP1 KO mouse model presents an unexpected discovery regarding promoting cancer growth in certain tissues, which suggests its tumor suppressor role. Our data analysis indicates that COP1 is highly elevated in many types of cancer and correlates with poor patient survival, which is consistent with its role as a negative regulator of tumor suppressor p27. More studies will be needed to resolve the discrepancy about COP1's role as a tumor suppressor.
Cytoplasmic distribution of COP1 and p27 is critical for their stability regulation.Citation35-37 The biological consequence of the link between 2 proteins' subcellular localization is presented in this study. CSN6's impact on p27 degradation is also related with p27 nuclear exporting, which is possibly mediated by COP1. Our study revealed that COP1 reduces the steady-state expression of p27 through nuclear exporting. The impact of COP1 on p27 subcellular localization is critical as p27 cytoplasmic localization changes the characteristic of p27. p27 is a nuclear protein with a tumor suppressive role, but oncogenic signal-mediated cytoplasmic retention and nuclear export of p27 results in enhanced oncogenic activity Citation28,38 or drug resistance.Citation26 Once translocated to the cytoplasm, p27 will gain the function of promoting cancer cell migration and invasiveness.Citation38 Thus, cytosolic p27 has a metastasis promoting function opposite to the tumor suppressor role of nuclear p27. The results that COP1 can reduce nuclear portion of p27 underscore the oncogenic potential of COP1 in the development of cancer. Also the COP1-p27 link is translated into the regulation of genes transcriptionally suppressed by p27, including mitotic Aurora kinase A,Citation39 providing explanation to a possible plethora of functions of CSN6-COP1 axis. Significantly, COP1 overexpression correlates with poor survival in cancer patients, attesting that CSN6/COP1 deregulation in cancer will lead to growth advantage by having p27 downregulation and subsequent alleviating p27-suppressed target genes.
Together, our findings provide important insight into the mechanisms by which CSN6 facilitates p27 degradation in cell. That CSN6 and COP1 collaborate to negatively regulate p27 stability and cause cytoplasmic mislocation indicates that blocking the CSN6-COP1 signaling axis is a feasible therapeutic strategy in cancers with downregulated p27.
Materials and Methods
Cell lines and reagents
HCT116 cells and HEK-293T (human embryonic kidney cell line) obtained from the ATCC. Cells were maintained in Dulbecco's modified Eagle's medium/nutrient F12 media (in-house supplier) supplemented with 10% (v/v) fetal bovine serum. Human RPMI8226 and 2008 cells (from ATCC) were maintained in RPMI-1640 medium supplemented with fetal bovine serum and antimicrobials as mentioned above. pcDNA6-Myc-COP1 and myc-CSN6 was constructed by our lab. pCMV5-Flag-COP1 was kindly provided by E. Bianchi. Wild-type (wt) GFP-COP1, GFP-COP1 NES, RFP-p27, RFP-p27 Flag-p27 (VPAA) mutant were constructed by PCR cloning. Flag-CSN6, Myc-CSN6 were previously described.Citation10 ShCSN6 RNA was constructed as previously described.Citation4–6Citation40 MG132 (C2211), cycloheximide (C4895) and Puromycin (P7255) were purchased from Sigma. Antibodies: CSN6 (ENZO life sciences), Flag (M2 monoclonal antibody, Sigma), Myc (abcam), Actin (Sigma).
Transfection and generation of stable transfectants
Transfection was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For the generation of CSN6 knockdown cells, HCT116 cells were infected with lentiviral shRNA transduction particles (GIPZ COPS6 shRNA Transfection kit, Thermo) containing either control shRNA or CSN6 shRNA. After infection, the cells were selected with 2 ug/ml puromycin for 2 weeks according to the manufacturer's protocols. The antibiotic-resistant colonies were then picked, pooled, and expanded for further analysis under selective conditions.
Immunoprecipitation and immunoblotting
Total cell lysates were solubilized in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium fluoride, 5 mM sodium orthovanadate, and 1 µg/ml each of aprotinin, leupeptin, and pepstatin) and processed as previously described.Citation41,42 Lysates were immunoprecipitated with indicated antibodies. Proteins were resolved by SDS-PAGE gels and proteins were transferred to polyvinylidene difluoride membranes (Millipore). The membranes were blocked with 5% nonfat milk for 1 hour at room temperature prior to incubation with indicated primary antibodies. Subsequently, membranes were washed and incubated for 1 hour at room temperature with peroxidase-conjugated secondary antibodies (Thermo Scientific). Following several washes, chemiluminescent images of immunodetected bands on the membranes were recorded on X-ray films using the enhanced chemiluminescence system (Millipore).
Ubiquitination assay
Ubiquitination was performed as previously described.Citation43–45 Basically, indicated cells cotransfected with indicated plasmids were used for the experiments. At 24 h post-transfection, cells were treated with 50 μg/ml of MG132 for 6 h. The ubiquitinated proteins were immunoprecipitated with anti-Flag or anti-p27. The protein complexes were then resolved by SDS-polyacrylamide gel and probed with anti-HA or anti-Ubi to visualize the level of ubiquitination.
Protein turnover assay
Protein turnover was performed as previously described.Citation43,44,46 The cells were transfected with the indicated plasmids and incubated at 37°C with 5% (vol/vol) CO2 for 24 h. Then cycloheximide was added into the media to a final concentration of 100 μg/ml. The cells were harvested at the indicated times after CHX treatment. The protein levels were analyzed by immunoblotting.
Immunofluorescence microscopy
Images of cells were captured with an Olympus FV300 microscope or Zeiss Axiovert 200 M microscope. The experiments were performed as previously described.Citation36
Quantitative PCR
We used primers for real-time quantitative PCR of p27 target genes as referenced in Primer Bank (http://pga.mgh.harvard.edu/primerbank/). Total RNA was extracted from cells using Trizol (Invitrogen); 1 µg RNA was used for producing cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative real-time PCR analyses were performed using iQ SYBR Green Super mix (Bio-Rad, 170-8882) and the iCycler iQ real-time PCR detection system. The genes' amplification folds were analyzed relative to controls.
Survival analysis
Samples were profiled using the U133A Plus 2.0 Gene Expression Arrays (Affymetrix). The CEL files were downloaded from Gene Expression Omnibus (GEO) and justRMA was used to compute expression values. For survival analysis, patients were grouped into percentiles according to COP1 expression. We assessed the relationship between COP1 expression and survival by choosing an optimal cutoff to split the samples into two groups. The log-rank test was employed to determine the significance of the association between COP1 expression and overall survival. The Kaplan-Meier method was used to generate survival curves. Microarray and clinical data were obtained from the gene expression profiles of the multiple myeloma dataset of Oncomine and GEO (GSE2658).Citation47
Xenograft experiment
Athymic (nu/nu) mice were housed in AAALAC-approved barrier facilities. CSN6-expressing or CSN6 knockdown HCT116 cells were harvested and injected into the flank of each mouse. At the end of the experiment, the tumors were removed for biochemical analysis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by grants from the National Institutes of Health (R01CA089266 to MHL), Directed Medical Research Programs (DOD SIDA BC062166 to SJY and MHL), Susan G. Komen Breast Cancer Foundation (KG081048 to SJY and MHL), and the Fidelity foundation to MHL. The University of Texas MD Anderson Cancer Center is supported by NIH core grant CA16672.
References
- Wei N, Deng XW. Making sense of the COP9 signalosome. A regulatory protein complex conserved from Arabidopsis to human. Trends Genet 1999; 15:98-103; PMID:10203806; http://dx.doi.org/10.1016/S0168-9525(98)01670-9
- Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, Gordon C, Naumann M, Dubiel W. A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J 1998; 12:469-78; PMID:9535219
- Zhang XC, Chen J, Su CH, Yang HY, Lee MH. Roles for CSN5 in control of p53/MDM2 activities. J Cell Biochem 2008; 103:1219-30; PMID:17879958; http://dx.doi.org/10.1002/jcb.21504
- Chen B, Zhao R, Su CH, Linan M, Tseng C, Phan L, Fang L, Yang HY, Yang H, Wang W, et al. CDK inhibitor p57 (Kip2) is negatively regulated by COP9 signalosome subunit 6. Cell Cycle 2012; 11:4633-41; PMID:23187808; http://dx.doi.org/10.4161/cc.22887
- Xue Y, Chen J, Choi HH, Phan L, Chou PC, Zhao R, Yang H, Santiago J, Liu M, Yeung GE, et al. HER2-Akt signaling in regulating COP9 signalsome subunit 6 and p53. Cell Cycle 2012; 11:4181-90; PMID:23095642; http://dx.doi.org/10.4161/cc.22413
- Choi HH, Gully C, Su CH, Velazquez-Torres G, Chou PC, Tseng C, Zhao R, Phan L, Shaiken T, Chen J, et al. COP9 signalosome subunit 6 stabilizes COP1, which functions as an E3 ubiquitin ligase for 14-3-3sigma. Oncogene 2011; 30:4791-801; PMID:21625211; http://dx.doi.org/10.1038/onc.2011.192
- Choi HH, Su CH, Fang LK, Zhang J, Yeung SC, Lee MH. CSN6 deregulation impairs genome integrity in a COP1-dependent pathway. Oncotarget 2015; in press
- Richardson KS, Zundel W. The emerging role of the COP9 signalosome in cancer. Mol Cancer Res 2005; 3:645-53; PMID:16380502; http://dx.doi.org/10.1158/1541-7786.MCR-05-0233
- Zhao R, Phan L, Chen B, Yang HY, Chen J, Che TF, Qiao Y, Zhang J, Yeung SC, Lee MH. Ubiquitination-mediated p57Kip2 Degradation by CSN5 Confers Cancer Cell Proliferation. Cancer Hallmarks 2014; 1:133-44; http://dx.doi.org/10.1166/ch.2013.1013
- Zhao R, Yeung SC, Chen J, Iwakuma T, Su CH, Chen B, Qu C, Zhang F, Chen YT, Lin YL, et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest 2011; 121:851-65; PMID:21317535; http://dx.doi.org/10.1172/JCI44111
- Chen J, Shin JH, Zhao R, Phan L, Wang H, Xue Y, Post SM, Ho Choi H, Chen JS, Wang E, et al. CSN6 drives carcinogenesis by positively regulating Myc stability. Nat Commun 2014; 5:5384; PMID:25395170; http://dx.doi.org/10.1038/ncomms6384
- Lee MH, Zhao R, Phan L, Yeung SC. Roles of COP9 signalosome in cancer. Cell Cycle 2011; 10:3057-66; PMID:21876386; http://dx.doi.org/10.4161/cc.10.18.17320
- Salmena L, Hakem R. From photomorphogenesis to cancer: a CSN journey. Cell Cycle 2013; 12:205-6; PMID:23287466; http://dx.doi.org/10.4161/cc.23422
- Wei W, Kaelin WG, Jr. Good COP1 or bad COP1? In vivo veritas. J Clin Invest 2011; 121:1263-5; PMID:21403396; http://dx.doi.org/10.1172/JCI57080
- Marine JC. Spotlight on the role of COP1 in tumorigenesis. Nature reviews Cancer 2012; 12:455-64; PMID:22673153; http://dx.doi.org/10.1038/nrc3271
- Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massague J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 1994; 78:59-66; PMID:8033212; http://dx.doi.org/10.1016/0092-8674(94)90572-X
- Leontieva OV, Demidenko ZN, Blagosklonny MV. Contact inhibition and high cell density deactivate the mammalian target of rapamycin pathway, thus suppressing the senescence program. Proc Natl Acad Sci U S A 2014; 111:8832-7.
- Yang HY, Zhou BP, Hung MC, Lee MH. Oncogenic signals of HER-2/neu in regulating the stability of the cyclin-dependent kinase inhibitor p27. J Biol Chem 2000; 275:24735-9; PMID:10859299; http://dx.doi.org/10.1074/jbc.C000147200
- Newman L, Xia W, Yang HY, Sahin A, Bondy M, Lukmanji F, Hung MC, Lee MH. Correlation of p27 protein expression with HER-2/neu expression in breast cancer. Mol Carcinog 2001; 30:169-75; PMID:11301477; http://dx.doi.org/10.1002/mc.1025
- Yang HY, Shao R, Hung MC, Lee MH. p27 Kip1 inhibits HER2/neu-mediated cell growth and tumorigenesis. Oncogene 2001; 20:3695-702; PMID:11439332; http://dx.doi.org/10.1038/sj.onc.1204472
- Zhang Y, Yang HY, Zhang XC, Yang H, Tsai M, Lee MH. Tumor suppressor ARF inhibits HER-2/neu-mediated oncogenic growth. Oncogene 2004; 23:7132-43; PMID:15273726; http://dx.doi.org/10.1038/sj.onc.1207918
- Yang H, Zhao R, Yang HY, Lee MH. Constitutively active FOXO4 inhibits Akt activity, regulates p27 Kip1 stability, and suppresses HER2-mediated tumorigenicity. Oncogene 2005; 24:1924-35; PMID:15688030; http://dx.doi.org/10.1038/sj.onc.1208352
- Liang J, Zubovitz J, Petrocelli T, Kotchetkov R, Connor MK, Han K, Lee JH, Ciarallo S, Catzavelos C, Beniston R, et al. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat Med 2002; 8:1153-60. Epub 2002 Sep 16; PMID:12244302; http://dx.doi.org/10.1038/nm761
- Lee MH, Yang HY. Negative regulators of cyclin-dependent kinases and their roles in cancers. Cell Mol Life Sci 2001; 58:1907-22; PMID:11766887; http://dx.doi.org/10.1007/PL00000826
- Caraballo JM, Acosta JC, Cortes MA, Albajar M, Gomez-Casares MT, Batlle-Lopez A, Cuadrado MA, Onaindia A, Bretones G, Llorca J, et al. High p27 protein levels in chronic lymphocytic leukemia are associated to low Myc and Skp2 expression, confer resistance to apoptosis and antagonize Myc effects on cell cycle. Oncotarget 2014; 5:4694-708; PMID:25051361
- Zhao H, Faltermeier CM, Mendelsohn L, Porter PL, Clurman BE, Roberts JM. Mislocalization of p27 to the cytoplasm of breast cancer cells confers resistance to anti-HER2 targeted therapy. Oncotarget 2014; 5:12704-14; PMID:25587029
- Nourse J, Firpo E, Flanagan WM, Coats S, Polyak K, Lee MH, Massague J, Crabtree GR, Roberts JM. Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature 1994; 372:570-3; PMID:7990932; http://dx.doi.org/10.1038/372570a0
- Yang H, Zhang Y, Zhao R, Wen YY, Fournier K, Wu HB, Yang HY, Diaz J, Laronga C, Lee MH. Negative cell cycle regulator 14-3-3sigma stabilizes p27 Kip1 by inhibiting the activity of PKB/Akt. Oncogene 2006; 25:4585-94; PMID:16532026; http://dx.doi.org/10.1038/sj.onc.1209481
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol 1999; 1:193-9; PMID:10559916; http://dx.doi.org/10.1038/12013
- Choi HH, Fang LK, Chen JS, Chou PC, Phan L, Su CH, Ivan C, Baggery K, Sood A, Yeung SC, et al. COP1 enhances ubiquitin-mediated degradation of p27Kip1 to promote cancer cell growth. Oncotarget 2015; in press
- Pippa R, Espinosa L, Gundem G, Garcia-Escudero R, Dominguez A, Orlando S, Gallastegui E, Saiz C, Besson A, Pujol MJ, et al. p27Kip1 represses transcription by direct interaction with p130/E2F4 at the promoters of target genes. Oncogene 2012; 31:4207-20; PMID:22179826; http://dx.doi.org/10.1038/onc.2011.582
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004; 429:86-92. Epub 2004 Apr 21; PMID:15103385; http://dx.doi.org/10.1038/nature02514
- Migliorini D, Bogaerts S, Defever D, Vyas R, Denecker G, Radaelli E, Zwolinska A, Depaepe V, Hochepied T, Skarnes WC, et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest 2011; 121:1329-43; PMID:21403399; http://dx.doi.org/10.1172/JCI45784
- Vitari AC, Leong KG, Newton K, Yee C, O'Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 2011; 474:403-6; PMID:21572435; http://dx.doi.org/10.1038/nature10005
- Dornan D, Shimizu H, Mah A, Dudhela T, Eby M, O'Rourke K, Seshagiri S, Dixit VM. ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 2006; 313:1122-6; PMID:16931761; http://dx.doi.org/10.1126/science.1127335
- Su CH, Zhao R, Velazquez-Torres G, Chen J, Gully C, Yeung SC, Lee MH. Nuclear export regulation of COP1 by 14-3-3sigma in response to DNA damage. Mol Cancer 2010; 9:243; PMID:20843328; http://dx.doi.org/10.1186/1476-4598-9-243
- Su CH, Zhao R, Zhang F, Qu C, Chen B, Feng YH, Phan L, Chen J, Wang H, Yeung SC, et al. 14-3-3sigma exerts tumor-suppressor activity mediated by regulation of COP1 stability. Cancer Res 2011; 71:884-94; PMID:21135113; http://dx.doi.org/10.1158/0008-5472.CAN-10-2518
- Wu FY, Wang SE, Sanders ME, Shin I, Rojo F, Baselga J, Arteaga CL. Reduction of cytosolic p27(Kip1) inhibits cancer cell motility, survival, and tumorigenicity. Cancer Res 2006; 66:2162-72; PMID:16489017; http://dx.doi.org/10.1158/0008-5472.CAN-05-3304
- Wu CC, Yang TY, Yu CT, Phan L, Ivan C, Sood AK, Hsu SL, Lee MH. p53 negatively regulates Aurora A via both transcriptional and posttranslational regulation. Cell Cycle 2012; 11:3433-42; PMID:22894933; http://dx.doi.org/10.4161/cc.21732
- Fang L, Yang Z, Zhou J, Yeung SC, Tung JY, Hsiao CD, Wang L, Deng Y, Wang P, Wang J, et al. Circadian clock gene CRY2 degradation is involved in chemoresistance of colorectal cancer. Molecular Cancer Therapeutics 2015; in press; PMID:25855785
- Laronga C, Yang HY, Neal C, Lee MH. Association of the cyclin-dependent kinases and 14-3-3 sigma negatively regulates cell cycle progression. J Biol Chem 2000; 275:23106-12; PMID:10767298; http://dx.doi.org/10.1074/jbc.M905616199
- Fuentes-Mattei E, Velazquez-Torres G, Phan L, Zhang F, Chou PC, Shin JH, Choi HH, Chen JS, Zhao R, Chen J, et al. Effects of obesity on transcriptomic changes and cancer hallmarks in estrogen receptor-positive breast cancer. J Natl Cancer Inst 2014; 106; PMID:24957076; http://dx.doi.org/10.1093/jnci/dju158
- Wen YY, Chou PC, Pham L, Su CH, Chen J, Hsieh YC, Xue Y-W, Qu C-J, Gully C, Parreno K, et al. DNA damage-mediated c-Myc degradation requires 14-3-3 sigma. Cancer Hallmarks 2013; 1:3-17; http://dx.doi.org/10.1166/ch.2013.1002
- Gully CP, Velazquez-Torres G, Shin JH, Fuentes-Mattei E, Wang E, Carlock C, Chen J, Rothenberg D, Adams HP, Choi HH, et al. Aurora B kinase phosphorylates and instigates degradation of p53. Proc Natl Acad Sci U S A 2012; 109:E1513-22; PMID:22611192; http://dx.doi.org/10.1073/pnas.1110287109
- Gully CP, Zhang F, Chen J, Yeung JA, Velazquez-Torres G, Wang E, Yeung SC, Lee MH. Antineoplastic effects of an Aurora B kinase inhibitor in breast cancer. Mol Cancer 2010; 9:42; PMID:20175926; http://dx.doi.org/10.1186/1476-4598-9-42
- Phan L, Shin JH, Zhou Z, Gully C, Phan L, Velazquez-Torres T, Fuentes-Mattei E, Yeung G, Su CH, Wang H, et al. The cell cycle regulator 14-3-3σ opposes and reverses cancer metabolic reprogramming Nat Commun 2015; in press
- Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B, et al. The molecular classification of multiple myeloma. Blood 2006; 108:2020-8; PMID:16728703; http://dx.doi.org/10.1182/blood-2005-11-013458
