Abstract
Cyclin-dependent kinase regulatory subunit 1 (CKS1) helps regulate the cell cycle to increase cell number. However, the hormonal regulation on CKS1 expression is not well understood. We report that CKS1 is involved in the promotion of cell proliferation with insulin regulation in the lepidopteran insect Helicoverpa armigera. CKS1 is expressed in various tissues during the larval feeding stage. CKS1 knockdown results in larval death, body weight decrease, pupation time delay, and small-sized pupa formation. The underlying mechanism involves the blocking of cell proliferation and the repression of gene expression in the insulin pathway after CKS1 knockdown. CKS1 overexpression in the epidermal cell line results in cell proliferation. The N45 amino acid asparagine in the CKS domain is essential for the function of CKS in cell proliferation. CKS1 is upregulated by insulin via an insulin receptor, but is repressed by a high level of steroid hormone 20-hydroxyecdysone (20E). Results suggest that CKS1 promotes cell proliferation and body growth in coordination with the regulatory actions of insulin and steroid hormone 20E.
Introduction
Animal body size (or body weight) is mainly determined by cell number and tissue sizes.Citation1,2 Cell proliferation from stem cells is the basis of cell growth and is a key process in determining the final body size. The insulin pathway is essential in regulating cell proliferation and body size and is conserved from human to insect.Citation1 Insulin initiates signaling by binding with its receptor (InR) and transmits the signal via protein kinase Akt/PKB and phosphatidyl inositol 3-kinase (PI3K).Citation3 Partial function loss of PI3K and Akt can lead to the developmental delay of Drosophila and formation of small organs because of the corresponding decrease in cell number and size.Citation4,5 Forkhead box class O (FOXO) serves a function that is opposite that of insulin; FOXO is inhibited by the insulin pathway.Citation6 FOXO may inhibit dMyc-induced cell growth and proliferation in Drosophila.Citation7
Insect body growth is also promoted by insulin.Citation8 However, steroid hormone 20-hydroxyecdysone (20E) may antagonize insulin's function to determine the development time and final size of Drosophila.Citation9,10 Insulin and ecdysone interplay regulates insect growth and developmental timing and determines the final body size.Citation11 Insulin regulates Drosophila growth up until a critical body weight, thereby facilitating the production of more steroid hormone 20E in the prothoracic gland and initiating the expression of the transcription factor Broad (Br) for metamorphosis.Citation12,13 These findings suggest the interaction of insulin and 20E in regulating insect body size.
Cyclin-dependent kinases (CDKs) and cyclin-dependent kinase regulatory subunits (CKSs) interact to regulate the cell cycle.Citation14 Two cyclin-dependent kinase regulatory subunits (CKSs) exist in mammalian cells, namely, CKS1 and CKS2, and these CKSs share 81% identity.Citation15 CKS1 and CKS2 both have a CDK interaction domain and an anion-binding domain in mammals; thus, they both participate in the cell's entrance into mitosis.Citation16 CKSs are essential in cell proliferation because the double mutation of CKS1 and CKS2 can be lethal in mouse.Citation17 However, the yeast CKS1 function cannot be replaced by CKS2, thereby indicating that CKS1 and CKS2 have different functions.Citation18 CKS1 knockout reduces mouse growth and results in small body size.Citation19 The Drosophila CKS1 ortholog CKS85A is essential for viability, and the null CKS1 mutation is lethal.Citation20 Despite the results from previous studies, the function and hormonal regulation of CKS1 in cell proliferation and body size control have not been investigated.
We found that CKS1 is expressed at high levels during the larval growth stage in the lepidopteran pest insect Helicoverpa armigera. Knockdown of CKS1 by RNAi delays pupation time, leads to the formation of small-sized pupae, results in high mortality and insulin pathway gene expression, and represses midgut cell proliferation. CKS1 overexpression in H. armigera epidermal cell line (HaEpi) promotes cell proliferation. CKS1 is upregulated by insulin and a low concentration of steroid hormone 20E, but is repressed by a high concentration of 20E. Our results suggest that insulin upregulates CKS1 expression for cell proliferation during larval growth. A high titer of 20E represses CKS1 expression during metamorphosis. The expression of CKS1 is regulated by both insulin and 20E in a coordinated manner.
Results
CKS1 was highly expressed during the larval growth stage
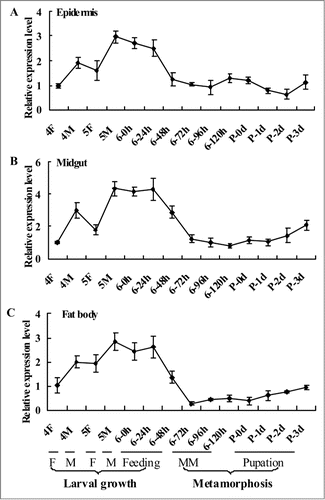
The expression profiles of CKS1 in the epidermis, midgut, and fat body tissues were analyzed by quantitative real-time reverse transcription PCR (qRT-PCR) from the fourth instar feeding stage to pupal stage (P-3 d) to determine the mRNA of CKS1 during insect development. CKS1 expression was higher during the larval growth stage prior to metamorphosis, and the highest expression was observed during the earlier stage of the sixth instar larvae either by feeding or molting (). This expression profile suggests that CKS1 serves major functions during larval growth.
Figure 1. qRT-PCR results showing the expression profile of CKS1 in various tissues. The expression profile of CKS1 in the epidermis (A), midgut (B), and fat body (C) with β-actin as the quantity and quality control. These experiments were repeated thrice and statistically analyzed. F: feeding stage; M: molting stage; MM: metamorphic molting.
CKS1 knockdown blocked larval growth and pupation
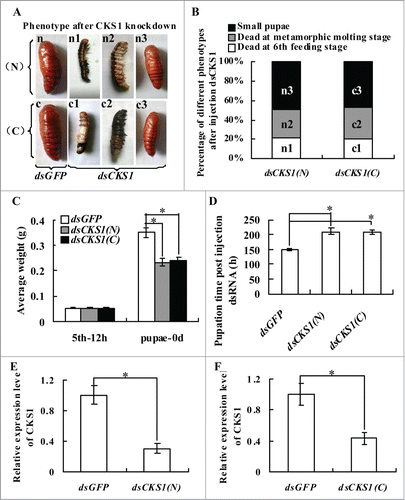
Fifth instar 12 h larvae were selected for CKS1 knockdown by injection of dsCKS1 (N) produced at the 5′ region and dsCKS1 (C) produced at 3′ region of CKS1 (Fig. S1) to determine the function of CKS1 in development. In the control group, which was injected with the same quantity of dsGFP, the larvae transformed to normal-sized pupae. However, in dsCKS1 (N)- and (C)-injected larvae, 21% of the larvae died at the sixth feeding stage, 30% of the larvae died at the metamorphic stage, and 49% of the larvae transitioned to small-sized pupae (). The body weight of the small-sized pupae decreased by 34% in the dsCKS1 (N)- and (C)-injected larvae compared with the dsGFP-injected control group (). In addition, pupation time was delayed by approximately 60 h after dsCKS1 (N) and (C) injection (). The knockdown of CKS1 was confirmed by injection of dsCKS1 (N) and dsCKS1 (C) (), and dsCKS1 (N) was used for later studies. These results suggest that CKS1 is important to insure larval growth and pupation.
Figure 2. CKS1 knockdown blocks larval growth, delays pupation, and forms small-sized pupae. Insect phenotype after CKS1 knockdown by injecting non-overlapping dsCKS1 produced by N-terminus (N) and C-terminus (C), respectively, to fifth instar 12 h larvae (500 ng/larva, 3 times in 48 h interval). n and c: Larvae from control groups injected with the same quantity of dsGFP; n1 and c1: dead larvae at the sixth feeding stage; n2 and c2: dead larvae at the sixth metamorphic molting stage; n3 and c3, small-sized pupae; (B) percentage of distribution of different phenotypes in (A); (C) statistical analysis of average body weight of pupae after CKS1 knockdown by injecting dsCKS1(N) and dsCKS1(C), with dsGFP injection as the control; (D) statistical analysis of pupation time after dsRNA injection. (E and F), qRT-PCR analyzing the specificity of RNAi by dsCKS1(N) and dsCKS1(C), 24 h post dsRNA injection in 5th 12 h larval midgut. These experiments were repeated thrice (30 larvae × 3) and statistically analyzed. The asterisks indicate significant differences from the control group (P < 0.05) by using the student's t test.
CKS1 expression determined body size
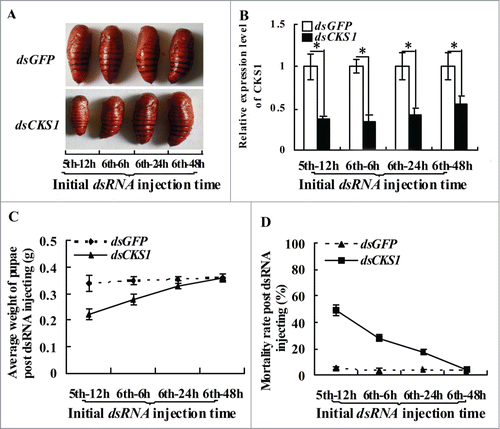
We injected dsRNA into the larvae at different developmental stages to examine the relationship between CKS1 and body size. In the control group injected with dsGFP, the larvae developed into normal-sized pupae, regardless of the dsGFP injection at fifth instar 12 h, sixth instar 6 h, sixth instar 24 h, or sixth instar 48 h stages. By contrast, in the dsCKS1-injected group, the larvae formed differentially sized pupae, and the variation in body size depended on the stage (from fifth instar 12 h to sixth instar 48 h) when dsCKS1 was injected (). Statistical analysis showed that injection at earlier larval stages resulted in the formation of smaller sized pupae and higher mortality, compared with the injection at the sixth instar 48 h stage (). These results suggest that CKS1 expression determines body size.
Figure 3. CKS1 expression determines body size. (A) phenotypes after dsCKS1 injection at different larval stages (fifth 12 h, sixth 6 h, sixth 24 h and sixth 48 h, 500 ng/larva, 24 h interval for 3 times injection), dsGFP injection at different larval stages with the same condition as the control group; (B) qRT-PCR to detect the effects of RNAi in the midgut, 24 h post the first dsRNA injection. The asterisks indicate significant differences from the control group (P < 0.05) by using the student's t test. (C) calculation of average pupae weight in (A); (D) calculation of average mortality rate in (A). The average was from 3 repeats (20 larvae × 3).
CKS1 participated in midgut cell proliferation
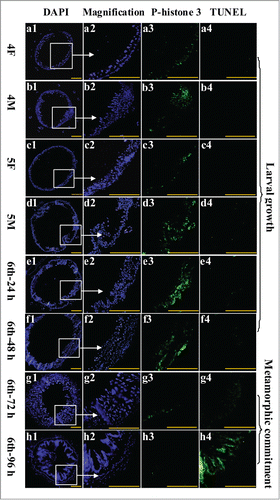
To study the function of CKS1 in cell proliferation, we examined midgut cell proliferation during larval development via immunohistochemistry analysis. During larval growth stages fourth instar 12 h (4F) to the sixth instar 48 h, midgut cell number and midgut size obviously increased, as detected by 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) staining (). Meanwhile, the cell proliferation signal was detected in larval midgut cells by the antibody anti-phospho-histone 3 from fourth instar 12 h to sixth instar 48 h (). However, a weak cell proliferation signal was detected in both larval midgut and imaginal midgut at the sixth instar 72 h and 96 h stages when the larvae became metamorphically committed (). However, the programmed cell death (PCD) signal was detected by terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) staining in the midgut at the sixth instar 96 h stage (). These results suggested that the larval midgut cells proliferate to increase the midgut cell number, but the imaginal midgut cells do not proliferate. The larval midgut cells stop proliferating and undergo PCD during metamorphosis.
Figure 4. Examination of midgut cell proliferation and PCD from 4th instar to 6th instar based on immunohistochemistry analysis. Panels a1 to h1, DAPI staining showing the nuclei of midgut cells; a2 to h2, magnification of a1 to h1; a3 to h3, phospho-histone 3 detection using the antibody and goat anti-mouse IgG Alexa-Fluor 488 (green) showing cell proliferation; a4 to h4, TUNEL staining showing the midgut PCD. Bar indicates 50 μm.
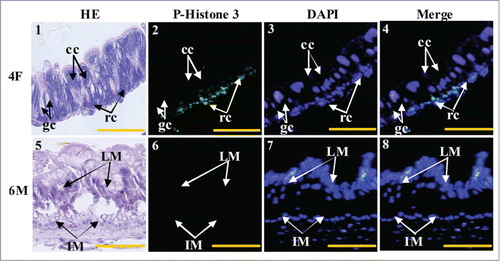
The cell types that proliferated in the larval midgut were further identified. Column cells, goblet cells, and regenerative cells were visualized via hematoxylin–eosin (HE) staining of the fourth instar feeding larvae (4F) stage (). The regenerative cells were phospho-histone 3-positive (). The imaginal midgut cells of the sixth instar 96 h larvae (6 M) were not stained by phospho-histone 3 antibody (). These results suggest that regenerative cells exist and proliferate during the larval growth stage, but not in the imaginal midgut at the metamorphic stage.
Figure 5. Midgut cell proliferation occurs in the larval regenerative cells. Panel 1, HE staining showing the midgut cells in fourth instar feeding larvae, cc: columnar cells, gc: goblet cells, rc: regenerative cells; panel 2, phospho-histone 3 antibody detection showing the proliferating cells; panel 3, DAPI staining showing the cell nucleus; panel 4, merged image of panels 2 and 3; panel 5, HE staining showing the midgut of sixth instar 96 h larvae, LM: larval midgut, IM: imaginal midgut; panel 6, phospho-histone 3 antibody detection; panel 7, DAPI staining; panel 8, merged image of panels 6 and 7. Bar indicates 20 μm.
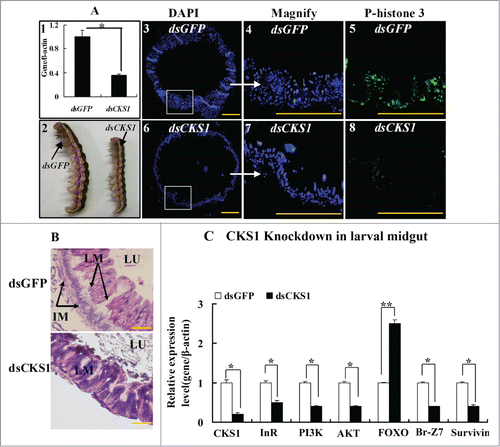
The involvement of CKS1 in midgut cell proliferation was examined in larvae using the RNAi technique. When CKS1 was knocked down by the injection of dsCKS1 into the fifth instar 12 h larvae (), larval body size decreased at sixth instar 24 h. However, the body size of the dsGFP-injected control larvae at the same sixth instar 24 h did not decrease (). In correlation, midgut cell proliferation occurred at sixth instar 24 h in the dsGFP-injected control larvae, as detected by anti-phospho-histone 3 primary antibody and goat anti-mouse IgG Alexa-Fluor 488 second antibody (). However, fewer midgut cells proliferated at sixth instar 24 h, as detected by phospho-histone 3 antibody in dsCKS1-injected larvae (). Statistical analysis of the cell numbers and phospho-histone 3 confirmed the significant difference (Fig. S2). Furthermore, imaginal midgut was formed when dsGFP-injected larvae developed to sixth instar 96 h. No imaginal midgut was formed, but larval midgut formed in dsCKS1-injected larvae when the larvae developed to sixth instar 96 h (). These results suggested that CKS1 regulates larval midgut cell proliferation and affects imaginal midgut formation. Meanwhile, the expression levels of InR, PI3K, Akt, Br, and survivin were repressed, whereas that of FOXO was upregulated by CKS1 knockdown in larval midgut (). These results indicate that CKS1 expression may positively promote insulin pathway gene expression, probably by promoting cell proliferation and nutrient absorption.
Figure 6. CKS1 knockdown represses cell proliferation and gene expression in the larval midgut. dsCKS1 was injected to the fifth instar 12 h larvae twice, 48 h interval, 500 ng/larva. dsGFP injection was the non-specific RNAi control. (A) Immunohistochemistry showing the repress of midgut cell proliferation by CKS1 knowckdown. Panel 1, efficacy of CKS1 knockdown examined at the sixth instar 24 h larvae; panel 2, phenotypes related to panel 1; panels 3 to 5, detection of cell proliferation after dsGFP injection by the antibody against phospho-histone 3 and goat anti-mouse IgG Alexa-Fluor 488 (green) second antibody; panels 6 to 8, detection of cell proliferation by anti-phospho-histone 3 primary antibody after dsCKS1 injection. Bar indicates 50 μm. The white arrows pointed to the amplification of the pictures. (B) HE-stained midguts at 6th 96 h after dsGFP or dsCKS1 was injected. LM: larval midgut, LU: lumen of midgut, IM: imaginal midgut. The scale bar is 50 μm. (C) qRT-PCR showing the gene expression after CKS1 knockdown in larval midgut. Asterisks indicate significant differences from the control group (P < 0.05) by using the student's t test based 3 independent repeats.
CKS1 overexpression in the HaEpi cells promoted cell proliferation
CKS1 was overexpressed as CKS1-GFP-His fusion protein in HaEpi cells, and PIEx-4-GFP-His vector was used to confirm its function in cell proliferation. The overexpression of GFP alone was used as the control parameter. The overexpressed GFP was distributed in the cytoplasm and in the nucleus, and phospho-histone 3 signal was not detected in GFP-expressing cells. By contrast, the overexpressed CKS1-GFP was mainly distributed in the nucleus, and phospho-histone 3 signals were detected in the cells (). These results suggest that overexpression of CKS1 promotes cell proliferation.
Figure 7. CKS1 overexpression in HaEpi cells leads to cell proliferation. (A) Detection of cell proliferation by the phospho-histone 3 antibody. Green fluorescence indicates overexpressed GFP or CKS1-GFP, and red fluorescence indicates the phospho-histone 3 detected by the phospho-histone 3 antibody and goat anti-mouse IgG Alexa-Fluor 568 (red). Blue indicates cell nucleus stained by DAPI. Merge is the overlapped red, green and blue. Panels 1 to 4, overexpressed-GFP; panels 5 to 8, overexpressed CKS1-GFP, bar indicates 20 μm; (B) statistic analysis of P-histone 3-staining cells according to the images in Figure S8A. (C) Western blot to confirm the expression and subcellular distribution of GFP and CKS1-GFP with His-taq that were detected by the monoclonal antibody anti-His-tag. Lanes 1 and 2, overexpressed GFP and CKS1-GFP, β-actin was used as the loading control; lanes 3 and 4, subcellular distribution of overexpressed GFP; lanes 5 and 6, subcellular distribution of overexpressed CKS1-GFP. SDS-PAGE gel with commassie Brilliant Blue staining was performed at the same time as loading control to normalize the protein quantity and quality in the cytosal and nucleus. cy: cytoplasm, Nu: nucleus. Bar indicates 20 μm. (D) Detection of cell proliferation by EdU. Green fluorescence indicates overexpressed GFP or CKS1-GFP, and red fluorescence indicates EdU staining; blue indicates cell nucleus stained by DAPI; merge is the overlapped red, green and blue. Bar: 20 μm. (E) Statistic analysis of EdU-staining cells according to the images in Figure S8B. Asterisks indicate significant differences between the groups (P < 0.05) by the student's t test based on 3 independent experiments.
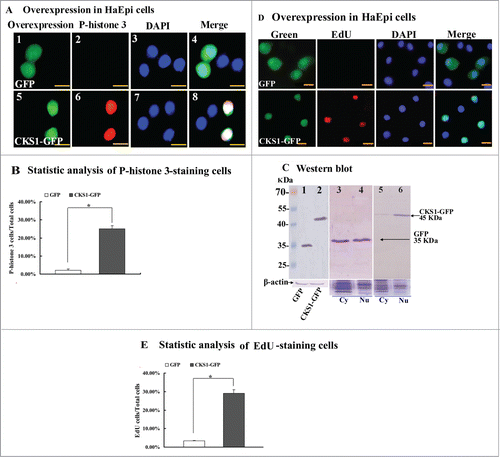
Western blot was performed using the mouse-anti-His-tag antibody to confirm the overexpression of CKS1 in the cells. GFP-His and CKS1-GFP-His were correctly expressed in the cells (Fig. S3), with molecular weights of 35 kDa (GFP-His) and 46 kDa (35 kDa of GFP-His plus 11 kDa of CKS1), respectively (, lanes 1 and 2). The overexpressed GFP-His was detected at equal levels in the cytoplasm and the nucleus (, lanes 3 and 4), whereas the overexpressed CKS1-GFP-His was mainly detected in the nucleus (, lanes 5 and 6). These results confirm the expression of CKS1 and indicate that CKS1 is mainly located in the nucleus.
To confirm the function of CKS1 in promoting cell proliferation, as detected by phospho-histone 3, 5-ethynyl-2´-deoxyuridine (EdU) staining was used. EdU is a novel thymidine analog that is incorporated into the DNA of dividing cells to indicate DNA synthesis.Citation21 EdU signals were detected from the cells that overexpressed CKS1-GFP-His, but fewer EdU signals were detected from the control cells that overexpressed GFP-His alone (). The EdU signals were significantly higher in CKS1-GFP-His-overexpressing cells than in GFP-His-overexpressing cells as confirmed by statistical analysis (). These results confirm that CKS1 promotes cell proliferation.
The amino acid N45 in the CKS domain had a critical effect on the function of CKS1
To address the critical motifs of CKS1 in promoting cell proliferation, 3 truncation mutations were constructed as follows: wild-type CKS1 with full length (b1), N-terminal truncation (b2), C-terminal truncation (b3), and N- and C-terminal double truncation (b4) (). The mutants were then overexpressed by the vector pIEx-GFP-His in HaEpi cells. All three truncation mutants (b2, b3, and b4) were localized in the nucleus and could lead to cell proliferation (). These results suggest that the N- or C-terminal sequences are not essential to CKS1 activity and that the CKS domain determines CKS1s activity in cell proliferation.
Figure 8. N45 in the CKS domain has a critical effect on the function of CKS1 in cell proliferation. (A) protein sequence of H. armigera CKS1; shadowed amino acids are the CKS domain; (B) models for truncation mutants of CKS1; (C) overexpression (OE) of mutants b2, b3, and b4; green color indicates overexpressed mutants; red color indicates phospho-histone 3 detected by the specific antibody and goat anti-mouse IgG Alexa-Fluor 568 (red); blue indicates the nucleus stained by DAPI; merge shows the overlapped green, red, and blue color; (D) OE of site mutation N45G (45N to G). Bar indicates 20 μm.
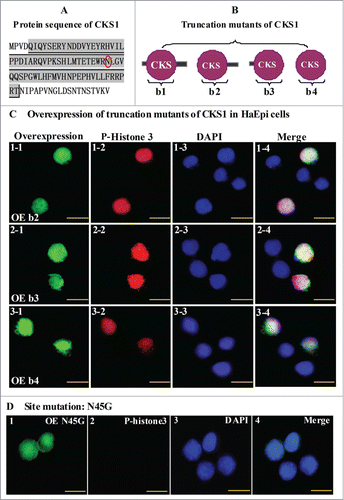
According to previous studies on humans, S41 serine and N45 asparagine mutants of CKS1 can notably decrease the function of CKS1 in cell proliferation.Citation15 The S41 mutation of CKS1 in mammals is varied to T41 in insects, but the N45 mutation of CKS1 is conserved from insects to mammals (Fig. S4). Hence, we selected N45 as the point mutation by replacing amino acid asparagine-45 (N) to glycine (G). No proliferation signal was detected by the phospho-histone 3 antibody in the N45G site in cells expressing the mutation, although the signal was localized in the nucleus (). These results suggest that amino acid N45 is the CKS1 domain's critical site for promoting cell proliferation.
CKS1 was upregulated by insulin, but was repressed by high 20E concentrations
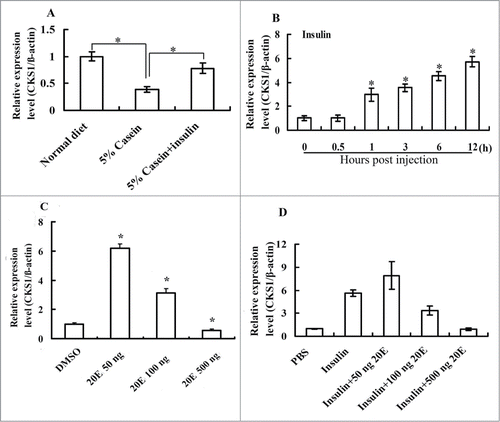
Considering the higher expression of CKS1 during feeding stages, we investigated the regulation of CKS1 by insulin and 20E by injecting hormones into the sixth instar 6 h larval hemocoel. CKS1 expression decreased under nutrition-deficient conditions, when the larvae were fed with 5% casein. However, CKS1 expression was partially rescued when insulin was injected (), thereby indicating that CKS1 expression is controlled by the insulin pathway. The CKS1 expression levels increased when insulin was injected from 1 h to 12 h (), confirming that CKS1 expression was regulated by insulin. A low concentration of 20E (50 ng) injected alone upregulated the expression of CKS1. However, high concentrations of 20E (100–500 ng) applied alone or injected with insulin repressed CKS1 expression in larval midgut (), thereby indicating that CKS1 expression is coordinately regulated by insulin and 20E.
Figure 9. CKS1 is upregulated by the insulin and repressed by 20E. (A) Starvation and rescue experiments in the larvae The sixth instar 6 h larvae were fed with normal diet, 5% casein (in 2% agarose, starvation), or 5% casein (24 h) plus insulin injection (2 μg/larva, rescue) for 6 h; (B) insulin (2 μg/larva) injection at the sixth instar 6 h larvae; (C) effect of 20E injection on CKS1 expression in sixth instar 6 h larvae for 6 h (50/100/500 ng/larva). (D) Effect of 20E (50/100/500 ng/larva) on insulin (2 μg/larva)-induced CKS1 expression in 6 h; The asterisks indicate significant differences from the control group (P < 0.05) by using the student's t test based triplicates.
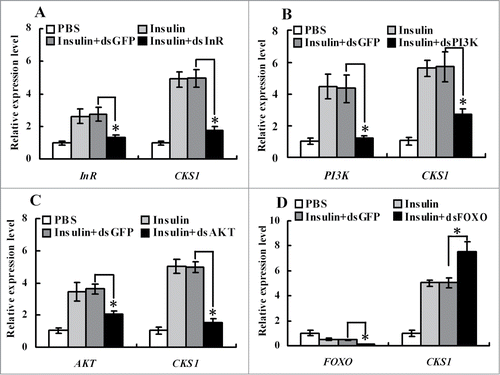
To demonstrate the pathway by which insulin upregulated CKS1 expression, insulin receptor InR, protein kinases PI3K and AKT, and transcription factor FOXO were knocked down in HaEpi cells. Knockdown of InR, PI3K, and AKT blocked insulin-induced CKS1 expression (), whereas knockdown of FOXO increased CKS1 expression (). These data indicated that CKS1 is upregulated by insulin via its receptor InR. Thus, we confirm that CKS1 serves its function downstream of InR, PI3K, AKT, and FOXO.
Figure 10. qRT-PCR showing the regulation of CKS1 by insulin via InR. (A) Knockdown of InR, (B) knockdown of PI3K, (C) knockdown of AKT, (D) knockdown of FOXO. 1*PBS is used as the solvent control for insulin, dsGFP as the non-specific dsRNA control for dsCKS1. These experiments were repeated 3 times and statistically analyzed. The asterisks indicate significant differences from the control group (P < 0.05) by using the student's t test.
Discussion
The mechanism involved in maintaining or switching off cell proliferation is not fully understood. Insect midgut presents a good model for studying the mechanisms underlying the regulation of cell proliferation by insulin and steroid hormone. During larval growth, insulin promotes CKS1 expression to increase midgut cell number and midgut size for nutrient absorption and imaginal midgut formation. During metamorphosis, high concentrations of 20E repress CKS1 expression, thereby stopping cell proliferation. CKS1 contributes to body size control by promoting cell proliferation under the coordinated regulation of insulin and 20E.
CKS1 promoted cell proliferation
Insect intestinal stem cells proliferate and differentiate to form the imaginal midgut during metamorphosis, as observed in the red flour beetle Tribolium castaneum.Citation22 Cell proliferation was observed in the regenerative cells located at the bottom of the larval midgut of H. armigera. However, cell proliferation occurred in regenerative cells during larval growth, but not in imaginal midgut cells at the metamorphic stage. This phenomenon might be the difference between lepidopterans and beetles. CKS1 knockdown not only repressed larval midgut cell proliferation but also repressed imaginal midgut formation. Therefore, CKS1 promotes midgut cell proliferation, thereby increasing larval midgut cell number and forming the imaginal midgut. CKS1 may also participate in cell proliferation in other tissues, including fat body and epidermis, because CKS1 is also expressed in these tissues.
CKS1 knockdown blocked larval growth and resulted in about 51% larval mortality. The mechanism underlying this phenomenon might involve CKS1 knockdown, which repressed expression of genes, including InR, PI3K, AKT, Br, and survivin, but increased FOXO expression. The repression of InR, PI3K, and AKT expression and the increase of FOXO by CKS1 knockdown indicated the repression of gene expression in the insulin pathway. The mechanism underlying CKS1s effect on gene expression in the insulin pathway might be due to CKS1s promotion of midgut cell proliferation and nutrient absorption. CKS1s affects gene expression in the insulin pathway via a positive feedback mechanism.
FOXO is a transcription factor that plays important roles in the nucleus. FOXO's function is opposite that of insulin, and high FOXO expression may repress the insulin pathway.Citation6 FOXO function is inhibited by the insulin pathway via AKT's phosphorylation of FOXO, thereby arresting FOXO in the cytosol.Citation6 FOXO may inhibit dMyc-induced cell growth and proliferation in Drosophila.Citation7 We observed that CKS1 knockdown resulted in the increase of FOXO mRNA. In turn, FOXO knockdown resulted in the increase of CKS1 mRNA, and FOXO overexpression repressed insulin-regulated CKS1 expression (Fig. S6). Thus, FOXO represses CKS1 expression, thereby repressing cell proliferation in the insulin pathway.
Br is known as a metamorphosis-initiating factor in the 20E pathway.Citation23,24 Insulin can regulate Br expression by promoting growth and 20E production in the prothoracic gland in Drosophila.Citation13 The decrease of Br expression by CKS1 knockdown indicates the repression of metamorphosis, which resulted in the delay of pupation.
Results of our RNAi experiments in larvae showed that the nuclei in the midgut cells did not appear smaller. The nuclei of CKS1-overexpressing HaEpi cells also did not appear enlarged. We further measured the area of the entire cell including the cytoplasm; the dsCKS1-treated cells had smaller area than the dsGFP-treated cells. The CKS1-GFP-overexpressing cells had larger area than the GFP-overexpressing cells. However, no significant difference was found, as confirmed by statistical analysis (Fig. S5).
N45 is the amino acid asparagine-45 in the cyclin-dependent kinase regulatory subunit (CKS) domain. In humans, N45 of CKS1 is the Skp2 ubiquitin ligase-binding site. Mutation of S41 and N45 of CKS1 cannotably decrease the function of CKS1 in cell proliferation.Citation15 CKS1–Skp2 complex promotes the association of Skp2 with CDK inhibitor p27 to promote the ubiquitinylation of p27 for degradation, thereby facilitating the function of CDK in cell proliferation.Citation18 We found that the amino acid asparagine N45 is the critical site of CKS1 in promoting cell proliferation in insect.
Midgut cell proliferation contributed to critical body weight control
Mammal intestinal epithelium cells are homeostatically maintained by the proliferation of intestinal stem cells.Citation25,26 Mammalian stem cells divide to renew themselves and to produce daughter cells or transient amplified (TA) cells, which are capable of further proliferating to intestinal epithelium cells.Citation27 However, Drosophila midgut cells directly divide from intestinal stem cells, and no TA cells are observed; thus, the regenerative cells are stem cells.Citation25 Drosophila intestinal stem cells are located at the posterior midgut region.Citation28,29 We detected cell proliferation in the midgut during the larval growth stage in Helicoverpa. Further study needs to be performed to determine whether these cells are stem cells or TA cells.
Critical body size-measure organs might be growing organs, such as imaginal discs. However, most imaginal discs grow after pupation in lepidopterans.Citation30 Fat body has been demonstrated as the critical organ for body size measurement in Drosophila because it increases in volume and stores nutrients.Citation31 In addition to fat body, the midgut is the second largest organ inside the insect's body. The metamorphosis of holometabolous insects is initiated in the absence of further nutritional input when the larvae reach a critical weight (species-specific size).Citation32 The midgut is the original nutrient-absorbing organ. The midgut increases its size by cell proliferation from regenerative cells. The increased midgut size enhances nutrient absorption to enable growth of other tissues, such as the fat body and the prothoracic gland. The midgut is also a nutrient storage organ. The larval midgut undergoes programmed cell death during metamorphosis and releases nutrients for imaginal midgut growth. Therefore, the midgut might be another critical organ for body size measurement. This study presents a model in which insulin regulates CKS1 expression to promote cell proliferation to reach the critical body weight.
The efficacy of RNAi from fifth instar 12 h to sixth instar 48 h is 40%–50%. However, the average pupal weight is lower, and the mortality rate is higher at fifth instar 12 h compared with the sixth instar 48 h larvae. These results suggested that the sixth instar 48 h larvae can tolerate CKS1 interference more than the fifth instar 12 h larvae possibly because the sixth instar 48 h larvae have reached the critical body weight for pupation.
CKS1 expression was coordinately regulated by insulin and 20E
Insulin upregulated CKS1 expression via InR, PI3K, and Akt, whereas starvation repressed CKS1 expression. Therefore, CKS1 expression is positively regulated by the insulin pathway. This finding explains the higher expression level of CKS1 during larval growth. In contrast, the lower expression level of CKS1 during metamorphosis might be due to the repression of the CKS1 expression by high 20E concentration. The prothoracic gland uses the target of rapamycin (TOR) nutrition pathway to grow and to produce ecdysone.Citation33 20E concentration greatly increases when larvae reach a critical body weight after metamorphosis commitment at the late larval stages.Citation34,35 We observed that low 20E concentration promoted CKS1 expression, whereas high 20E concentration repressed insulin-induced CKS1 expression in HaEpi cells. CKS1 expression level was high during larval growth because of dominant insulin pathway regulation with sufficient 20E concentration. By contrast low CKS1 expression level was observed during metamorphosis when insulin pathway was repressed and 20E concentration was high.
Because the midgut cell line has not yet been established, HaEpi cells were used for this study. The HaEpi cell line was cultured to a permanent cell line, and the characters of the cell line might have normalized because of the loss of in vivo environment. We used this cell line to study the gene distribution in other tissues,Citation24 and the results from HaEpi cells are well-correlated with the results from the larval experiments. Insulin regulates the phosphorylation of Akt for 1 h, which is the critical indicator of insulin-induced signaling in HaEpi cells (Fig. S7). The 20E-induced cell responses have been reported in our previous studies.Citation36
In summary
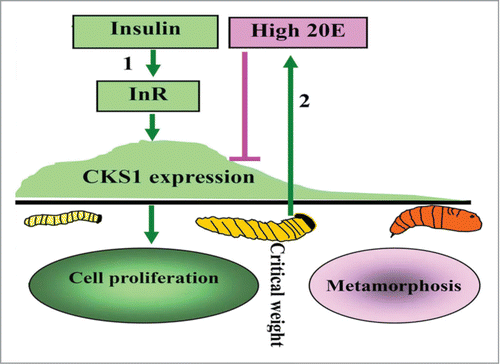
CKS1 promotes midgut regenerative cell proliferation during the larval growth stage to increase midgut cell number, thereby increasing midgut size for nutrient absorption and larval growth. Insulin (via InR, PI3K, and AKT) promotes CKS1 expression during larval growth. High 20E concentration during metamorphosis represses CKS1 expression ().
Figure 11. Illustration of the function and hormonal regulation of CKS1 in insect development. (1) insulin via its receptor InR promotes CKS1 expression for cell proliferation and larval growth to reach to critical body weight; (2) the last instar larvae produces high titer of 20E to repress CKS1 expression.
Materials and Methods
Insects
H. armigera was selected as the laboratory experiment animal. H. armigera was obtained from the Wuhan Institute of Virology (Chinese Academy of Science, Wuhan, China). The bollworms were reared with an artificial diet mainly composed of wheat and soybean at 27 ± 1°C in a 14 h light/10 h dark cycle.Citation37 The entire life span of the cotton bollworm consists of 4 developmental stages: egg, larva, pupa, and adult. The larval stage consists of 6 instars. The larvae stopped feeding at sixth instar 48 h then began to metamorphose by molting.
Quantitative real-time reverse transcription PCR (qRT-PCR)
The total RNA of insects from the fourth instar 12 h to pupae (P-3 d) was extracted using Unizol reagent (Biostar, Shanghai, China). The extract was reverse-transcribed into first strand cDNA, which was used as the template for qRT-PCR with primers CKS1-QF and CKS1-QR (Table S1). The conditions of qRT-PCR were as follows: 94°C for 3 min for initial denaturation, followed by 94°C for 20 s, 60°C for 20 s, 72°C for 20 s, 2 s for 40 cycles of plate read, and a melt curve from 65°C to 95°C. qRT-PCR was performed using a 10 μL mixture, which was composed of 5 μL of SsoFastTM EvaGreen Supermix (BioRad, California, USA), 1 μL of cDNA (1:50 diluted), 2 μL of 1 μmol/L forward primer, and 2 μL of 1 μmol/L reverse primer. β-actin was used for quantity and quality control. Data were analyzed using the formula: R = 2−[ΔCtsample − ΔCtcontrol], where R is the relative expression level, ΔCt sample is the difference between the Ct of the gene and the average β-actin in the treatment sample, and ΔCt control is the difference between the Ct of the gene and the average β-actin in the control sample.
CKS1 knockdown by dsRNA injection into the larvae
dsRNA of CKS1 was synthesized using the primer pairs CKS1-N-Fi and CKS1-N-Ri and CKS1-C-Fi and CKS1-C-Ri (Table S1) with MEGAscript™ RNAi kit (Ambion, Austin, TX, USA). dsRNAs of InR, Akt, PI3K, and FOXO were prepared using the method described above (primers are in Table S1). Fifth instar 12 h larvae were used for dsRNA injection (500 ng/larva). Three independent experiments (30 larvae × 3) were performed and analyzed. Larvae injected with the dsRNA of green fluorescence protein (dsGFP) at the same quantity were used as the nonspecific dsRNA control sample. The total RNA of the epidermis, midgut, and fat body tissues from fifth instar molting stage larvae (fifth instar 36 h) were extracted to determine the effects of RNAi-induced CKS1 knockdown by qRT-PCR. The experiments were independently performed thrice, and the results were statistically analyzed.
Hematoxylin eosin (HE) staining
Dewaxed and gradient-rehydrated histologic sections were washed thrice with a phosphate buffer saline (PBS, 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4) and subsequently stained with hematoxylin for 10 min at room temperature. After washing with running water, the tissue sections were stained with Scott's Bluing reagent (0.35 g of NaHCO3 and 2 g of MgSO4 in 100 mL of deionized H2O) for 1 min, followed by 1% hydrochloric acid–alcohol differentiation liquid for 20 s. The tissue sections were then stained with 0.5% water soluble eosin dye solution for 30 s. Finally, the stained histologic sections were washed under running water and observed under an Olympus BX51 fluorescence microscope (Shinjuku-ku, Tokyo, Japan).
Phospho-histone 3 and 5-ethynyl-2´-deoxyuridine (EdU) detection
The overexpression of CKS1 was achieved using PIEx-4-GFP-His vector in HaEpi cells. After HaEpi cells were cultured for 48 h, the cell culture medium was discarded, and the cells were washed thrice with PBS for 5 min. Subsequently, the cells were fixed in 4% paraformaldehyde for 10 min at room temperature. The cells were washed with PBS and then permeabilized with 0.2% TritonX-100 in PBS at 37°C for 30 min. After being washed with PBS thrice, the cells were blocked with a blocking buffer (2% bovine serum albumin (BSA) in PBS) for 30 min at 37°C. The mouse monoclonal antibody anti-phospho-histone 3 (phosphor S10) (Abcam Inc.., Hongkong, China) was used to incubate the cells (1:1000 in blocking buffer) at 37°C for 3 h. After washing with PBS thrice, the cells were incubated with the antibody goat anti-mouse IgG Alexa-Fluor 568 (red, 1:1000 in blocking buffer) at 37°C for 1 h. The nuclei were stained with 1 μg/mL 4–6-diamidino-2-phenylindole dihydrochloride (DAPI, AnaSpec Inc.., San Jose, CA) for 10 min at room temperature. Finally, the cells were washed thrice with PBS and examined under a fluorescence microscope. The midgut was dissected in ice-cold PBS separately and fixed with 4% paraformaldehyde in PBS at 4°C for 16 h. After being dehydrated and cleared by dimethylbenzene, the tissue slices were digested using protein kinase (50 μg/mL) at 37°C for 10 min. After washing with PBS thrice, the tissue slices were blocked with buffer for 30 min at 37°C. The mouse monoclonal antibody anti-phospho-histone 3 was used to incubate the tissue slices at 37°C for 3 h. After washing with PBS thrice, the tissue slices were incubated with the antibody goat anti-mouse IgG Alexa-Fluor 488 (green, 1:1000 in blocking buffer) at 37°C for 1 h. The nuclei were stained with DAPI for 10 min at room temperature. Finally, the tissue slices were washed thrice with PBS and examined under a fluorescence microscope. An EdU kit was purchased from RIBOBIO, China, and the staining method used was in accordance to the supplier's protocol. HaEpi cells were cultured to 80% confluence at 27°C in Grace's medium supplemented with 10% fetal bovine serum (FBS). CKS1-GFP was overexpressed in cells using PIEx-4-GFP-His vector, and GFP-His was overexpressed as the tag control. Cells were incubated in 100 μL EdU substrate (50 μM, Grace's:buffer A = 1000:1) for 2 h. After washing with Dulbecco's Phosphate Buffered Saline (DPBS; 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, and 8 mM Na2HPO4) twice for 5 min, the cells were fixed using 100 μL 4% paraformaldehyde for 30 min at room temperature, incubated with 100 μL 2 mg/mL glycine for 5 min with shaking, washed with DPBS twice for 5 min, incubated with 1× Apollo staining reaction liquid for 30 min with shaking in the dark, washed with DPBS twice for 10 min, incubated with 1× Hoechst 33342 staining reaction (deionized water : Hoechst 33342 = 100 : 1) for 30 min with shaking in the dark, and then washed with DPBS twice for 5 min.
Terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) assay
The histologic sections were dewaxed and gradient-rehydrated using the above-described procedure. The sections were subsequently digested using proteinase K (20 μM) for 10 min at 37°C. An In Situ Cell Death Detection Kit (Roche, Mannheim, Germany) was used to detect the programmed cell death (PCD) of the tissues based on the manufacturer's instructions as follows: cells or histologic sections were fixed and permeabilized, washed with PBS, incubated with a mixture of enzyme and label solutions (1:10) for 1 h at 37°C, and finally, the nuclei were stained using DAPI (1 μg/mL in PBS) at room temperature for 10 min. After washing with PBS thrice, the histologic sections were observed under an Olympus BX51 fluorescence microscope.
Overexpression of CKS1 in HaEpi cells
The full sequence of CKS1 used for overexpression was amplified using primers CKS1-OE-F and CKS1-OE-R (Table S1) and then inserted into the overexpression vector pIEx-4-GFP-His. The recombinant plasmid was used for overexpression in HaEpi cells, which were established from the fifth instar larval epidermis of H. armigera and named HaEpi cells.Citation38 The HaEpi cells were cultured on a cover glass in a 24-well tissue culture plate with 500 μL of 10% FBS containing Grace's complete culture medium at 27 °C until the coverage ratio reached 70%–80%. DNAfectin Transfection Reagent (Tiangen, Beijing, China) was used for the transfection of the recombinant overexpression plasmid pIEx-CKS1-GFP. The details of the transfection process are as follows: a total of 2 μL of Cellfectin II was diluted in 25 μL of Grace's medium without antibiotics and serum, gently mixed, and incubated at room temperature for 10 min. In a separate cover glass, 0.5 μg of the recombinant plasmid was diluted in 25 μL Grace's medium without antibiotics and serum, gently mixed, and incubated at room temperature for 10 min. Two parts each of the above solutions were gently mixed and incubated for 15 min at room temperature. The DNA–lipid mixture was subsequently added into the well with 200 μL Grace's medium without antibiotics and incubated in the serum for 16 h. The medium was subsequently replaced with 10% FBS containing Grace's complete culture medium and cultured for 48 h.
Construction of mutant overexpression plasmids
The N-terminus deletion mutant was obtained via PCR by using primers CKS1-OE-domain-F and CKS1-OE-R. The C-terminus deletion mutant sequence was obtained using specific primers CKS1-OE-F and CKS1-OE-domain-R. The N- and C-terminal double mutant (domain overexpression sequence) sequences were amplified using primers CKS1-OE-domain-F and CKS1-OE-domain-R (Table S1). For point mutations, primers CKS1-OE-F and CKS1-OE-N45G-R were used to amplify the forward sequence, whereas primers CKS1-OE-N45G-F and CKS1-OE-R were used to amplify the backward sequence; the two obtained sequences were used to amplify the point mutant sequence via PCR using the primers CKS1-OE-F and CKS1-OE-R. The mutant sequences were inserted into the overexpression vector pIEx-GFP-His. The recombinant overexpression plasmids were transfected into the HaEpi cells and observed under the Olympus BX51 fluorescence microscope, as described above.
Starvation and hormonal induction in larvae
Insulin (human insulin) was diluted in PBS and stored at −20°C at a storage concentration of 10 mg/mL. The stored insulin was diluted to 500 ng/μL with PBS before injection into the larvae. 20E (Sigma, Steinheim, Germany) was dissolved in DMSO (10 mg/mL, 20 mM). The sixth instar 6 h larvae were fed with normal diet, 5% casein in 2% agarose, or 5% casein plus insulin injection (Sigma, Steinheim, Germany). For insulin injection, 2 μg was injected into each larva (500 ng/μL, for a total of 4 μL), and the control group was injected with an equal volume of PBS. For the 20E injection, 500 ng 20E was used for each larva (100 ng/μL, for a total of 5 μL),Citation39,40 with DMSO as the solvent control.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Author Contributions
Chun-Yan Liu performed the experiments and drafted the manuscript; Wen-Li Zhao did EdU staining and analysis; Jin-Xing Wang directed the studies; Xiao-Fan Zhao designed the studies and edited the manuscript.
1053664_supplemental_files.doc
Download MS Word (10.3 MB)Funding
This work was supported by the grants from the National Basic Research Program of China (973 Program, No. 2012CB114101) and the National Natural Science Foundation of China (No. 31230067) and Ph.D. Programs Foundation of Ministry of Education of China (No. 20120131110025).
References
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol 2001; 11:213-21; PMID:11250149; http://dx.doi.org/10.1016/S0960-9822(01)00068-9
- Nijhout HF. The control of body size in insects. Dev Biol 2003; 261:1-9; PMID:12941617; http://dx.doi.org/10.1016/S0012-1606(03)00276-8
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001; 414:799-806; PMID:11742412; http://dx.doi.org/10.1038/414799a
- Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, Hafen E, Sun H, Xu T. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development 1999; 126:5365-72; PMID:10556061
- Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol 1999; 1:500-6; PMID:10587646; http://dx.doi.org/10.1038/70293
- Puig O, Mattila J. Understanding Forkhead box class O function: lessons from Drosophila melanogaster. Antioxid Redox Signal 2011; 14:635-47; PMID:20618068; http://dx.doi.org/10.1089/ars.2010.3407
- Demontis F, Perrimon N. Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 2009; 136:983-93; PMID:19211682; http://dx.doi.org/10.1242/dev.027466
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 1999; 97:865-75; PMID:10399915; http://dx.doi.org/10.1016/S0092-8674(00)80799-0
- Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carre C, Noselli S, Leopold P. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 2005; 310:667-70; PMID:16179433; http://dx.doi.org/10.1126/science.1119432
- McBrayer Z, Ono H, Shimell M, Parvy JP, Beckstead RB, Warren JT, Thummel CS, Dauphin-Villemant C, Gilbert LI, O'Connor MB. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell 2007; 13:857-71; PMID:18061567; http://dx.doi.org/10.1016/j.devcel.2007.11.003
- Orme MH, Leevers SJ. Flies on steroids: the interplay between ecdysone and insulin signaling. Cell Metab 2005; 2:277-8; PMID:16271526; http://dx.doi.org/10.1016/j.cmet.2005.10.005
- Mirth C, Truman JW, Riddiford LM. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr Biol 2005; 15:1796-807; PMID:16182527; http://dx.doi.org/10.1016/j.cub.2005.09.017
- Mirth CK, Riddiford LM. Size assessment and growth control: how adult size is determined in insects. Bioessays 2007; 29:344-55; PMID:17373657; http://dx.doi.org/10.1002/bies.20552
- Patra D, Dunphy WG. Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase- promoting complex at mitosis. Genes Dev 1998; 12:2549-59; PMID:9716407; http://dx.doi.org/10.1101/gad.12.16.2549
- Sitry D, Seeliger MA, Ko TK, Ganoth D, Breward SE, Itzhaki LS, Pagano M, Hershko A. Three different binding sites of Cks1 are required for p27-ubiquitin ligation. J Biol Chem 2002; 277:42233-40; PMID:12140288; http://dx.doi.org/10.1074/jbc.M205254200
- Ghorbani M, Vasavan B, Kraja E, Swan A. Cks85A and Skp2 interact to maintain diploidy and promote growth in Drosophila. Dev Biol 2011; 358:213-23; PMID:21827746
- Martinsson-Ahlzen HS, Liberal V, Grunenfelder B, Chaves SR, Spruck CH, Reed SI. Cyclin-dependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol Cell Biol 2008; 28:5698-709; PMID:18625720
- Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat Cell Biol 2001; 3:321-4; PMID:11231585
- Spruck C, Strohmaier H, Watson M, Smith AP, Ryan A, Krek TW, Reed SI. A CDK-independent function of mammalian Cks1: targeting of SCF(Skp2) to the CDK inhibitor p27Kip1. Mol Cell 2001; 7:639-50; PMID:11463388
- Swan A, Barcelo G, Schupbach T. Drosophila Cks30A interacts with Cdk1 to target Cyclin A for destruction in the female germline. Development 2005; 132:3669-78; PMID:16033797; http://dx.doi.org/10.1242/dev.01940
- Chehrehasa F, Meedeniya AC, Dwyer P, Abrahamsen G, Mackay-Sim A. EdU, a new thymidine analogue for labelling proliferating cells in the nervous system. J Neurosci Methods 2009; 177:122-30; PMID:18996411
- Parthasarathy R, Palli SR. Proliferation and differentiation of intestinal stem cells during metamorphosis of the red flour beetle, Tribolium castaneum. Dev Dyn 2008; 237:893-908; PMID:18297733
- Zhou B, Riddiford LM. Hormonal regulation and patterning of the broad-complex in the epidermis and wing discs of the tobacco hornworm, Manduca sexta. Dev Biol 2001; 231:125-37; PMID:11180957
- Cai MJ, Liu W, Pei XY, Li XR, He HJ, Wang JX, Zhao XF. Juvenile hormone prevents 20-hydroxyecdysone-induced metamorphosis by regulating the phosphorylation of a newly identified broad protein. J Biol Chem 2014; 289:26630-41; PMID:25096576
- Wang P, Hou SX. Regulation of intestinal stem cells in mammals and Drosophila. J Cell Physiol 2010; 222:33-7; PMID:19739102
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007; 449:1003-7; PMID:17934449; http://dx.doi.org/10.1038/nature06196
- Marshman E, Booth C, Potten CS. The intestinal epithelial stem cell. Bioessays 2002; 24:91-8; PMID:11782954; http://dx.doi.org/10.1002/bies.10028
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 2006; 439:475-9; PMID:16340959; http://dx.doi.org/10.1038/nature04371
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 2006; 439:470-4; PMID:16340960; http://dx.doi.org/10.1038/nature04333
- Stern D. Body-size control: how an insect knows it has grown enough. Curr Biol 2003; 13:R267-9; PMID:12676103
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell 2003; 114:739-49; PMID:14505573; http://dx.doi.org/10.1016/S0092-8674(03)00713-X
- Mirth CK, Truman JW, Riddiford LM. The ecdysone receptor controls the post-critical weight switch to nutrition-independent differentiation in Drosophila wing imaginal discs. Development 2009; 136:2345-53; PMID:19515698; http://dx.doi.org/10.1242/dev.032672
- Layalle S, Arquier N, Leopold P. The TOR pathway couples nutrition and developmental timing in Drosophila. Dev Cell 2008; 15:568-77; PMID:18854141; http://dx.doi.org/10.1016/j.devcel.2008.08.003
- Riddiford LM, Hiruma K, Zhou X, Nelson CA. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem Mol Biol 2003; 33:1327-38; PMID:14599504; http://dx.doi.org/10.1016/j.ibmb.2003.06.001
- Kumar D, Subrahmanyam B, Sharan SK, Mishra PK, Singh BMK, Suryanarayana N. Endogenous 20-hydroxyecdysone levels in the haemolymph of non-diapause-destined and diapause-destined generations of tasar silkworm, Antheraea mylitta (Lepidoptera: Saturniidae) and associated developmental changes. Eur J Entomol 2008; 105:591-8; http://dx.doi.org/10.14411/eje.2008.079
- Liu W, Cai MJ, Zheng CC, Wang JX, Zhao XF. Phospholipase cgamma1 connects the cell membrane pathway to the nuclear receptor pathway in insect steroid hormone signaling. J Biol Chem 2014; 289:13026-41; PMID:24692553; http://dx.doi.org/10.1074/jbc.M113.547018
- Zhao XF WJ, Wang YC. Purification and characterization of a cysteine proteinase from eggs of the cotton boll worm. Insect Biochem Mol Biol 1998; 28:6
- Shao HL, Zheng WW, Liu PC, Wang Q, Wang JX, Zhao XF. Establishment of a new cell line from lepidopteran epidermis and hormonal regulation on the genes. PLoS One 2008; 3:e3127; PMID:18769621; http://dx.doi.org/10.1371/journal.pone.0003127
- Parthasarathy R, Palli SR. Developmental and hormonal regulation of midgut remodeling in a lepidopteran insect, Heliothis virescens. Mech Dev 2007; 124:23-34; PMID:17107775; http://dx.doi.org/10.1016/j.mod.2006.09.002
- Jiang J, Ge X, Li Z, Wang Y, Song Q, Stanley DW, Tan A, Huang Y. MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform B in the silkworm, Bombyx mori. Insect Biochem Mol Biol 2013; 43:692-700; PMID:23707601; http://dx.doi.org/10.1016/j.ibmb.2013.05.002
