Abstract
The pericentromere chromatin protrudes orthogonally from the sister-sister chromosome arm axis. Pericentric protrusions are organized in a series of loops with the centromere at the apex, maximizing its ability to interact with stochastically growing and shortening kinetochore microtubules. Each pericentromere loop is ∼50 kb in size and is organized further into secondary loops that are displaced from the primary spindle axis. Cohesin and condensin are integral to mechanisms of loop formation and generating resistance to outward forces from kinesin motors and anti-parallel spindle microtubules. A major unanswered question is how the boundary between chromosome arms and the pericentromere is established and maintained. We used sister chromatid separation and dynamics of LacO arrays distal to the pericentromere to address this issue. Perturbation of chromatin spring components results in 2 distinct phenotypes. In cohesin and condensin mutants sister pericentric LacO arrays separate a defined distance independent of spindle length. In the absence of Smt4, a peptidase that removes SUMO modifications from proteins, pericentric LacO arrays separate in proportion to spindle length increase. Deletion of Smt4, unlike depletion of cohesin and condensin, causes stretching of both proximal and distal pericentromere LacO arrays. The data suggest that the sumoylation state of chromatin topology adjusters, including cohesin, condensin, and topoisomerase II in the pericentromere, contribute to chromatin spring properties as well as the sister cohesion boundary.
Introduction
Mitosis is the stage of the cell cycle where condensed and tethered sister chromatids are aligned by the spindle apparatus in preparation for segregation into daughter cells. Microtubules nucleated from spindle pole bodies are attached to the kinetochore at centromere DNA. Biorientation of sister chromatids is a consequence of pulling forces from the spindle microtubules. The pericentric chromatin (30–50 kb surrounding the centromere) emanates from the sister cohesion axis, adopts a looped organization, and resists pulling forces with an inward non-linear spring force.Citation1 The chromatin spring is composed of cohesin, condensin, and pericentric chromatin Citation2 as well as topoisomerase 2 (Top2).Citation3,4 Tension generated by the opposing forces of the outward-directed pulling microtubules and the inward-directed chromatin spring is sensed at the kinetochore to ensure faithful segregation.Citation5 It remains to be elucidated how tension is transduced through pericentric chromatin that spans ˜800 nm between 2 sister kinetochores to ensure proper sensing.
Visualization of LacO/LacI-GFP sequences proximal to the budding yeast centromere has revealed key features of the pericentromere and how it responds to tension. The first major observation was sister foci separation and alignment along the spindle axis when kinetochores become bioriented on the spindle.Citation6-9 Second was the organization of cohesin into a barrel structure around the spindle microtubules.Citation10 Cohesin and condensin are enriched in the 30–50 kb surrounding the centromere, generating a chromatin loop as deduced by 3C technologies.Citation10 The loops emanate from the sister chromatid arm axis and orient toward the spindle pole bodies of the bipolar spindle. Outward microtubule based forces are balanced by the pericentromere chromatin spring.Citation2 Depletion of key components of the spring, including cohesin, condensin, and topoisomerase results in pericentromere LacO arrays stretching asymmetrically along the spindle axis as well as spindle fluctuations beyond the values observed in wild type spindles.Citation1,2, 4 Mathematical modeling of yeast microtubule spindle dynamics revealed that a non-linear chromatin spring with a threshold extension faithfully captures experimental asymmetric stretching, spindle length fluctuations, and declustering of the 16 kinetochores.Citation1 The evidence for secondary pericentric loops is further supported by the probability distribution heat maps of pericentric LacO arrays and the non-overlapping positions of condensin (350 nm dia. along the spindle axis) and cohesin (500 nm dia. radially displaced from the spindle axis) in the pericentromere ().Citation2,11,12 The behavior of chromatin loops and how they respond to changes in spindle length in these and several other mutants is depicted in . Any model of the pericentromere is incomplete without understanding the organization of the chromatin spring and critically, the boundary between the pericentromere and the sister chromatid arm axis.
Figure 1. Hypothesized structure of the pericentromere. (A) Wild type mitotic spindle drawn to scale with a single sister chromatid pair. (B) Hypothesized compaction of 25 kb of the pericentromere from the sister cohesion axis to the centromere. The 25 kb is separated into 7 kb extended along the spindle tension axis and 2 cohesin/condensin-based loops each 8–10 kb in size. (C) The hypothesized pericentromere compaction accounts for size increases in H3 repressed spindles via loss of half the nucleosomes/compaction and cohesin or condensin mutants in which asymmetric stretching occurs. Scale bar = 1 μm.
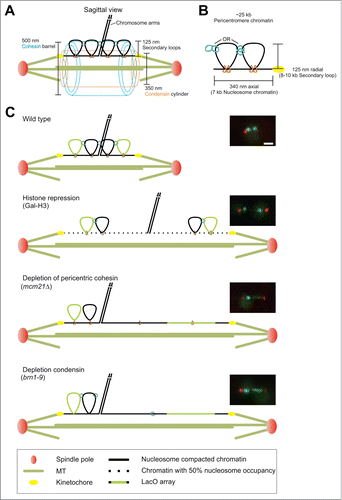
Both condensin and the SUMO deconjugation peptidase Smt4/Ulp2 Citation13 have been implicated in maintaining the sister cohesion boundary adjacent to the pericentromere. Recently, it was found that condensin, Cbf5/dyskerin, and Lrs4/monopolin display both separation and aberrant alignment of a pericentric distal LacO array (13.2 kb from CEN 11).Citation14 Smt4 and Top2 have been shown to affect separation of distal pericentromere.Citation4 Furthermore, Top2-SUMO is recruited to the yeast pericentromere,Citation15 Xenopus inner centromere,Citation16 and human centromere.Citation17 However, it is unclear what is functionally distinct about Top2 with and without the SUMO post-translational modification in the pericentromere.Citation3 In addition, Smt4 could affect many essential chromatin proteins considering that Smt4 and condensin genetically interact Citation18 and both TOP2 and SMC proteins contain sumoylation sites and are sumoylated.Citation19
The boundary between sister chromatid cohesion and the pericentromere cohesin barrel is critical to mechanisms that focus tension at the kinetochore for proper sensing via the spindle checkpoint. We used a distal pericentromere label (13.2 kb) from CEN to measure the position and dynamic behavior in both wild type and mutants. We find that upon depletion of chromatin proteins (histone, pericentric cohesin, condensin) distal pericentromere separation remains constant over a large change in spindle length. This behavior is exemplified by a constant force spring and suggests there is a boundary to sister-sister cohesion adjacent to the pericentromere that resists separation. Furthermore, condensin and cohesin are not responsible for dictating the arm boundary adjacent to the pericentromere. Depletion of the desumoylation peptidase Smt4 results in a linear separation distance between sister pericentromere loci and spindle length. The sumoylation pathway is key to the boundary complex and aids in focusing tension to the kinetochore by resisting chromosome arm separation and stretching of the chromatin spring at both centromere proximal and pericentromere distal loci.
Results
Condensin tethers the distal pericentromere to the spindle tension axis
Depletion of condensin results in the separation and loss of alignment along the spindle axis for loci distal to the centromere.Citation14 Condensin tethers chromatin to the spindle axis leading to the hypothesis that pericentromeres from different chromosomes are cross-linked via condensin's interaction with dyskerin and pericentric tRNA genes.Citation14 This hypothesis predicts that the pericentromere from one chromosome will be detached upon removal of all tDNA genes throughout the chromosome.Citation14 Toward this end, tDNA genes were removed from chromosome III, the essential tDNA genes relocated to other chromosomes to ensure survival of the mutant strain.Citation14 Probability distribution heat maps were generated for separated 11.5 kb distal pericentromere LacO arrays relative to their respective spindle pole bodies Citation2,11,20 for wild type and Chr3-tDNAΔ strains. Separated wild type distal pericentromere loci at 11.5 kb displayed clustering around the spindle axis similar to proximal pericentromere arrays and had an average radial distribution from the spindle axis of 143 ± 8 nm (standard error, n = 166, ). Removal of tDNA sites resulted in a significant increase in the radial distance from the spindle axis to 196 ± 11 nm (standard error, n = 188, p < 0.01, ). The probability distribution map is shown in . Reduced condensin at a single pericentromere via deletion of tDNA genes results in the radial displacement of the distal pericentromere from the spindle axis ().Citation14
Figure 2. Condensin tethers the distal pericentromere to the spindle axis. (A, B) Probability distribution map of a separated LacO array inserted at 11.5 kb from CEN III in wild type and tDNAΔ chromosome 3. (C) Graph of average radial distance from spindle axis in a LacO array 11.5 kb from CEN III or LacO array 13.2 kb from CEN XI. Error bars represent standard error. (*) Asterisks denotes significant increase compared to wild type, P< 0.01.
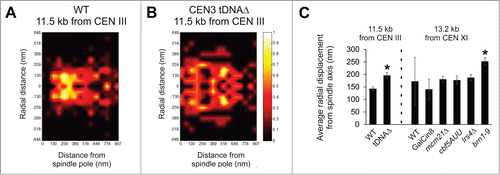
To determine if condensin is partially or solely responsible for distal pericentromere radial position, we imaged a distal LacO array in wild type and various mutants. Using a LacO array at 12.6 kb (centroid 13.2 kb, 1.2 kb array) we measured the radial displacement of separated LacO arrays. In wild type cells the distal pericentromere LacO was radially displaced 172 ± 95 nm from the spindle axis. An increase in spindle length without perturbing the chromatin spring can be accomplished by overexpression of the kinesin motor protein Cin8.Citation21 Spindle length increased ˜50% (1.5 μm to 2.2 μm) and the radial position of the distal pericentromere LacO decreased slightly to 140 ± 42 nm (GalCin8, ), suggesting that increased spindle length does not increase radial positon. Depletion of pericentric cohesin has been reported to be responsible for the radial position of the centromere proximal LacO arrays.Citation2 We depleted the pericentromere of cohesin via mcm21Δ which resulted in a longer spindle, increased percentage of mitotic cells with separated LacO, but no significant change in the radial position of the distal pericentromere LacO arrays (181 ± 12 nm, p > 0.05, n = 158, ). Taken together these data suggest that increased spindle length, increased LacO separation, and centromere proximal confinement dictated by pericentric cohesin are not necessary to maintain the radial position of distal pericentromere loci.
Depletion of dyskerin or monopolin results in a 40–60% depletion of pericentric condensin and a loss of alignment along the spindle axis.Citation14 The proximity of the distal pericentromere LacO to the spindle axis did not change upon their depletion (177–187 nm, n = 50 and 86, p > 0.05, ). However, depletion of a subunit of condensin (brn1-9) results in the distal pericentromere LacO becoming significantly displaced from the spindle axis, increasing from 172 nm in wild type to 252 ± 15 nm (n = 118, p < 0.01, ). This is contrary to condensin's role in the centromere proximal pericentromere where condensin dictates axial but not radial position.
To quantitate the radial position of pericentric chromatin, we used centromere proximal 10 kb LacO arrays inserted at 1.8 kb (centroid at 6.8 kb) from CEN XV which have been shown to stretch asymmetrically. A stretched (linear fluorescence signal) and its sister LacO that remains as a compact foci are indicative of the differential distribution of tension between sister chromatids (Fig. S1A). There is higher tension along the spindle axis (stretched chromatin) vs. radially displaced foci (compact chromatin). In wild type spindles the stretched LacO resides on average at 82 nm from the spindle axis while its un-stretched sister resides 143 nm (Fig. S1B). In chromatin mutants depleted of histone, pericentric cohesin, or condensin, this trend persists. However, during asymmetric stretching the focus becomes further radially displaced from the spindle axis (˜150 nm to >225 nm in the mutants, Fig. S1B). The displacement of one focus is likely due to the loss of tension once its sister LacO array decompacts and stretches along the spindle axis. The average radial displacement for stretched LacO arrays are significantly less than that of a focus (blue vs. red bars, Fig. S1). The radial positions of stretched pericentromere under tension and its sister not under tension mirror the change in radial displacement seen at distal pericentromere loci from spindles with condensin or depleted of condensin (brn1-9, ). The radial increase of distal pericentromere loci in condensin mutant brn1-9 could result from a similar mechanism via loss of tethering and tension from the spindle axis and misalignment of the sister spot.
Distal pericentromere radial position is dictated by tension via histone and condensin compaction while SMT4 resist tension based stretching
Condensin tethering of the distal pericentromere may also dictate the transition from the chromosome arms to the pericentromere. In order to determine the separation behavior of distal pericentromere chromatin we imaged the 1.2 kb LacO/LacI-GFP array at 12.6 kb from CEN XI (centroid at 13.2 kb) in wild type and mutant spindles along with spindle pole bodies (Spc29-RFP). Wild type cells show infrequent separation of distal pericentromere (5%, n = 61, ) in agreement with previous findings.Citation8 To determine if the distal pericentromere separates upon elongation of the metaphase spindle without perturbing the chromatin spring we overexpressed the kinesin motor protein Cin8 (GalCin8). Mean spindle length increased from 1.5 μm to 2.2 μm while separation of sister 13.2 kb LacO arrays increased from 5% to 11% (n = 57, ). When the chromatin spring is perturbed via depletion of pericentric cohesin (mcm21Δ) or condensin (brn1-9) mean spindle length increased to 2.1 and 2.6 μm, and the incidence of 13.2 kb LacO separation increased to 45% and 62% respectively (). cbf5-AUU/dyskerin mutant and lrs4Δ/monopolin mutants displayed a similar mean spindle length as wild type (1.5 μm) but showed a significant increase in distal pericentromere percent separation (52% and 25%). The increase in separation percentage in dyskerin mutants is consistent with its role in recruiting condensin to the mid-spindle.Citation14 Thus, sister chromatin loci distal to the centromere resist tension in wild type or upon elongation of the spindle via kinesin generated force, but separate upon perturbation of the chromatin spring. The ability to differentially effect sister chromatid separation is indicative of the different mechanistic contributions of microtubule-based motor proteins vs. chromatin topology adjusters.
Figure 3. Compaction dictates axial alignment while SMT4 and TOP2 resist axial strain. (A and B) Metaphase spindles were imaged with distal pericentromere labeled via LacO/LacI-GFP at 13.2 kb from CEN XI and spindle poles bodies Spc29-RFP in wild type and mutants GalCin8, GalH3 (23 kb), mcm21Δ, brn1-9, cbf5-AUU, lrs4Δ, top2-4, and smt4Δ. (A) Metaphase spindles were scored for the percentage of cells displaying separated 13.2 kb LacO/LacI-GFP foci. (B) Separated 13.2 kb LacO foci were classified as axial (blue), perpendicular to the spindle (orange) or stretching/splitting (green). Representative images show spindle pole bodies (purple) and 13.2 kb LacO (green). Scale bar = 1 μm.
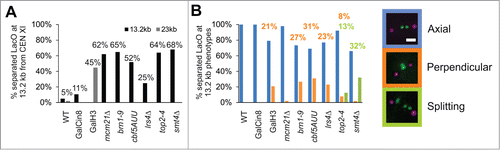
Pericentric cohesin and condensin both maintain the chromatin spring, but there are distinct mechanistic differences.Citation2 In order to gain insight into functional specialization we analyzed LacO arrays 13.2 kb distal pericentromere loci. Separated 13.2 kb LacO arrays in wild type and GalCin8 overexpression mutants remained aligned along the spindle axis, denoted as axial (blue, ). Similarly, depletion of pericentric cohesin results in separation and axial alignment along the spindle axis (mcm21Δ, ). In contrast, depletion of condensin or its recruiting complexes (dyskerin, monopolin) results in separated 13.2 kb foci that are misaligned relative to the spindle axis. The separated foci lie perpendicular to the spindle axis in ˜30% of spindles (orange, ). Thus, condensin is responsible for both the separation and the proper alignment of distal pericentromere chromatin along the spindle axis.Citation14
Pericentric condensin both compacts and resists stretching of the pericentric chromatin proximal to the centromere. This was evidenced by imaging LacO 6.8 kb from CEN XV in a condensin temperature sensitive mutant (brn1-9).Citation2 LacO foci distance from the spindle pole increased (decompaction) and LacO/LacI-GFP foci were imaged as a line signal more frequently (stretching). Distal pericentromere label 13.2 kb does not show any evidence of stretching or splitting upon depletion of condensin (). Decompaction of centromere proximal loci (6.8 kb) was also accomplished by depletion of histones via GalH3 repression with no increase in pericentromere stretching.Citation2 Decompaction of the pericentromere via histone depletion results in similar increases in spindle size (2.4 μm) and distal LacO separation percentage as observed in condensin mutants (GalH3 45% separation of 23 kb LacO from CEN, ). Furthermore, decompaction of the pericentromere via histone depletion results in aberrant perpendicular alignment of separated distal pericentromere LacO to levels comparable to percentages in condensin, dyskerin, or monopolin (˜30% perpendicular separated 13.2 kb or 23 kb, ). Thus, loss of histone or condensin based chromatin compaction results in a separated distal pericentromere that fail to properly align relative to the spindle axis.
Stretching of centromere proximal pericentromere LacO arrays was reported by Bachant and colleagues when cells were depleted of Top2 or desumoylation enzyme Smt4.Citation3,4 To determine if these components also increase separation of distal pericentromere loci, we generated mutants in strains containing the LacO array 13.2 kb from CEN. Temperature sensitive Top2 mutant (top2-4) and deletion of Smt4 (smt4Δ) resulted in similar increases in spindle length (1.5 μm WT to 2.2–2.5 μm) and increased percentage of separated 13.2 kb LacO arrays (˜60%, ), similar to previous findings.Citation3 In both top2-4 and smt4Δ, stretching (line signal) or splitting (a single sister foci breaking into multiple foci) of the LacO array was observed in a fraction of spindles (green, ). This is the first reported stretching of distal pericentromere LacO arrays and LacO arrays of such a small size (1.2 kb, 360 nm B-form DNA). This unique stretching/splitting phenotype was also reported for top2-4, smt4Δ, and non-sumoylated top2 (top2-SMN) mutants containing centromere proximal LacO arrays.Citation3,4 Thus, distal pericentromere LacO at 13.2 kb resists stretching in a Smt4 and Top2-dependent mechanism.
Distal pericentromere separation distance is dictated by the desumoylation enzyme SMT4
The distal pericentromere could be separating to a defined distance dictated by the pericentromere or in a linear function related to spindle length. To test these 2 possibilities we measured percent of separated sister loci at 13.2 kb from CEN as well as the distance between separated sister loci in spindles of increasing length. All mutants showed relatively linear increases in percentage of 13.2 kb LacO separation for increasing spindle lengths (). However, the distance between separated LacO spots vs. increasing spindle length bins revealed a more complex situation. For wild type spindles, separation only occurred at higher spindle lengths and for the small number of events the average separation distance was ˜0.5 μm (n = 2, ). A 0.5–0.8 μm 13.2 kb LacO separation distance occurred irrespective of spindle length for mutants with decreasing histone compaction (GalH3), depleted of pericentric cohesin (mcm21Δ), condensin (brn1-9, cbf5-1 and lrs4Δ), or overexpression of the kinesin motor Cin8 (GalCin8; , Table S1). These data suggest that there is a tether/boundary of sister chromatid arm cohesion adjacent to the pericentromere that limits the physical length of distal pericentromere separation. This boundary is independent of pericentric cohesin, condensin enrichment, or histone compaction.
Figure 4. Distal pericentromere separates a defined distance irrespective of spindle length and is controlled by Smt4. (A and B) Metaphase spindles were imaged with distal pericentromere labeled via LacO/LacI-GFP at 13.2 kb from CEN XI and spindle poles bodies Spc29-RFP in wild type and mutants GalCin8, GalH3 (23 kb), mcm21Δ, brn1-9, cbf5-AUU, lrs4Δ, top2-4, and smt4Δ. (A) Percentage of spindles with separated 13.2 kb LacO foci and (B) distance (μm) between separated foci for increasing spindle length bins. Linear line fitting data is reported in Table 1. (C) Cartoon depiction of the results where deletion of SMT4 results in a linear increase/proportional response of separation distance between the distal pericentromere and increase in spindle length. Error bars represent standard error.
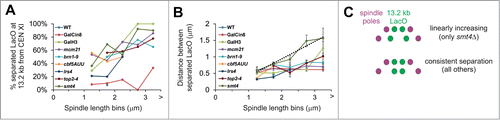
It has been hypothesized that Top2 and the sumoylation pathway dictate the interchange from the sister-sister cohesion boundary relative to the orthogonal pericentromere loops.Citation3,4 Depletion of Top2 (top2-4) displays a similar constant distance of separation relative to spindle length. However, deletion of the desumoylation peptidase Smt4 results in a linear increase in 13.2 kb LacO foci separation and spindle length (). Linear fit to the data reveals a slope of 0.5 with a good fit (R2 = 0.94, Table S1) indicating that the chromatin spring length on average increases uniformly with half the increase coming between the labeled foci and half the increase coming from outside the labeled foci to account for the increase in spindle length. Thus, the sumoylation pathway and specifically desumoylation peptidase Smt4 dictates the sister-sister cohesion boundary adjacent to the pericentromere chromatin spring.
Smt4 is also component of the pericentromere spring
Smt4 is responsible for cohesion of the arms at the boundary of the pericentromere. To determine if Smt4 is also responsible for centromere proximal force resistance, we deleted Smt4 in a strain with LacO at 6.8 kb from CEN XV. An increase in 6.8 kb LacO pericentromere stretching was measured for increasing spindle length bins (), suggesting Smt4 contributes to both the proximal and distal portions of the pericentromere. This is expected from the demonstrated sumoylation of cohesin, condensin, and topoisomerase 2.Citation19 The stretching behavior is similar to other known chromatin spring components cohesin, condensin, and Top2.Citation2,3 Interestingly, the probability of stretching for 6.8 kb LacO and 13.2 kb LacO are similar in smt4Δ mutants (41% vs. 32%; ) while stretching is not seen at the 13.2 kb for other chromatin spring proteins (). Thus, Smt4 aids force resistance in both proximal and distal pericentromere.
Figure 5. Deletion of Smt4 results in pericentromere stretching and spindle length fluctuations. (A) Mitotic spindle smt4Δ were scored for one focus (green), 2 foci (blue), asymmetric stretching (red), and symmetric stretching (purple) of LacO/LacI-GFP arrays with centroid at 6.8 kb from CEN XV (n = 88). Scale bar = 1 μm. (B) Graph of proximal 6.8 kb LacO and distal 13.2 kb LacO percentage stretching in smt4Δ mutants. Error bar represents standard deviation. (C) Line graph of spindle length over time for a representative mitotic spindle for wild type (gray) and smt4Δ mutant (black). (D) Bar graph of average change in length over time (ΔL) for wild type and smt4Δ. Error bars represent standard error for n = 177 and 169 respectively.
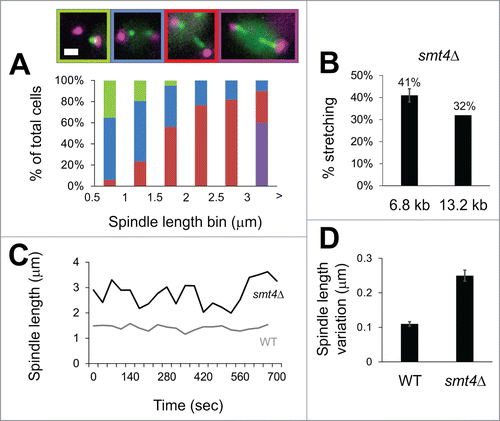
Perturbation of the chromatin spring results in increased pericentromere stretching coupled with spindle fluctuations. To determine whether smt4 mutants display a similar phenotype, we measured spindle length fluctuations in smt4Δ spindles. Mutant smt4Δ spindles show increased spindle fluctuations (). The average change in length (ΔL) increased from 110 nm in wild type to 250 nm in smt4Δ (, n = 177 and 169 respectively). Thus, Smt4 is both a major component of the chromatin spring as well as functioning at the boundary between the sister-sister cohesion axis and the pericentromere.
Smt4 aids condensin clustering in the pericentromere
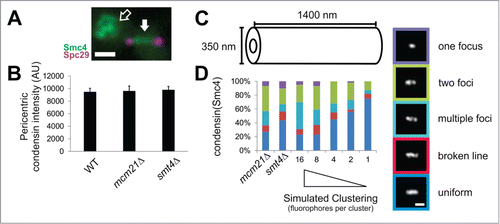
Since Smt4 is a desumoylation enzyme it is unlikely that the protein itself maintains the chromatin spring but instead controls the sumoylation of a chromatin spring component. It has been found that sumoylation can keep proteins from aggregating or clustering.Citation22-25 Sumoylation sites have been found on Top2, cohesin, and condensin.Citation19 Previously, we have shown that condensin clusters in the pericentromere.Citation12 To determine if loss of Smt4 disrupts condensin clustering in the pericentromere, we deleted Smt4 in a strain with condensin Smc4-GFP and spindle pole bodies Spc29-RFP (). In wild type spindles Smc4-GFP forms either a focus, 2 foci, or a uniform line signal with equal probability.Citation2 Simulations accounting for the number of condensin molecules and the size of the pericentromere and condensin structure revealed that clustering of 8–16 molecules of condensin are required to recapitulate the experimental pattern of fluorescence.Citation12 If a molecule is dispersed (not clustered) it should highlight the entire structure (line signal) while clustering causes a smaller signal or non-uniform (single foci or 2 foci). To account for the similar increase in spindle length of mcm21Δ and smt4Δ mutants (from 1.45 to 2.15 μm), we increased the size of the simulated condensin cylinder from 700 nm in WT to 1400 nm (). Upon lengthening of the spindle in mcm21Δ spindles, condensin total fluorescence remains unchanged ( andCitation12) while uniform signal remains in the minority (40% uniform or broken line signal) and clustered remains the majority (60% one focus, 2 foci, or multiple foci), similar to wild type. Mutant smt4Δ also maintain condensin pericentromere enrichment (), but display a more uniform Smc4-GFP distribution (60 % uniform vs. 40% clustered, ). Modeling of condensin molecules at different levels of clustering reveals that mcm21Δ maintains a high level of condensin clustering (8–16 molecules per cluster) while the increase in uniform labeling for smt4Δ best matches a decline in clustering (2–4 molecules per cluster). Thus, the loss of desumoylation results in declustering of condensin which likely perturbs of the chromatin spring.
Figure 6. Smt4 is partially responsible for clustering of condensins in the mitotic spindle. (A) An example image of condensin (Smc4-GFP, green) in a mitotic spindle with labeled spindle pole bodies (purple). The pericentric condensin is between the spindle poles (solid white arrow) and the rDNA condensin is off the spindle axis (hollow arrow). (B) Graph of condensin intensities measured in the pericentromere (between the spindle poles) and corrected for background for WT, mcm21Δ and smt4Δ (n = 20, 18, 32 respectively; p > 0.6). Error bars represent standard error. (C) Size measurements for simulated condensin cylinder in mutants mcm21Δ and smt4Δ. Wild type condensin cylinder best fit to a simulated cylinder 700 nm in length with a 1.45 μm spindle length.Citation12 Simulated condensin cylinder length was increased to 1400 nm to account for increase in spindle length to 2.15 μm for mcm21Δ and smt4Δ mutants. (D) Line scans drawn through experimental images of condensin fluorescence along the spindle axis in mcm21Δ and smt4Δ mitotic spindles to determine classification of one focus, 2 foci, multiple foci, broken line, or uniform. The measured number of condensins ˜240 Citation12 were randomly placed inside the cylinder as groups of 16, 8, 4, 2, or 1 to simulate a gradient of clustering to individual molecules. Scale bars = 1 μm.
Discussion
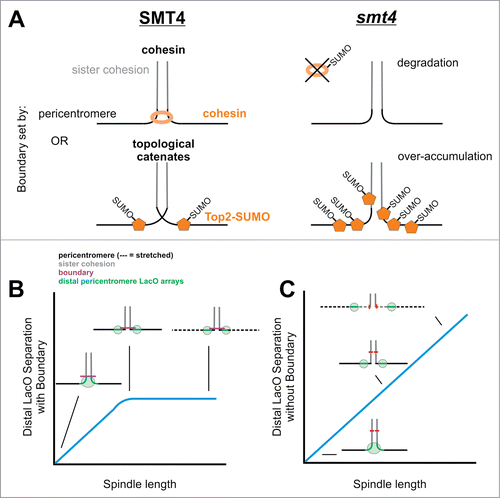
The pericentric chromatin in budding yeast is organized into an intramolecular loop with the centromere at the apex of the primary loop and condensin at the base of secondary loops. The pericentric loops are restricted to 30–50 kb surrounding each centromere. Outside the pericentric region, sister chromatid arms are linked in an intermolecular fashion through the action of cohesin. The boundary between these 2 structural modes is critical to understanding how tension is generated between sister centromeres. A widely cited model is that tension is focused on the intermolecular cohesion between sister chromatids. Based on the intramolecular pericentric loops, this would have to be outside the pericentromere, along the chromosome arms. Alternatively, tension could be generated within the pericentromere, through the packaging and confinement of chromatin loops.Citation1,2 In either scenario, there is a transition between intra- and inter-molecular sister chromatid linkages. The identity and function of boundary factors and how they might establish an inflection zone has not been elucidated. One of the mitigating factors is that the transition zone may vary among the 16 replicated chromosomes in sequence and spatially relative to the spindle. Using LacO/LacI-GFP arrays inserted at distal pericentromere sites we have identified a boundary that restricts separation of the pericentric chromatin. This boundary is dictated by the Smt4 SUMO peptidase that deconjugates SUMO, a post translational modification known to occur on many essential chromatin proteins such as cohesin, condensin, and topoisomerases. In the absence of the Smt4, there is a linear increase in centromere DNA separation as a function of spindle length. This phenotype is unique to Smt4 and defines its functional role in maintenance of the boundary. Furthermore, Smt4 contributes to the mechanical properties of the pericentromere.
Upon deletion of the SUMO deconjugation enzyme Smt4 additional chromatin from the arms is brought into the spindle resulting in increased distance between LacO arrays (). Several components enriched in the pericentromere including cohesin, condensin, and topoisomerase II are sumoylated. It is likely that the transition zone is dictated by one or more of these sumoylated chromatin proteins. Condensin sumoylation determines clustering in the pericentromere (discussed later), but evidence is lacking for a role in determining the boundary. On the other hand, either cohesinCitation26 or topoisomerase IICitation27 could act to maintain sister cohesin at the boundary ().
Figure 7. Summary of boundary between sister cohesion and the pericentromere. (A) The boundary could be set by cohesin or topological catenates. Loss of boundary in the desumoylation peptidase Smt4 deletion mutant occurs due to over sumoylation of either cohesin which leads to degradation or Top2 which recruits to the boundary to relieve all catenates (see discussion). (B) The boundary between sister cohesion and the pericentromere ensures tension is focused to the pericentromere and kinetochore. (C) Upon loss of the boundary more chromatin comes into the spindle (see red mark on sister cohesion chromatin move into the spindle) and tension is dispersed across more chromatin not allowing for proper transduction of force through the pericentromere to the kinetochore.
Cohesin's main function is to promote sister-sister cohesionCitation28,29 and sumoylation of cohesin has been shown to cause degradation of cohesin. Siz1 and 2 –dependent sumoylation of cohesin in preparation for degradation is antagonized by Pds5 or overexpression of SMT4 which can suppress pds5 mutants.Citation30 Alternatively, sumoylation of Pds5 itself has been reported to lead to an inability to protect cohesin from degradation or dissolution of sister cohesion.Citation31 Here we showed that loss of pericentric cohesin enrichment does not result in linear separation, suggesting cohesin at the sister cohesion boundary is not recruited by the kinetochore COMA complex. In addition, cohesin enrichment proximal to the centromere via ChIP in smt4Δ mutants remains similar to wild type.Citation3 Therefore, additional research is required to clearly determine cohesin's possible role in defining the transition between intra- and intermolecular sister chromatid cohesion.
Of the essential proteins in the pericentromere, sumoylated topoisomerase II is specifically recruited to the pericentromere during metaphase in many eukaryotes.Citation15-17 Pericentric Top2-SUMO is required to relax topological stress when sister centromeres are bioriented. Topological stress is induced upon nucleosome release from stretched pericentric chromatin on the spindle axis.Citation2,4,32 Similarly, Top2 sumoylation aids in the relief and redistribution of mechanical stress as in meiotic crossover interference.Citation33 It is possible that topological catenates outside the pericentromere determine the boundary between sister cohesion and the pericentromere. Topological catenates are sufficient to maintain sister chromatid cohesion even when cohesin is depleted.Citation34,35 Thus, over accumulation of Top2-SUMO in the pericentromere without the SUMO peptidase Smt4 could result in removing catenates responsible for setting the boundary, allowing sister cohesion chromatin to transition into pericentric chromatin. This idea is supported by the partial suppression of the separation of LacO arrays at the distal pericentromere in smt4Δ mutants by a non-sumoylatable version of Top2 (top2-SMN).Citation4
The requirement for sumoylation at the boundary between intra- and intermolecular looping provides insights into how tension is transmitted to the centromere/kinetochore (). A second unique phenotype of the Smt4 deletion is that distal pericentromere LacO arrays stretch as frequently as the proximal pericentromere (). One possibility is the transition zone is ill-defined, and there is more pericentric chromatin proximal to the spindle (). The increase in crowding could increase tension as evidenced by stretching of distal chromatin (). In addition, the increase in sumoylated proteins could perturb the mechanical properties of the chromatin spring.
The distal pericentric LacO array is a monitor of tension throughout the pericentromere. One of the major phenotypes of separated distal pericentromere LacO arrays is perpendicular alignment to the spindle axis (). This occurred upon decompaction of the chromatin spring via histone repression (GalH3), condensin depletion (brn1-9, lrs4Δ, cbf5AUU), or mutation in topoisomerase II (top2-4). These foci are not displaced from the spindle axis, except for depletion of condensin brn1-9 (). Rather, they lose their alignment relative to the primary microtubule spindle axis. The defect in alignment might reflect disorganization of the pericentric chromatin network surrounding the central spindle. Condensin functions to cross-link pericentromeres of neighboring chromosomes to the central spindle axis.Citation14,36 However, depletion of nucleosomes does not disrupt inter-pericentromere cross-linking. Thus, disruption of the cross-linked pericentromere network is more likely to result in displacement of LacO from the spindle axis as seen only in condensin mutants (). An alternative is that perpendicular alignment is the result of reduced tension between sisters. This is evident in asymmetric stretching of centromere proximal LacO arrays in condensin mutants (60%).Citation2 While one pericentromere strand is proximal to the spindle and decompacted, its sister chromatid remains compact and displaced from the spindle axis (see Fig. S1). The displaced foci often moves closer to the spindle pole suggesting tension at this locus is lost (Fig. S1). In mammalian cells, condensin mutants also fail to properly transduce tension to the kinetochore for proper sensing, leading to chromosome misalignment and inability to correct improper attachments.Citation37,38
The role of condensin in cooperative condensation/clustering has been noted in a number of recent studies. It was first reported that condensin condenses chromatin cooperatively.Citation39,40 Theoretical work described how cooperativity and clustering of condensin complexes would result in condensation.Citation41 We have found that condensin forms either one foci, 2 foci or a line signal in the pericentromere with equal frequency. This behavior can be simulated by clustering of condensin complexes using the experimentally determined size of the condensin cylinder in the pericentromere and the number of condensin molecules.Citation12 Sun and Marko have found similar condensin clusters in human mitotic chromosomes both in the pericentromere and in the arm via antibody labeled fluorescence of isolated sister chromatids (Sun et al., data unpublished). The mechanism of clustering may reside in the binding to tRNA genes.Citation14,42
It has been reported that sumoylation can modulate the clustering or cooperative binding of proteins.Citation22-25 Our evidence suggest a similar mechanism for the control of condensin clustering as excess sumoylation (smt4Δ) results in pericentric condensin declustering (). Clustered condensins compact the pericentromere into loops and cross-link different pericentromeres to coordinate movement and resist tension from outward spindle forces. Deletion of Smt4 does not disrupt the enrichment of cohesinCitation3 nor condensin () but does result in decompaction of pericentromere loops and stretching along the spindle axis viewed by imaging centromere LacO arrays. While the phenotype is complex, the increased concentration of SUMO in the spindle perturbs the network in a way that disrupts the coordination among the pericentromeres. This leads to asymmetries in the spatial distribution of force within the pericentric, manifested as asymmetric and off-axis stretching. The appropriate balance of sumoylated complexes in the mitotic spindle is essential to maintaining the pericentromere chromatin spring as well as its boundary with sister arm cohesion.
The modulation of the boundary between the pericentromere and sister arm cohesion as well as the pericentromere chromatin spring is dictated by sumoylation and the SUMO deconjugation peptidase Smt4. The inability to set a boundary between the pericentromere and sister arm cohesion results in premature sister chromatid separation and dramatic increase in stretching/decompaction at distal pericentromere loci. Furthermore, lack of desumoylation results in condensin declustering and increased stretching of centromere proximal chromatin that leads to spindle length fluctuations. These perturbations results in the inability to properly sense tension and correct errors.Citation43,44 Thus, the desumoylation peptidase Smt4 is a key regulator of the transition between intermolecular sister chromatid cohesion and the intramolecular pericentric chromatin spring.
Materials and methods
Cell preparations
Cells were incubated in YPD (2% glucose, 2% peptone, and 1% yeast extract) at 32°C for WT strains. Temperature-sensitive strains containing brn1-9, lrs4Δ and top2-4 were grown at 24°C. Temperature-sensitive strains brn1-9 and top2-4 were grown into early log phase at 24°C and then shifted to restrictive temperature at 37°C for 3 h before filming. Temperature-sensitive stains were then viewed at RT for no longer than 45 min. Gal-Cin8 strains were grown in YPD (2% glucose) then shifted to YPG (2% galactose) for 3 hours before imaging. Gal-H3 strains were α factor arrested in YPG (2% galactose), washed, and then released into YPD (2% glucose) for 3–4 h before viewing, to deplete H3 histone as outlined in Bouck and Bloom.Citation45 LacO/lacI-GFP strains were grown in SD-His media to induce lacI-GFP under the HIS promoter as outlined by Goshima and YanagidaCitation6 and Pearson et al.Citation8
Microscopy
Wide-field microscope images were acquired at RT (25°C) using a microscope stand (Eclipse TE2000-U; Nikon) with a 100× Plan Apo 1.4 NA digital interference contrast oil emersion lens with a camera (Orca ER; Hamamatsu Photonics). MetaMorph 6.1 software (Molecular Devices) was used to acquire unbinned z series image stacks with a z step size of 300 nm. Imaging was performed in water on ConA-coated coverslips. Image exposure times were between 300 and 800 ms.
LacO array analysis
Population images of metaphase cells were acquired of strains with 13.2 kb LacO arrays (1.2kb array inserted at 12.6 kb from CEN XI; Citation8) in WT, GalCin8, GalH3, mcm21Δ, brn1-9, cbf5-AUU, lrs4Δ, top2-4, and smt4Δ mutants. LacI-GFP is a fusion protein of GFP-LacI-NLS driven by the HIS3 promoter, with the LacI gene deleted for 11 C-terminal amino acids to prevent tetramerization.Citation46 Images were captured using unbinned acquisitions and were analyzed in MetaMorph. Orientation relative to the spindle axis was determined as axial if both foci lie parallel to the spindle axis, whereas perpendicular orientation was determined by foci lying at a 90° angle to the axis as outlined in Snider et al., .14 In MetaMorph line scans were used to determine focus versus stretching or splitting LacO spots.Citation2 Focus LacO spots were determined by a Gaussian distribution line scan through the brightest pixel. Oppositely, stretched LacO was determined by a non-Gaussian distribution or broadening of the line scan with a ˜0.5 decrease in brightest pixel fluorescence signal compared with a focus. LacO was classified as splitting if the line scan revealed to bright pixels >250 nm apart for a single sister loci.
Using Metamorph the brightest pixel was logged into Excel (Microsoft) for each spindle pole body (Spc29-RFP) and LacO/LacI-GFP. In Excel data was analyzed to measure spindle length and/or LacO separation distance in micrometers. Distances were measured in 2 dimensions using the Pythagorean Theorem. Excel was used to apply geometrical rotations to each spindle and respective LacO coordinates to a defined y-axis then report the radial displacement, distance in y, of the LacO coordinate from the spindle axis.
Probability distribution heatmaps
For heat mapping, images of 1.7-kb LacO/LacI-GFP were rotated and aligned relative to the spindle axis (determined by Spc29-RFP) using MATLAB. The LacO array is a 1.2-kb 32-mer described in LacO array analysis, inserted at Met14 1.1 kb from the centromere on chromosome XI.Citation8 The distance in x and y from the spindle pole was measured for the brightest pixel of each LacO and logged in Excel. The number of occurrences of a LacO occupying a position within one quadrant of the spindle was recorded and mirrored across the x axis. This information was transferred to MATLAB, in which an image was generated using the black body radiation spectrum to depict probability of the LacO position.
Spindle length and variation
Spindle lengths were measured by logging the coordinates of the brightest pixel of each spindle pole body, marked by Spc29-RFP, using MetaMorph. Coordinates of pixel position were measured in triplicate. The coordinates of sister spindle poles were transferred to Excel and converted into distance spindle length in micrometers. Spindle lengths were measured in 2 dimensions and 3 dimensions using the Pythagorean theorem. Time-lapse videos were performed on single cells using Acquire Timelapse in MetaMorph to take a z series every 35 s for 20 time points, equaling 11.67 min. Change in spindle length, denoted as variation, was calculated by the absolute value of the difference between spindle length at each time point and the mean spindle length of the time lapse. All metaphase spindle lengths and time lapses were taken in spindles of at least 1.1 µm, with separated Nuf2 kinetochore foci and spindles not exhibiting linearly increasing anaphase spindle behavior.
Analyzing Smc4-GFP fluorescence
MATLAB (MathWorks Inc.) was used to rotate the spindle axis of MetaMorph images horizontally using Spc29-RFP as markers of spindle ends. Horizontally rotated images could then be analyzed in MetaMorph with the brightest pixel of both spindle pole bodies along the same y coordinates. Line scans 1 pixel in width were drawn along the spindle axis through the brightest pixel. The data of pixel position and intensity of Smc4-GFP were transferred to Excel (Microsoft) and graphed to determine the classification (one focus, 2 foci, or uniform signal) of condensin enrichment between the spindle poles. Simulated fluorescence images were analyzed in the same manner.
Condensin enrichment was measured as reported in Snider et al.,.Citation14 In ImageJ a box the size of 15 × 27 was used to measure the integrated intensity of the pericentric condensin and a box 25 × 37 was used to correct for background. Integrated intensity of Smc4 (FSmc4) was then determined by the following formula in Excel: FSmc4 = FI - Fbackground, in which Fbackground = (FO - FI) × (area of inner region - area between perimeter of inner and outer regions), FI = integrated intensity of inner region, and FO = integrated intensity of outer region.
Model convolution
The experimental PSF of our microscope was determined by imaging a 100-nm fluorescent bead with 100-nm z-steps spanning 1 μm above and below the brightest plane. Five different z-series of 100-nm fluorescent beads were aligned and averaged to generate the average PSF of our microscope.Citation47 The experimental PSF z-stack was imported into Microscope Simulator 2.0.0 for use. The Microscope Simulator 2.0.0 software program (CISMM UNC-Chapel Hill; http://cismm.cs.unc.edu/downloadsCitation48) was used to generate geometrical cylinder models with length, inner diameter, and outer diameter. Cylinders were filled randomly with a designated number of fluorophores to represent clustering (fewer fluorophores = more clustered, more fluorophores = lack of clustering). The experimentally measured number of condensin fluorophores is ˜240.Citation12 Each fluorophore in the cylinder was convolved with the experimental PSF. The convolution of the entire cylinder matrix is the summarization of the contributions of the fluorescence from each simulated fluorophore position in x, y, and z to the image plane.Citation47,49 Image(x,y) = Σ cylinder matrix (x,y,x) · PSF(x,y,z). All simulated images were focused on the midplane of the cylinder. Random Gaussian noise (cohesin − mean 250, SD 3; condensin − mean 220, SD 2.6) matching the noise from experimental images was added to the generated images. Maximum intensity from simulated images was scaled to reflect the maximum intensity observed in experimental images. Cylinders were randomly placed (±65 nm) relative to the x,y coordinates for each output-simulated image to avoid aliasing.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1046656_Supplemental_Material.zip
Download Zip (756.4 KB)Acknowledgments
The authors thank members of the Bloom laboratory for critical reading of the manuscript. We thank Will Lewis for technical assistance.
Funding
This work was supported by grants R37 GM32238 to K. Bloom and AHA postdoctoral fellow grant #14POST20490209 to AD Stephens.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Stephens AD, Haggerty RA, Vasquez PA, Vicci L, Snider CE, Shi F, Quammen C, Mullins C, Haase J, Taylor RM, 2nd, et al. Pericentric chromatin loops function as a nonlinear spring in mitotic force balance. J Cell Biol 2013; 200:757-72; PMID:23509068; http://dx.doi.org/10.1083/jcb.201208163
- Stephens AD, Haase J, Vicci L, Taylor RM, 2nd, Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. JCell biol 2011; 193:1167-80; PMID:21708976; http://dx.doi.org/10.1083/jcb.201103138
- Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell 2002; 9:1169-82; PMID:12086615; http://dx.doi.org/10.1016/S1097-2765(02)00543-9
- Warsi TH, Navarro MS, Bachant J. DNA topoisomerase II is a determinant of the tensile properties of yeast centromeric chromatin and the tension checkpoint. Mol Biol Cell 2008; 19:4421-33; PMID:18701701; http://dx.doi.org/10.1091/mbc.E08-05-0547
- Bloom K, Yeh E. Tension management in the kinetochore. Curr Biol 2010; 20:R1040-8; PMID:21145023; http://dx.doi.org/10.1016/j.cub.2010.10.055
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 2000; 100:619-33; PMID:10761928; http://dx.doi.org/10.1016/S0092-8674(00)80699-6
- He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 2000; 101:763-75; PMID:10892747; http://dx.doi.org/10.1016/S0092-8674(00)80888-0
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol 2001; 152:1255-66; PMID:11257125; http://dx.doi.org/10.1083/jcb.152.6.1255
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell 1999; 98:847-58; PMID:10499801; http://dx.doi.org/10.1016/S0092-8674(00)81518-4
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol 2008; 18:81-90; PMID:18211850; http://dx.doi.org/10.1016/j.cub.2007.12.019
- Haase J, Stephens A, Verdaasdonk J, Yeh E, Bloom K. Bub1 kinase and Sgo1 modulate pericentric chromatin in response to altered microtubule dynamics. Curr Biol 2012; 22:471-81; PMID:22365852; http://dx.doi.org/10.1016/j.cub.2012.02.006
- Stephens AD, Quammen CW, Chang B, Haase J, Taylor RM, 2nd, Bloom K. The spatial segregation of pericentric cohesin and condensin in the mitotic spindle. Mol Biol Cell 2013; 24:3909-19; PMID:24152737; http://dx.doi.org/10.1091/mbc.E13-06-0325
- Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 2000; 20:2367-77; PMID:10713161; http://dx.doi.org/10.1128/MCB.20.7.2367-2377.2000
- Snider CE, Stephens AD, Kirkland JG, Hamdani O, Kamakaka RT, Bloom K. Dyskerin, tRNA genes, and condensin tether pericentric chromatin to the spindle axis in mitosis. J Cell Biol 2014; 207:189-99; PMID:25332162; http://dx.doi.org/10.1083/jcb.201405028
- Baldwin M, Warsi T, Bachant J. Analyzing Top2 distribution on yeast chromosomes by chromatin immunoprecipitation. Methods Mol Biol 2009; 582:119-30; PMID:19763946; http://dx.doi.org/10.1007/978-1-60761-340-4_10
- Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. TEMBO J 2005; 24:2172-82; PMID:15933717; http://dx.doi.org/10.1038/sj.emboj.7600700
- Diaz-Martinez LA, Gimenez-Abian JF, Azuma Y, Guacci V, Gimenez-Martin G, Lanier LM, Clarke DJ. PIASgamma is required for faithful chromosome segregation in human cells. PloS one 2006; 1:e53; PMID:17183683
- Strunnikov AV, Aravind L, Koonin EV. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics 2001; 158:95-107; PMID:11333221
- Takahashi Y, Yong-Gonzalez V, Kikuchi Y, Strunnikov A. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics 2006; 172:783-94; PMID:16204216; http://dx.doi.org/10.1534/genetics.105.047167
- Anderson M, Haase J, Yeh E, Bloom K. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell 2009; 20:4131-9; PMID:19656849; http://dx.doi.org/10.1091/mbc.E09-05-0359
- Saunders W, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell 1997; 8:1025-33; PMID:9201713; http://dx.doi.org/10.1091/mbc.8.6.1025
- Janer A, Werner A, Takahashi-Fujigasaki J, Daret A, Fujigasaki H, Takada K, Duyckaerts C, Brice A, Dejean A, Sittler A. SUMOylation attenuates the aggregation propensity and cellular toxicity of the polyglutamine expanded ataxin-7. Hum mol Genet 2010; 19:181-95; PMID:19843541; http://dx.doi.org/10.1093/hmg/ddp478
- Kolesar P, Sarangi P, Altmannova V, Zhao X, Krejci L. Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic acids Res 2012; 40:7831-43; PMID:22705796; http://dx.doi.org/10.1093/nar/gks484
- Krumova P, Meulmeester E, Garrido M, Tirard M, Hsiao HH, Bossis G, Urlaub H, Zweckstetter M, Kugler S, Melchior F, et al. Sumoylation inhibits alpha-synuclein aggregation and toxicity. J Cell Biol 2011; 194:49-60; PMID:21746851; http://dx.doi.org/10.1083/jcb.201010117
- Zhang YQ, Sarge KD. Sumoylation of amyloid precursor protein negatively regulates Abeta aggregate levels. Biochem Biophys Res Commun 2008; 374:673-8; PMID:18675254; http://dx.doi.org/10.1016/j.bbrc.2008.07.109
- Guacci VA. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle 2009; 8:3811-2; PMID:19887903; http://dx.doi.org/10.4161/cc.8.23.10383
- Baldwin ML, Julius JA, Tang X, Wang Y, Bachant J. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle 2009; 8:3406-19; PMID:19823017; http://dx.doi.org/10.4161/cc.8.20.9911
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 1998; 12:1986-97; PMID:9649503; http://dx.doi.org/10.1101/gad.12.13.1986
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 1997; 91:35-45; PMID:9335333; http://dx.doi.org/10.1016/S0092-8674(01)80007-6
- D'Ambrosio LM, Lavoie BD. Pds5 prevents the PolySUMO-dependent separation of sister chromatids. Curr Biol 2014; 24:361-71; PMID:24485833; http://dx.doi.org/10.1016/j.cub.2013.12.038
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol 2003; 163:729-41; PMID:14623866; http://dx.doi.org/10.1083/jcb.200305080
- Verdaasdonk JS, Gardner R, Stephens AD, Yeh E, Bloom K. Tension-dependent nucleosome remodeling at the pericentromere in yeast. Mol BiolCell 2012; 23:2560-70; PMID:22593210; http://dx.doi.org/10.1091/mbc.E11-07-0651
- Zhang L, Wang S, Yin S, Hong S, Kim KP, Kleckner N. Topoisomerase II mediates meiotic crossover interference. Nature 2014; 511:551-6; PMID:25043020; http://dx.doi.org/10.1038/nature13442
- Toyoda Y, Yanagida M. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol Biol Cell 2006; 17:2287-302; PMID:16510521; http://dx.doi.org/10.1091/mbc.E05-11-1089
- Vagnarelli P, Morrison C, Dodson H, Sonoda E, Takeda S, Earnshaw WC. Analysis of Scc1-deficient cells defines a key metaphase role of vertebrate cohesin in linking sister kinetochores. EMBO Rep 2004; 5:167-71; PMID:14749720; http://dx.doi.org/10.1038/sj.embor.7400077
- Stephens AD, Snider CE, Haase J, Haggerty RA, Vasquez PA, Forest MG, Bloom K. Individual pericentromeres display coordinated motion and stretching in the yeast spindle. J Cell Biol 2013; 203:407-16; PMID:24189271; http://dx.doi.org/10.1083/jcb.201307104
- Samoshkin A, Arnaoutov A, Jansen LE, Ouspenski I, Dye L, Karpova T, McNally J, Dasso M, Cleveland DW, Strunnikov A. Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PloS one 2009; 4:e6831; PMID:19714251; http://dx.doi.org/10.1371/journal.pone.0006831
- Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol 2009; 184:383-90; PMID:19188492; http://dx.doi.org/10.1083/jcb.200811028
- Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol 1998; 142:1595-604; PMID:9744887; http://dx.doi.org/10.1083/jcb.142.6.1595
- Strick TR, Kawaguchi T, Hirano T. Real-time detection of single-molecule DNA compaction by condensin I. Curr Biol 2004; 14:874-80; PMID:15186743; http://dx.doi.org/10.1016/j.cub.2004.04.038
- Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res 2012; 40:11202-12; PMID:23074191; http://dx.doi.org/10.1093/nar/gks925
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 2008; 22:2204-14; PMID:18708579; http://dx.doi.org/10.1101/gad.1675908
- Era S, Abe T, Arakawa H, Kobayashi S, Szakal B, Yoshikawa Y, Motegi A, Takeda S, Branzei D. The SUMO protease SENP1 is required for cohesion maintenance and mitotic arrest following spindle poison treatment. Biochem Biophys Res Commun 2012; 426:310-6; PMID:22943854; http://dx.doi.org/10.1016/j.bbrc.2012.08.066
- Lee MT, Bachant J. SUMO modification of DNA topoisomerase II: trying to get a CENse of it all. DNA repair 2009; 8:557-68; PMID:19230795; http://dx.doi.org/10.1016/j.dnarep.2009.01.004
- Bouck DC, Bloom K. Pericentric chromatin is an elastic component of the mitotic spindle. Curr Biol 2007; 17:741-8; PMID:17412588; http://dx.doi.org/10.1016/j.cub.2007.03.033
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol 1996; 6:1599-608; PMID:8994824; http://dx.doi.org/10.1016/S0960-9822(02)70783-5
- Sprague BL, Pearson CG, Maddox PS, Bloom KS, Salmon ED, Odde DJ. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys J 2003; 84:3529-46; PMID:12770865; http://dx.doi.org/10.1016/S0006-3495(03)75087-5
- Quammen CW, Richardson AC, Haase J, Harrison BD, Taylor RM, 2nd, Bloom KS. FluoroSim: a visual problem-solving environment for fluorescence microscopy. Eurographics Workshop Vis Comput Biomed 2008; 2008:151-8; PMID:20431698
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol 1989; 30:353-77; PMID:2494418; http://dx.doi.org/10.1016/S0091-679X(08)60986-3
