Abstract
JADE1 belongs to a small family of PHD zinc finger proteins that interacts with histone acetyl transferase (HAT) HBO1 and is associated with chromatin. We recently reported JADE1 chromatin shuttling and phosphorylation during G2/M to G1 transition, which was sensitive to Aurora A inhibition. In the current study we examined mechanisms of the cell cycle regulation by the small isoform of JADE1 protein, JADE1S, and report data showing that JADE1S has a novel function in the regulation of cytokinesis. Using FACS assays, we show that, JADE1S depletion facilitated rates of G1-cells accumulation in synchronously dividing HeLa cell cultures. Depletion of JADE1S protein in asynchronously dividing cells decreased the proportion of cytokinetic cells, and increased the proportion of multi-nuclear cells, indicative of premature and failed cytokinesis. In contrast, moderate overexpression of JADE1S increased the number of cytokinetic cells in time- and dose- dependent manner, indicating cytokinetic delay. Pharmacological inhibition of Aurora B kinase resulted in the release of JADE1S-mediated cytokinetic delay and allowed progression of abscission in cells over-expressing JADE1S. Finally, we show that JADE1S protein localized to centrosomes in interphase and mitotic cells, while during cytokinesis JADE1S localized to the midbody. Neither JADE1L nor partner of JADE1, HAT HBO1 was localized to the centrosomes or midbodies. Our study identifies the novel role for JADE1S in regulation of cytokinesis and suggests function in Aurora B kinase-mediated cytokinesis checkpoint.
Introduction
The small family of JADE proteins includes JADE1, JADE2, and JADE3 homologs.Citation1-Citation3 The JADE1 (gene for apoptosis and differentiation in epithelia-1) protein, also known as PHF17 (PHD zinc finger factor 17), is expressed in a variety of cultured epithelial cells and in epithelial cell lining in vivo (unpublished data, MP lab).Citation4 JADE1 contains one canonical Cys4HisCys3 plant homeo domain (PHD) followed by a non-canonical extended PHD domain, which are zinc-binding motifs.Citation5
JADE1 mRNA gives rise to 2 protein products: a full-length JADE1L consisting of 842 amino acids and a truncated splice variant, JADE1S, that lacks a large C-terminal fragment of 333 amino acids. The molecular and cellular function of the short isoform of JADE1 is the most described so far by us and others.Citation4,6-12 The JADE1 protein is associated with chromatin and is a candidate transcription factor.Citation7 JADE1 promotes histone H4 acetylation by associating with a histone H4-specific endogenous HAT in cultured cells and in vitro.Citation7 In the context of chromatin, the histone acetylation activity of JADE1 requires intact PHD zinc fingers, suggesting a chromatin-targeting role for PHD zinc fingers in live cells.Citation6,7 JADE1 is a part of the HAT HBO1 complex which is the most studied protein partner.Citation4,6-8,10,13-15
HBO1 (MYST2, KAT7Citation16) was originally identified in a yeast 2-hybrid screen as a HAT binding origin recognition complex-1 (Orc1).Citation17-Citation19 Histone H4-specific HAT HBO1 has been implicated in a positive role in the pre-replication complex assembly, DNA synthesis, transcriptional regulation as well as linked to the cellular stress response and carcinogenesis.Citation14,17,19-25 The cooperative interactions of JADE1 with the components of the HBO1 complex have been established.Citation6 JADE1 promotes acetylation of histone H4 by associating with HBO1 in a chromatin context.Citation6,7 JADE1 deficiency led to the downregulation of HBO1 protein and diminished chromatin recruitment of replication factors during the cell cycle.Citation4 In addition to JADE1, the cellular activities of the HBO1 complex might be controlled by the presence of other PHD zinc finger bearing partners.Citation10,26 Other protein partners of JADE1 have been reported.Citation1,7,11,27
Although the cellular role of JADE1 has been under investigation, the mechanism of JADE1 action remains elusive. Moreover, based on published studies, the activities of the 2 JADE1 isoforms in cell growth and apoptosis described thus far do not readily reconcile.Citation9,13,28 Recent reports from our laboratory demonstrated a role for JADE1 in the cell cycle.Citation4,8 In cultured cells, depletion of JADE1 proteins by siRNA decreased rates of thymidine incorporation.Citation4 Agreeing with this, the silencing of a novel long non-coding RNA, lncRNA-JADE1 resulted in JADE1 downregulation and decreased cell proliferation.Citation28 Our most recent study identifies intracellular chromatin shuttling of JADE1 and HBO1 during G2/M- to G1-phase transition linked to phosphorylation of JADE1S by a mitotic kinase.Citation8 According to this study, during the G2 gap JADE1S is phosphorylated and dissociated from chromatin, while in early G1, JADE1S is dephosphorylated, re-associated with chromatin, and localized to the nucleus. Six phosphorylated amino acid residues in a mitotic specie of JADE1S were identified by Mass Spectroscopy analysis. The chromatin dissociation and phosphorylation of JADE1S were prevented by the pharmacological inhibitor of Aurora A kinase, which is one of the mitotic master kinases.Citation8 The functional role of JADE1S during mitosis has not been addressed.
Little is known about the biological role of JADE1. In the mice model of acute kidney injury and regeneration, JADE1S and JADE1L proteins were upregulated in tubular epithelial cells during regeneration.Citation4 In the course of tubular regeneration, the activation of JADE1S isoform expression correlated with the exit of the epithelial cells from quiescent state and activation of the cell cycle, suggesting the JADE1 role in the cell cycle progression.Citation4,8
In contrast, other studies using cultured cell models found a proapoptotic and growth inhibitory function of JADE1.Citation9,13 Transient overexpression of JADE1 resulted in decreased cell growth rates, apoptosis, and activation of p21, based on which the JADE1 role in apoptosis was suggested.Citation9,13 Interestingly, efforts to create cell lines stably over-expressing JADE1 were not successful which was interpreted as being a result of JADE1 proapoptotic activity.Citation9,13
The dynamic properties of JADE1 during the cell cycle progression were evident during the G2/M to G1 transition, a time which includes cytokinesis.Citation8 Cytokinesis is a final stage of the cell division that follows mitosis and yields 2 daughter cells. Cytokinesis consists of a sequence of events including chromosome segregation and cell membrane furrowing, actin cytoskeleton remodeling, formation of the intracellular bridge and assembly of the midbody. The final separation of the 2 daughter cells, termed abscission, occurs near the midbody region which may take up to 2 hours and has been actively studied in recent years. Like other phases of the cell cycle, cytokinesis and final abscission are events tightly controlled by regulatory protein complexes, including checkpoint proteins. A number of cytokinesis-regulatory proteins associated with midbody has been identified and reported.Citation29-Citation37 For example, recently it has been reported that Abscission/NoCut Checkpoint Regulator (ANCHR) protein mediates Aurora-B-dependent abscission checkpoint control in HeLa cells. Manipulating ANCHR protein expression greatly affected the number of cytokinetic cells, as well as rates of cytokinesis.Citation38 These and other results of the study demonstrated that ANCHR protein is a negative regulator of cytokinesis and final abscission.Citation38
In the current study we investigated the role of JADE1 in the cell cycle and report the novel function which is specific to JADE1S isoform. We examined the effects of JADE1S on the rates of cell cycle progression, specifically focusing on cytokinesis. We analyzed the effects of JADE1S on the fraction of cytokinetic and multi-nuclear cells and sensitivity to the pharmacological inhibitor of Aurora B mitotic kinase. In addition, we investigated the subcellular localization of JADE1S protein in interphase, mitosis, and cytokinesis. Our results implicate that JADE1S has a novel cellular function in the regulation of the final step of cell division, cytokinesis.
Results
JADE1S regulates cell cycle progression
Our recent study has identified a functional link between cell cycle-mediated JADE1S chromatin shuttling during mitosis and JADE1S phosphorylation by a mitotic kinase.Citation8 To investigate the role of JADE1S in the cell cycle, we examined the effects of JADE1S depletion on the rates of G2/M- to G1- phase transition by FACS.
HeLa cells were synchronized by arresting the cell cycle in late G1 by treatment with mimosine. The cell cycle was released by removing the drug and adding fresh media (0 hours). Cells were collected at intervals to follow synchronization and cell cycle progression by FACS analysis (). Gated fractions of G1-, S-, and G2/M- cell populations were quantitated from the corresponding DNA profiles and plotted against time to monitor cell cycle kinetics. Rates of G2/M- to G1- cell cycle progression were followed for up to 22 hours after the cell cycle release ().
Figure 1. Depletion of JADE1S protein facilitates kinetics of G1 cell accumulation in synchronously dividing cells. (A) Experimental design: HeLa cells were transduced with non-silencing shRNA or shJADE1S as described in Methods. Cells were synchronized by arresting cell cycle in late G1 with mimosine. At 0 hours cell cycle was released and cells were collected at intervals (0–22 hours). Cell cycle progression was monitored by analysis of DNA profiles (FACS). (B, C). Kinetic analysis of G1- and G2/M- cells in synchronized cultures. Gated fractions of G1-, S*-, and G2/M- cells were plotted against time to monitor cell cycle kinetics. Non-silencing shRNA (shControl, blue line); JADE1S-specific shRNA (shJADE1S, red line). (B) G1-cell fraction. (C) G2/M-cell fraction. (D) Whole cell lysates analyzed for endogenous JADE1S, JADE1L, and β-Actin by western blots. Note that transduction of shJADE1S resulted in depletion of JADE1S protein, but not JADE1L, indicating specificity of shRNA tool. *See Fig S1.
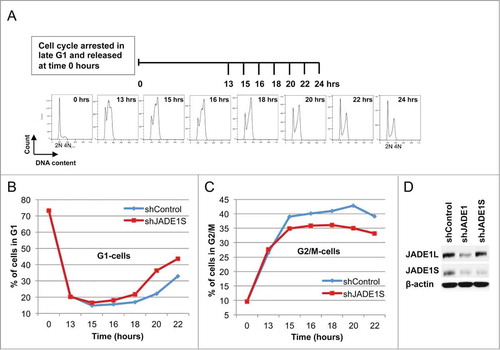
Mimosine removal resulted in release of the cell cycle from G1 arrest. This was followed by cells entering the S-phase which is evident by the sharp decrease of the G1- and increase of the S-phase cell fractions ( and Fig. S1, shControl, 0–13 hrs). Subsequently the gradual increase of G2/M cells revealed the population of cells which completed the S-phase (, shControl). The G2/M cell fraction reached a plateau between 15 hours and 18 hours, indicating progression to the G2 gap (, shControl). Finally, the increase of the nascent fraction of G1 cells at 18 hours indicated that G2/M cells progressed through mitosis and entered the G1 gap of the next cell cycle round (, shControl).
Using this model, we examined the effects of JADE1S depletion on rates of G1 cells accumulation. Lentiviral-based transduction provided uniform efficiency and resulted in a substantial decrease of endogenous JADE1S protein expression (). The silencing shRNA sequence targeted only JADE1S and did not affect expression levels of JADE1L protein (). To our surprise, depletion of JADE1S protein facilitated rates of G1-cells accumulation between 18 hours to 22 hours of the cell cycle release (, compare slopes for shControl and shJADE1S).
In order to confirm the above results, we assessed the effects of JADE1S depletion in an experimental protocol with double cell cycle synchronization. Cells were treated as in , except nocodazol was added at 8.5 hours after mimosine removal (). After conditioning for total of 12 hours, nocodazole was removed (0 hours, start of recovery, ) and the cell cycle progression was monitored at different time points by FACS analysis (). Parallel samples were used to assess endogenous JADE1S expression by western blot analysis in shControl and shJADE1S samples (). JADE1S downregulation was evident in all time points examined (). Note that, in agreement with our previous report, at 0 hours of recovery JADE1S appears phosphorylated and dissociated from chromatin fraction (, 0 hours, shControl-upper band and seeCitation8). After 4 hours of the cell cycle recovery, the JADE1S protein re-associated with chromatin and de-phosphorylated (, 4 hours, shControl, and seeCitation8). De-phosphorylation and chromatin re-association of endogenous JADE1S was accompanied by the plateauing of the G1-cells curve which indicated that newly divided cells are entering the G1 gap (, shControl, >4 hours).
Figure 2. JADE1S protein depletion facilitates kinetics of the nascent G1 cells accumulation after G2/M arrest and release of the cell cycle. (A) Experimental design: HeLa cells were arrested in late G1 by treatment with mimosine. Cell cycle was released by refreshing the media, and after 8.5 hrs of the cell cycle recovery, nocodazole was added to arrest cells in metaphase. At time 0 hours the drug was removed and the cell cycle progression was monitored by FACS analysis of DNA profiles. (B, C) Gated fractions of G1- and G2/M- cells were plotted against time to monitor cell cycle kinetics. Non-silencing shRNA (shControl, blue line); JADE1S-specific shRNA (shJADE1S, red line). (B) G1-cell fraction; (C) G2/M cell fraction; (D) Whole cell lysates of Control shRNA and JADE1S shRNA at different time points of the cell cycle release was analyzed for endogenous JADE1S and β-actin by western blots.
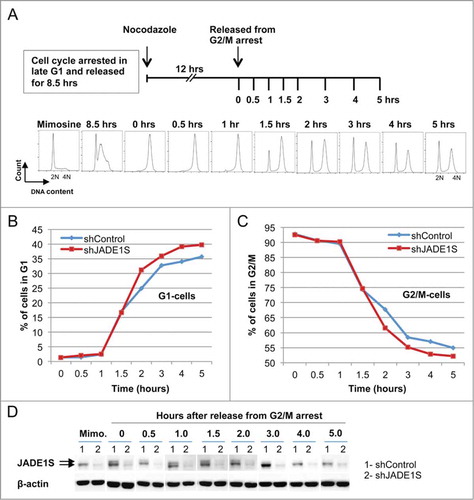
Importantly, supporting the result shown in , JADE1S depletion resulted in faster kinetics of nascent G1 cells accumulation (). Note that G1 cells accumulation by FACS measures rates of cells splitting, which is a final step of cell division. In both assays based on FACS analysis the effects of JADE1S depletion was moderate and transient, and suggested the role of JADE1S in G2/M to G1 progression, possibly cytokinesis.
JADE1S is localized to centrosomes and midbody in dividing cells
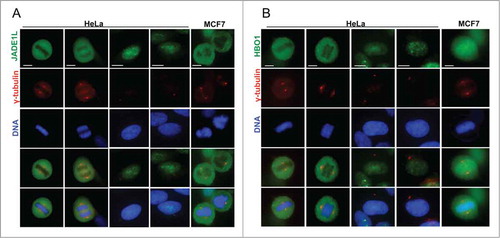
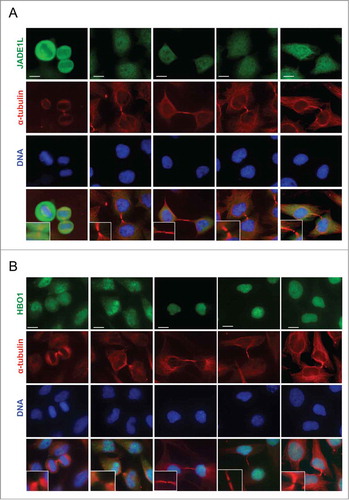
To further investigate the function of JADE1 in the cell cycle, we thoroughly examined subcellular localization of endogenous JADE1 in interphase, mitotic, and cytokinetic cells, using whole cell immunofluorescence assays. Specifically, we sought to determine whether JADE1S, JADE1L and their partner, HBO1 are localized to the key regulatory structures of the cell division, such as centrosomes and midbody. A set of non-cross-reacting antibodies specific to each of these proteins has been verified by multiple assays and described previously in detail.Citation4,8 The immunostaining revealed that in interphase cells, a fraction of endogenous JADE1S protein was concentrated around the centrosome marker gamma-tubulin, suggesting physical association with pericentrosomal material ( and Fig. S2). Similar results suggesting centrosomal association of JADE1 in interphase cells has been reported recently by others.Citation11 Interestingly, centrosomal JADE1S was evident also in prophase, metaphase and telophase of mitosis () which to our knowledge has not been shown before. In fact, as cells proceed through mitosis, JADE1S localization appeared to condense and fully co-localize with gamma-tubulin, suggesting dynamic properties (, inserts and B). In addition to gamma-tubulin, JADE1S also co-localized with other markers of centrosome, pericentrin and Aurora A (). Centrosomal localization of JADE1S protein was also confirmed in other cell lines (). Thus, our findings show that JADE1S is a novel centrosome protein. Ectopically expressed JADE1S protein recapitulated the endogenous counterpart and localized to the centrosome, supporting functionality of the overexpressed protein (not shown). Surprisingly, unlike JADE1S, JADE1L isoform did not localize to the centrosome in all cell lines examined ( and data not shown). Similarly, HBO1, which is a partner of both JADE1 isoforms, also did not localize to the centrosomes ().
Figure 3. JADE1S localizes to the centrosomes in interphase and mitotic cells. Asynchronously dividing cells were processed for immunofluorescence and co-stained with antibodies for centrosome markers: (A, B and D) γ-tubulin, (C) Aurora-A and pericentrin. (A) Representative confocal images showing the localization of endogenous JADE1S protein to the centrosomes in interphase and during mitosis (white arrows). (B) Magnified confocal images of centrosomes showing JADE1S localizing to the ‘pericentriolar’ region in interphase cells and co-localizing with γ-tubulin in mitotic centrosomes. Scale bars: 10 µm.
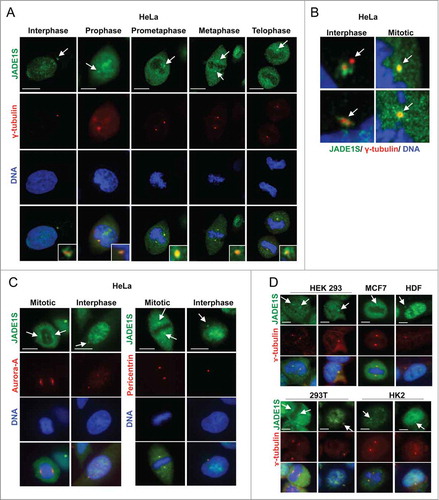
Figure 4. Endogenous JADE1L and HBO1 proteins do not localize to the centrosomes in dividing cells. (A, B) Asynchronously dividing HeLa and MCF7 cells were processed for immunofluorescence and stained with indicated antibodies. Representative confocal images showing the localization of endogenous JADE1L and HBO1 proteins in interphase and mitotic cells. γ-tubulin antibody (red) marks the centrosomes. Scale bars: 10 µm.
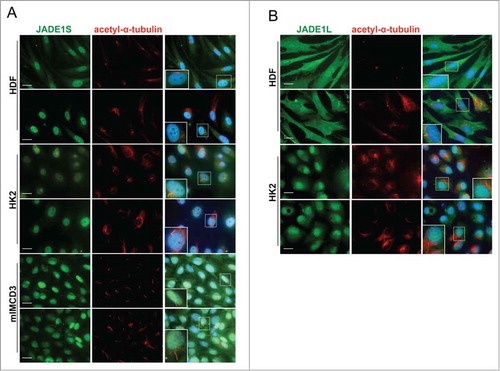
In specific conditions, centrioles and pericentriolar material of many quiescent cells are known to differentiate into a basal body that nucleates to form cilia, an organelle characteristic of a more differentiated cellular state. We investigated whether JADE1S might be associated with cilia structures in several types of cells known to grow mature cilia upon appropriate culturing conditions. Judging by the presence of acetylated α-tubulin, cilia were robustly induced and became readily detectable in conditions promoting differentiation (). In contrast, we found no evidence of JADE1 protein associations with basal body or cilia. Judging by this criteria, both JADE1S and JADE1L were absent from the cilia structures ().
Figure 5. JADE1S and JADE1L are absent from the cilia structures. For robust cilia production HDF and HK2 cells were cultured in 0.1% sera for 48 hours. IMCD3 cells were cultured for 3 d in serum-depleted followed by serum-free media for 16 hours. Cells processed for IF with indicated antibodies. Acetylated-α-tubulin antibody was used to mark cilia. Note that, (A) JADE1S and (B) JADE1L1 do not co-localize with acetylated-α-tubulin. Cells boxed are magnified as inserts. Scale Bars: 20 µm.
Next, we examined confocal images of the cells undergoing cytokinesis. Interestingly, we found that during cytokinesis a fraction of endogenous JADE1S became clearly associated with the midzone and subsequently with the midbody of cells preparing for the final abscission. Midbody was assessed by its localization in reference to the empty mid-space visualized with α-tubulin staining (). Similar results were obtained with ectopic expression of FLAG-tagged JADE1S (). Note that, unlike JADE1S, neither HBO1 nor JADE1L associated with the midzone and the midbody ().
Figure 6. JADE1S localizes to the midbody during cytokinesis. Asynchronously dividing HeLa cells were processed for immunofluorescence and co-stained with antibodies indicated. Representative confocal images showing the dynamic character of JADE1S protein localization during cytokinesis. (A) Endogenous JADE1S and (B) FLAG-JADE1S protein localizes to the midbody in early and late cytokinesis. Note that, in addition to the midbody, in early cytokinesis a fraction of JADE1S localizes to the cytoplasm, while at later stages JADE1S re-associated with chromatin. Enlarged images of the midbody are in inserts. Scale bars: 10 µm.
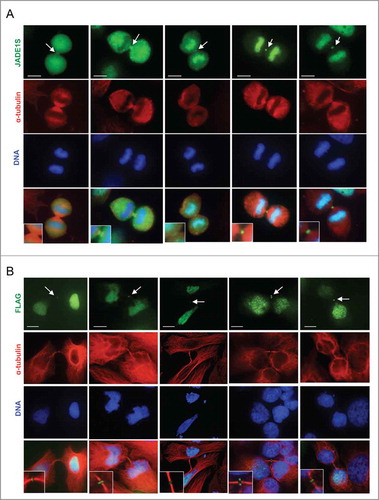
Figure 7. JADE1L and HBO1 proteins do not localize to the midbody during cytokinesis. Asynchronously dividing HeLa cells were processed for immunofluorescence and co-stained with antibodies indicated. Representative confocal images showing the localization of endogenous (A) JADE1L and (B) HBO1 proteins during various stages of cytokinesis. Enlarged images of the midbody are in inserts. Scale bars: 10 µm.
To summarize, so far we show that 1) JADE1S depletion modestly but reproducibly facilitated the accumulation of G1-cells in synchronously dividing cells; 2) JADE1S is localized to the centrosomes of interphase and mitotic cells; and 3) JADE1S is localized to the midbody of the cells undergoing cytokinesis. Taken together, our data suggested the JADE1S role in cytokinesis.
JADE1S is a negative regulator of cell cytokinesis
Cytokinesis is a final stage of the cell division that follows mitosis and yields 2 daughter cells. Cytokinesis consists of a sequence of events including chromosome segregation and cell membrane furrowing, actin cytoskeleton remodeling, formation of the intracellular bridge and assembly of the midbody.Citation29-Citation34 The final separation of the 2 daughter cells, termed abscission, occurs near the midbody region which may take up to 2 hours and has been actively studied in recent years. Like other phases of the cell cycle, cytokinesis and final abscission are events tightly controlled by regulatory protein complexes, including checkpoint proteins. A number of cytokinesis-regulatory proteins associated with midbody has been identified and reported.Citation35-Citation37 Thus, recently it has been reported that Abscission/NoCut Checkpoint Regulator (ANCHR) protein mediates Aurora-B-dependent abscission checkpoint control in HeLa cells. In that study, manipulating ANCHR protein expression greatly affected the number of cytokinetic cells, as well as rates of cytokinesis.Citation38
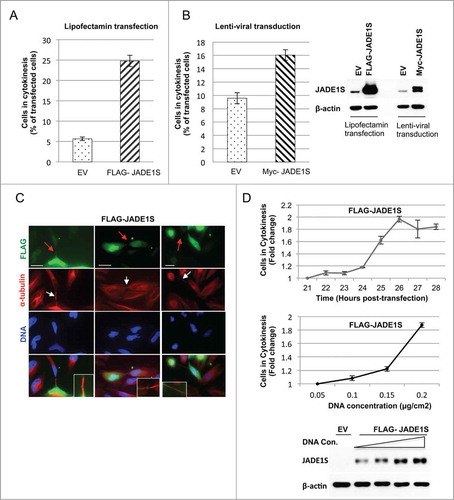
In order to test the role of JADE1S during cytokinesis we examined whether manipulating JADE1S protein expression affects the proportion of cells undergoing cytokinesis in asynchronously dividing cells. Strikingly, a 3-fold downregulation of JADE1S protein by shRNA resulted in a 1.5-fold decrease in fraction of cells in cytokinesis (). Note that the specific shRNA used targeted only JADE1S, but not JADE1L protein (). The effect was evident in cells either transfected or transduced with shJADE1S construct (). In contrast, JADE1S overexpression elevated the fraction of cells in cytokinesis (). Interestingly, while modest physiological levels of ectopically expressed Myc- tagged JADE1S caused a 1.7-fold increase of fraction of cells in cytokinesis (), the higher levels of Flag- tagged JADE1S resulted in a dramatic 4-fold increase of fraction of cells in cytokinesis (). The observed increase in number of cells undergoing cytokinesis might be due to a delay in abscission caused by JADE1S overexpression. Notethat FLAG-JADE1S-expressing cells developed the long and stable cytokinesis bridges which indicate the delay in completing cytokinesis (). To further support the JADE1S role in cytokinesis, we performed dose- and time-dependent studies with ectopically expressed JADE1S. Indeed, the proportion of cells in cytokinesis increased by 2-fold between 21 hours and 26 hours of cell culturing (, upper panel). Further supporting a role in cytokinesis delay, JADE1S protein increased the proportion of cells in cytokinesis in a dose-dependent manner (Fig. 9D, lower panel and western blot).
Figure 8. Depletion of JADE1S decreases fraction of cells in cytokinesis and increases fraction of multinucleated cells. (A and B) HeLa cells transfected or transduced with JADE1S shRNA (GFP-tagged) were processed for immunofluorescence 26 hours post-transfection or 5-days post-transduction. GFP-positive cells were scored manually to quantify cells in cytokinesis and multinucleated cells. Values are given as a percentage of the total transfected or transduced cells from the 3 independent experiments ± s.d., >100 cells per experiment. Error bars represents standard deviation. JADE1S expression was assessed by western blots of whole cell lysates from parallel samples using JADE1S and β-actin antibodies. (C) Representative confocal images showing multinucleated cells in shJADE1S-positive cells (GFP). (D) Cell cycle profiles of cells dividing asynchronously for 3-, 4-, 5-, and 6 d post-transduction (upper panel). The sub-G1 cell population peak reaches maximum at 5-days post-transduction in shJADE1S sample DNA profile (lower panel). Representative dot-plots depict the sub-G1 population. Note distinctive G1 and sub-G1 cell populations in the right insert. Scale bars: 10 µm.
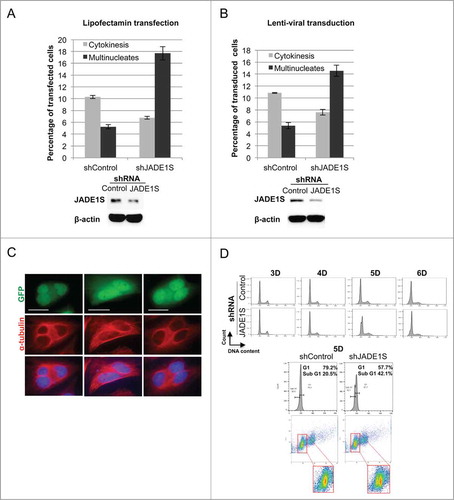
Figure 9. Ectopic expression of JADE1S increased fraction of cells in cytokinesis in time and dose-dependent manner. Effects of JADE1S were examined in lentiviral and lipofectamine-based overexpression models. Twenty six hours post-transfection and 5 d post-transduction HeLa cells were processed for immunofluorescence with FLAG and α-tubulin antibodies. (A and B) Bar graphs illustrate the effects of JADE1S overexpression on fraction of cells in cytokinesis. FLAG-JADE1S-positive cells in cytokinesis were scored manually under the fluorescence microscope, and normalized to the total JADE1S-positive cells in 3 independent experiments, ± s.d., >150 cells counted in each experiment. Error bars represents standard deviation. (C) Representative confocal images showing transfected cells arrested in cytokinesis. Red arrows indicate JADE1S localization to the midbody. White arrows points at the long bridges indicating the delay in cytokinesis abscission. Scale bars; 10 µm. (D) JADE1S increases cytokinesis scores in (D, upper panel) time- and (D, lower panel) dose-dependent manner. Transfected HeLa cells were scored manually for cells in cytokinesis as in A and B. Cytokinesis cell percentages were normalized to the value of the initial time point to get the cytokinesis fold change.
Depletion of factors regulating cytokinesis often results in either failed or premature cytokinesis manifested by the accumulation of polyploidy or aneuploidy cells. Hence, we determined the effects of JADE1S depletion on the proportion of bi- and multi-nucleated cells by manual counting of cells in samples either transfected or transduced with shJADE1S. Indeed, the decrease in cytokinetic cell fraction was associated with the 3.6-fold increase in multinucleated cell fraction (), further supporting the JADE1S role in the regulation of cytokinesis.
In addition to multi-nuclear phenotype, deregulation of cytokinesis may lead to aneuploidy. Since aneuploidy is not visible for manual assessment, we used FACS analysis and scrutinized the DNA profiles of cells depleted of JADE1S protein, after cells were cultured for increasing lengths of time. Of note, depending on the quantity, the aneuploidy cells in a given sample may not be detectable due to FACS sensitivity. Despite that, we were able to detect a sub-G1 peak in JADE1S-depleted cells, suggesting accumulation of aneuploidy cells. The sub-G1 cell population increased in a time-dependent manner (). The dot-plot of gated G1-cells shows the presence of 2 clear cell populations in the shJADE1, but not in the shControl sample (, see the insert). Thus, based on our data, JADE1S plays a role in the regulation of cytokinesis and might function in control of final abscission during cytokinesis.
JADE1S effects on cytokinesis might be mediated by Aurora B kinase pathway
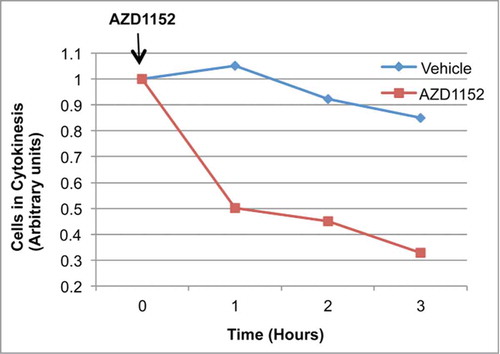
One of the master regulators of the cytokinesis and the abscission checkpoints is Aurora B, a kinase known to phosphorylate multiple targets.Citation39,40,41,42 We investigated whether JADE1S-mediated increase of cytokinesis scores is dependent on Aurora-B activity. Twenty six hours after transfection with FLAG-JADE1S cDNA, cells were treated with the pharmacological inhibitor of Aurora B, AZD1152 or vehicleCitation43 and the proportion of cells in cytokinesis was calculated by manual counting in a time course experiment. Clearly, in untreated samples the fraction of FLAG-JADE1S-positive cytokinesis cells remained largely unchanged (). In contrast, treatment with Aurora B inhibitor resulted in a time-dependent decrease of FLAG-JADE1S-positive cytokinetic cells, indicating release from cytokinetic arrest and progression through abscission. Thus, these data further support a role of JADE1S in cytokinesis and suggest that JADE1S-mediated arrest in cytokinesis depends on Aurora-B signaling.
Figure 10. Pharmacological inhibition of Aurora B kinase resulted in the release of JADE1S-mediated delay in cytokinesis. HeLa cells transiently transfected with FLAG-JADE1S cDNA. Twenty five hours post-transfection cells were treated with 1 µm AZD1152 or vehicle for 0, 1, 2 or 3 hours. Slides were processed for IF and stained with α-tubulin and Flag antibodies to score manually for cells in cytokinesis as described in . Values are given as a percentage of total transfected cells from n = 3 independent experiments ± s.d., number of cells >100 per experiment.
Discussion
JADE1 belongs to a small family of PHD zinc finger proteins that also includes JADE2 and JADE3 homologs. Our previous work has shown cell-cycle mediated chromatin shuttling of JADE1 coupled to phosphorylation, suggesting a regulatory function.Citation8 In this study we examined mechanisms of cell cycle regulation by the small isoform of JADE1 protein, JADE1S. We report a novel function of JADE1S in the regulation of cytokinesis and suggest a role in cell abscission. JADE1S depletion facilitated G2/M- to G1 progression, decreased the proportion of cells undergoing cytokinesis, and increased the proportion of multi-nuclear cells. In contrast, moderate overexpression of JADE1S increased the number of cytokinesis cells in time- and dose- dependent manner, indicating cytokinetic delay. An inhibitor of Aurora B kinase was able to release JADE1S-mediated cytokinesis arrest and allowed progression of abscission. Finally, we show that JADE1S protein localized to centrosomes in interphase and mitotic cells, while during cytokinesis JADE1S localized to the midbody. Our study identifies the novel role for JADE1S in the cytokinesis phase of the cell cycle.
The final step of cell division, cytokinesis, is initiated by signaling from anaphase spindles during mitosis and is characterized by a dramatic remodeling of dividing cells. The initial ingression of the cleavage furrow is mediated by contraction of the actomyosin ring around the spindle midzone and eventually forms the intracellular cytoskeletal bridge. Ultimately, the assembly of the midbody initiates signaling of the final stage of cytokinesis, the abscission.Citation29,32,33 The orderly recruitment of structural, remodeling and regulatory factors to the midzone ensures proper chromosome segregation and prevention of ploidy. Although early cytokinesis events have been broadly studied, the molecular mechanisms regulating the later steps of cytokinesis and abscission remain obscure. Recent studies identified the existence of an abscission checkpoint (NoCut) which includes a series of events largely mediated by the Aurora B kinase.Citation39,40,44-47
The most recently identified protein, ANCHR is a novel abscission checkpoint NoCut factor.Citation38 Interestingly, cellular responses to the manipulations of JADE1S expression levels were reminiscent of that reported for the ANCHR protein. First, similar to ANCHR, depletion and overexpression of JADE1S protein in HeLa cell cultures resulted in decrease and increase of cytokinesis frequency, respectively. Second, the effects of JADE1S over-expression on cytokinesis frequency were dose- and time- dependent. Third, as with ANCHR, depletion of JADE1S resulted in an increased frequency of multinucleates. Fourth, an inhibitor of Aurora B kinase decreased the frequency of cytokinesis cells in a time-dependent manner, suggesting release from JADE1S-mediated cytokinesis arrest. These functional data along with subcellular localization of JADE1S to the centrosome and midbody during mitosis and cytokinesis identifies JADE1S as a novel regulator of cytokinesis. Moreover, our data suggest that JADE1S functions in abscission delay mediated by Aurora B kinase pathway.
The novel finding that JADE1S protein functions in cytokinesis is exciting and raises a set of questions. First, is the known activity of JADE1S in promoting protein acetylation required for the function in cytokinesis? Acetylation of multiple targets during cytokinesis has been documented,Citation48 and this might involve JADE1S. While JADE1S partner, HBO1 did not localize to either centrosomes or midbody, other HATs might cooperate in the novel subcellular localization of JADE1S. For example, HAT TIP60 has been reported to interact with JADE1S and would be a candidate.Citation7
Second, it appears that at least 2 pools of JADE1S exist in a cell at any given cell cycle stage. Nuclear and centrosomal JADE1S is found in interphase, cytoplasmic and centrosomal JADE1S in late G2 through mitosis, and finally chromatin, nuclear and midbody-associated JADE1S pools during early and late cytokinesis. The question arises, what are the mechanisms of the specific targeting? It is highly likely that phosphorylation by specific kinases guide JADE1S localization and function. Indeed, we reported results suggesting that JADE1S dissociation from chromatin in late G2 required its phosphorylation by Aurora A kinase pathway.Citation8 We identified that 6 amino acid residues in JADE1S polypeptide are phosphorylated in a mitosis-dependent manner. Some of these sites might be involved in the targeting of JADE1S to centrosome and midbody. Other phosphorylation sites for JADE1 were reported and might play a role (http://www.phosphosite.org).Citation12
Of particular interest is the finding that the cytokinesis function appears to be specific to the small isoform of JADE1, JADE1S. JADE1S, but not JADE1L protein, consistently localized to the centrosome and midbody. We do not exclude the possibility that the antibody used did not recognize centrosomal JADE1L due to potential posttranslational modification of JADE1L protein. This, however, is unlikely since the JADE1L-specific antibody recognizes the C-terminal peptide within the JADE1L molecule which does not contain putative sites for post-translational modifications. Nevertheless, more assays are required to exclude this possibility. Assuming that the function of regulation of cell abscission is specific to JADE1S, it would be interesting to learn whether JADE1L functions in the cell cycle or has other distinctive cellular roles.
Examples of transcription factors, histone and DNA modifiers with predominantly nuclear localization that have been found to function in cytokinesis and abscission include Brd4 and WDR5.Citation49,50 Others include the FYVE and RING finger proteins ANCHR.Citation38 JADE1S bears a tandem of 2 PHD zinc fingers, including PHD2 domain which is extended and may have structural similarities with the RING finger. PHD zinc fingers of JADE1S are required for chromatin association and are candidate epigenetic code readers.Citation14,51 It would be interesting to find out whether PHD zinc fingers play a role in the novel JADE1S activities during cytokinesis and potentially abscission.
It is well known that ectopically expressed high, non-physiological levels of cell cycle regulatory proteins are likely to inflict negative, even deleterious cellular effects. In fact, it has been our observation that vastly over-expressed JADE1 proteins are highly toxic to the cell and must be avoided. We found that transfection of JADE1S cDNA in quantities suggested by the manufacturers typically resulted in many dead cells, cells with abnormal phenotype with large, multi-lobular nuclei, and diminished the apparent transfection efficiency, most likely due to cell death (Fig. S3, and not shown). Considering this, for this functional study we used approaches of either JADE1S knockdown or ectopic expression of physiologically relevant protein quantities. The role of JADE1S in apoptosis has been proposed in earlier studies which were often reliant on overexpression approaches.Citation9,13 While role in signaling of apoptosis is quite possible, these studies and their interpretations may need to be re-visited.
In conclusion, the results of this study uncover a novel function for JADE1S protein in the regulation of cytokinesis of the cell cycle. In addition to the ability to promote chromatin associations of a HAT complex, the specific pool of JADE1S protein associates with centrosome and midbody and function to negatively regulate cytokinesis. Regulation of cytokinesis including NoCut checkpoint provides faithful division of the cellular content of the 2 daughter cells. The final stage of the cell division, cytokinesis and abscission, has been studied actively during recent years and more proteins were found to play a role. Specific molecular and cellular mechanisms of JADE1S activities require more studies and are yet to be uncovered.
Materials and Methods
Cell culture
HeLa, HEK-293, MCF7, HDF, 293T/17 and mIMCD-3 were obtained from ATCC. All cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) Penicillin-Streptomycin (Cellgro), at 37°C in a humidified incubator with 5% CO2 atmosphere. Sub-confluent cells grown in 60-, or 100-mm dishes were either transiently transfected with various cDNA constructs using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol or transduced using lentiviral particles produced by co-transfecting 293T cells with lentiviral packaging plasmids and transfer vector. Puromycin selection was done 24 hours post-transduction.
Pharmacological treatments
To synchronize cells in G1/S phase, L-mimosine (250 μM) was added to cell cultures for 16 hours. Cell cycle was released by replacing the conditioning media with fresh media. When indicated, nocodazole (50 ng/ml) was added to the conditioning media at 8.5 hours after mimosine removal to further synchronize cell cycle in G/2M phase. After release into fresh media, the cells were collected at different time points for FACS analysis or immunoblotting. For robust cilia production HDF and HK2 cells were cultured in 0.1% sera for 48 hours. Mouse IMCD-3 cells were cultured for 3 d without changing the media (serum depletion) followed by culturing in serum free media for 16 hours. Aurora-B inhibitor AZD1152 (1 µM) was added to the conditioning media at 25 hours post-transfection for 0, 1, 2 or 3 hours or left untreated.
Protein extraction from cultured cells
Cells were collected in cold PBS, and centrifuged at 1000 rpm for 5 min at 4°C (accuSpin Micro 17R; Fisher Scientific). The pellets were re-suspended in 50 mM HEPES (pH 7.8) buffer containing 50 mM KCl, 5 mM MgCl2, 0.2% Triton X-100, 420 mM NaCl, protease and phosphatase inhibitors and PMSF and centrifuged at 13,300 rpm for 5 min at 4°C (Centrifuge 5810 R; rotor A-4–81; Eppendorf) .The supernatant (whole cell lysates) was analyzed using western blot analysis.
Antibodies and chemicals
Mouse monoclonal antibodies for FLAG M5 (1:1000; F1804), α-tubulin (1:1000; T9026) and β-actin (1:250; A1978), as well as rabbit polyclonal antibodies for FLAG (1:1000; F7425), were from Sigma-Aldrich. Mouse monoclonal antibodies for Pericentrin (1:700; ab28144) and Aurora-A (1:2000; ab13824), rabbit polyclonal antibodies for acetylated α-tubulin (1:1000; ab24610), α-tubulin (1:1000, ab18251), HBO1 (1:250; ab70183) were from Abcam. Mouse monoclonal antibody for gamma-tubulin (1:300; MA1–850) was from Thermo Scientific. Mouse anti-c-Myc antibody (1:250; sc-40), Goat anti-mouse and anti-rabbit IgG-horseradish peroxidase conjugates (1:2000; sc-2005 and sc-2004) were from Santa Cruz Biotechnology. Alexa Fluor dye-labeled secondary antibodies (1:500; A31627, A31623, A31619, A31631) were from Life Technologies. Protease inhibitor cocktail and phosphatase inhibitor cocktail was from Roche Diagnostics. Nocodazole (M1404), L- Mimosine (M0253) and Puromycin (P8833) were from Sigma-Aldrich, AZD1152-HQPA (S1147) was from Selleckchem. Rabbit polyclonal sera and affinity-purified antibodies specific for JADE1S, JADE1L, and HBO1 were custom generated (Proteintech Group).
Constructs
FLAG-JADE1S was described previously.Citation6 MGC (Mammalian Genome Collection) fully sequenced human PHF17 (JADE1L) cDNA cloned in pCMV-SPORT6 vector (clone 5111727) was obtained from Open Biosystems. PHF17 cDNA was then cloned into pcDNA3.1 vector modified with an N-terminal FLAG-tag and into lentiviral vector pMA3211 (Addgene; plasmid 46879Citation52) modified with an N-terminal Myc-tag using NotI and XbaI sites. Constucts were verified by sequencing (MCLAB, http://www.mclab.com). GIPZ Lentiviral shRNA constructs for JADE1S (V2LHS_176747, V3LHS_404943) and non-silencing control were from Dharmacon Research, Inc.. Second generation lentiviral packaging plasmid psPAX2 (plasmid 12260) and envelop expressing plasmid pMD2.G (plasmid 12259) were purchased from Addgene (Didier Trono, unpublished).
Flow cytometry (FACS)
Cells were washed in PBS, trypsinized, and collected by centrifugation at 1000 rpm for 10 min (accuSpin Micro 17R; Fisher Scientific). Cells resuspended in PBS were fixed in cold 70% ethanol. Cells collected by centrifugation at 1500 rpm for 10 min were stained with BD PharMingen™ PI/RNase staining buffer as recommended. Equal numbers of cells (10,000–30,000) were subjected to flow cytometry at Flow cytometry core facility (BUSM, http://www.bu.edu/cores) using either BD FACSCalibur or FACScan flow cytometer (BD Biosciences; San Jose, CA) and was analyzed using either CellQuest (BD Biosciences, San Jose, CA) or FlowJo (FlowJo LLC, Ashland, OR).
Immunofluorescent labeling of whole cells (IF)
IF was done as described.Citation4,7 Cells grown on chamber slides were rinsed with PBS, and then fixed with 4% paraformaldehyde for 20 min at room temperature. After incubation, cells were rinsed with PBS 3 times and then permeabilized with 0.5% Triton X-100 for 15 min at room temperature. Blocking was performed with 3% horse serum and 2% BSA for 1 hour at room temperature. Primary antibodies in 2% BSA, 0.05% Tween-PBS, were incubated overnight at 4°C and secondary antibodies in 2% BSA, 0.05% Tween-PBS were incubated for 1 hour at room temperature. Coverslips were mounted using Vectashield (VECTOR) mounting medium with DAPI. Images were analyzed on Olympus IX 70 inverted microscope with DSU spinning disk confocal system housed with a wide band mercury halide illumination system (Olympus, Tokyo, Japan) and Leica TCS SP5 confocal system with an acousto-optic beam splitter for rapid switching between lasers and conventional point-scan as well as high-speed resonance scanning (Leica Microsystems) at the Cellular Imaging Core in Boston University School of Medicine using 40X and 60X magnification lens. Images were edited in ImageJ 1.45S (NIH, Bethesda, MD) software. All experiments were repeated at least 2 times and in 3 different types of cells specified, yielding essentially the same results.
Quantitation of cytokinetic and multinucleates cells
Transfected or transduced HeLa cells processed for immunofluorescence with antibodies against Myc-tag, FLAG-tag and α-tubulin. In JADE1S knockdown experiment, cells transfected with shJADE1 were visualized by direct fluorescence for GFP expression. Cells were scored by manual counting under the fluorescence microscope (Olympus BX60) to quantify the cytokinetic, bi- and multinucleated cells as percentage of total transfected or transduced cells. All experiments were repeated 3 times and data are presented as average ± SD. To calculate the fold change the percentage of cytokinetic cells at each data point were normalized to the value of the first data point.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1068476_supplemental_figures_and_legends.zip
Download Zip (3 MB)Funding
This work was supported by National Institutes of Health Grant RO1 DK087910 (MVP) and from Boston University CTSI grant UL1-TR000157 (MVP and NSS).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Zhou MI, Wang H, Ross JJ, Kuzmin I, Xu C, Cohen HT. The von Hippel-Lindau tumor suppressor stabilizes novel plant homeodomain protein Jade-1. J Biol Chem 2002; 277:39887-98; PMID:12169691; http://dx.doi.org/10.1074/jbc.M205040200
- Tzouanacou E, Tweedie S, Wilson V. Identification of Jade1, a gene encoding a PHD zinc finger protein, in a gene trap mutagenesis screen for genes involved in anteroposterior axis development. Mol Cell Biol 2003; 23:8553-2; PMID:14612400; http://dx.doi.org/10.1128/MCB.23.23.8553-8562.2003
- Szelei J, Soto AM, Geck P, Desronvil M, Prechtl NV, Weill BC, Sonnenschein C. Identification of human estrogen-inducible transcripts that potentially mediate the apoptotic response in breast cancer. J Steroid Biochem Mol Biol 2000; 72:89-102; PMID:10775800; http://dx.doi.org/10.1016/S0960-0760(00)00025-X
- Havasi A, Haegele JA, Gall JM, Blackmon S, Ichimura T, Bonegio RG, Panchenko MV. Histone acetyl transferase (HAT) HBO1 and JADE1 in epithelial cell regeneration. Am J Pathol 2013; 182:152-62; PMID:23159946; http://dx.doi.org/10.1016/j.ajpath.2012.09.017
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 1995; 20:56-9; PMID:7701562; http://dx.doi.org/10.1016/S0968-0004(00)88957-4
- Foy RL, Song IY, Chitalia VC, Cohen HT, Saksouk N, Cayrou C, Vaziri C, Côté J, Panchenko MV. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem 2008; 283:28817-26; PMID:18684714; http://dx.doi.org/10.1074/jbc.M801407200
- Panchenko MV, Zhou MI, Cohen HT. von Hippel-Lindau partner Jade-1 is a transcriptional co-activator associated with histone acetyltransferase activity. J Biol Chem 2004; 279:56032-41; PMID:15502158; http://dx.doi.org/10.1074/jbc.M410487200
- Siriwardana NS, Meyer R, Havasi A, Dominguez I, Panchenko MV. Cell cycle-dependent chromatin shuttling of HBO1-JADE1 histone acetyl transferase (HAT) complex. Cell Cycle 2014; 13:0-1; http://dx.doi.org/10.4161/cc.28759
- Zhou MI, Foy RL, Chitalia VC, Zhao J, Panchenko MV, Wang H, Cohen HT. Jade-1, a candidate renal tumor suppressor that promotes apoptosis. Proc Natl Acad Sci U S A 2005; 102:11035-40; PMID:16046545; http://dx.doi.org/10.1073/pnas.0500757102
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Côté V, Selleck W, Lane WS, Tan S, Yang XJ, Côté J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell 2006; 21:51-64; PMID:16387653; http://dx.doi.org/10.1016/j.molcel.2005.12.007
- Borgal L, Habbig S, Hatzold J, Liebau MC, Dafinger C, Sacarea I, Hammerschmidt M, Benzing T, Schermer B. The ciliary protein nephrocystin-4 translocates the canonical Wnt regulator Jade-1 to the nucleus to negatively regulate β-catenin signaling. J Biol Chem 2012; 287:25370-80; PMID:22654112; http://dx.doi.org/10.1074/jbc.M112.385658
- Borgal L, Rinschen MM, Dafinger C, Hoff S, Reinert MJ, Lamkemeyer T, Lienkamp SS, Benzing T, Schermer B. Casein kinase 1 α phosphorylates the Wnt regulator Jade-1 and modulates its activity. J Biol Chem 2014; 289:26344-56; PMID:25100726; http://dx.doi.org/10.1074/jbc.M114.562165
- Avvakumov N, Lalonde ME, Saksouk N, Paquet E, Glass KC, Landry AJ, Doyon Y, Cayrou C, Robitaille GA, Richard DE, et al. Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol Cell Biol 2012; 32:689-703; PMID:22144582; http://dx.doi.org/10.1128/MCB.06455-11
- Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Côté V, et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell 2009; 33:257-65; PMID:19187766; http://dx.doi.org/10.1016/j.molcel.2009.01.007
- Lalonde ME, Avvakumov N, Glass KC, Joncas FH, Saksouk N, Holliday M, Paquet E, Yan K, Tong Q, Klein BJ, et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev 2013; 27:2009-24; PMID:24065767; http://dx.doi.org/10.1101/gad.223396.113
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell 2007; 131:633-6; PMID:18022353; http://dx.doi.org/10.1016/j.cell.2007.10.039
- Iizuka M, Matsui T, Takisawa H, Smith MM. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol 2006; 26:1098-108; PMID:16428461; http://dx.doi.org/10.1128/MCB.26.3.1098-1108.2006
- Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem 1999; 274:23027-34; PMID:10438470; http://dx.doi.org/10.1074/jbc.274.33.23027
- Burke TW, Cook JG, Asano M, Nevins JR. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J Biol Chem 2001; 276:15397-408; PMID:11278932; http://dx.doi.org/10.1074/jbc.M011556200
- Georgiakaki M, Chabbert-Buffet N, Dasen B, Meduri G, Wenk S, Rajhi L, Amazit L, Chauchereau A, Burger CW, Blok LJ, et al. Ligand-controlled interaction of histone acetyltransferase binding to ORC-1 (HBO1) with the N-terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1-dependent coactivation of transcription. Mol Endocrinol 2006; 20:2122-40; PMID:16645042; http://dx.doi.org/10.1210/me.2005-0149
- Johmura Y, Osada S, Nishizuka M, Imagawa M. FAD24 acts in concert with histone acetyltransferase HBO1 to promote adipogenesis by controlling DNA replication. J Biol Chem 2008; 283:2265-74; PMID:18029353; http://dx.doi.org/10.1074/jbc.M707880200
- Sharma M, Zarnegar M, Li X, Lim B, Sun Z. Androgen receptor interacts with a novel MYST protein, HBO1. J Biol Chem 2000; 275:35200-8; PMID:10930412; http://dx.doi.org/10.1074/jbc.M004838200
- Stedman W, Deng Z, Lu F, Lieberman PM. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J Virol 2004; 78:12566-75; PMID:15507644; http://dx.doi.org/10.1128/JVI.78.22.12566-12575.2004
- Iizuka M, Takahashi Y, Mizzen CA, Cook RG, Fujita M, Allis CD, Frierson HF Jr, Fukusato T, Smith MM. Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene 2009; 436:108-14; PMID:19393168; http://dx.doi.org/10.1016/j.gene.2009.01.020
- Wang WZ, Liu HO, Wu YH, Hong Y, Yang JW, Liu YH, Wu WB, Zhou L, Sun LL, Xu JJ, et al. Estrogen receptor α (ERalpha) mediates 17beta-estradiol (E2)-activated expression of HBO1. J Exp Clin Cancer Res 2010; 29:140; http://dx.doi.org/10.1186/1756-9966-29-140
- Mishima Y, Miyagi S, Saraya A, Negishi M, Endoh M, Endo TA, Toyoda T, Shinga J, Katsumoto T, Chiba T, et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 2011; 118:2443-53; PMID:21753189; http://dx.doi.org/10.1182/blood-2011-01-331892
- Chitalia VC, Foy RL, Bachschmid MM, Zeng L, Panchenko MV, Zhou MI, Bharti A, Seldin DC, Lecker SH, Dominguez I, et al. Jade-1 inhibits Wnt signalling by ubiquitylating β-catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol 2008; 10:1208-16; PMID:18806787; http://dx.doi.org/10.1038/ncb1781
- Wan G, Hu X, Liu Y, Han C, Sood AK, Calin GA, Zhang X, Lu X. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. Embo J 2013; 32:2833-47; PMID:24097061; http://dx.doi.org/10.1038/emboj.2013.221
- Agromayor M, Martin-Serrano J. Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends Cell Biol 2013; 23:433-41; PMID:23706391; http://dx.doi.org/10.1016/j.tcb.2013.04.006
- Schiel JA, Prekeris R. Membrane dynamics during cytokinesis. Curr Opin Cell Biol 2013; 25:92-8; PMID:23177492; http://dx.doi.org/10.1016/j.ceb.2012.10.012
- Schiel JA, Childs C, Prekeris R. Endocytic transport and cytokinesis: from regulation of the cytoskeleton to midbody inheritance. Trends Cell Biol 2013; 23:319-27; PMID:23522622; http://dx.doi.org/10.1016/j.tcb.2013.02.003
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol 2012; 28:29-58; PMID:22804577; http://dx.doi.org/10.1146/annurev-cellbio-101011-155718
- Steigemann P, Gerlich DW. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol 2009; 19:606-16; PMID:19733077; http://dx.doi.org/10.1016/j.tcb.2009.07.008
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci U S A 2011; 108:4846-51; PMID:21383202; http://dx.doi.org/10.1073/pnas.1102714108
- Fabbro M, Zhou BB, Takahashi M, Sarcevic B, Lal P, Graham ME, Gabrielli BG, Robinson PJ, Nigg EA, Ono Y, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell 2005; 9:477-88; PMID:16198290; http://dx.doi.org/10.1016/j.devcel.2005.09.003
- Hu CK, Coughlin M, Mitchison TJ. Midbody assembly and its regulation during cytokinesis. Mol Biol Cell 2012; 23:1024-34; PMID:22278743; http://dx.doi.org/10.1091/mbc.E11-08-0721
- Gromley A, Jurczyk A, Sillibourne J, Halilovic E, Mogensen M, Groisman I, Blomberg M, Doxsey S. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J Cell Biol 2003; 161:535-45; PMID:12732615; http://dx.doi.org/10.1083/jcb.200301105
- Thoresen SB, Campsteijn C, Vietri M, Schink KO, Liestøl K, Andersen JS, Raiborg C, Stenmark H. ANCHR mediates Aurora-B-dependent abscission checkpoint control through retention of VPS4. Nat Cell Biol 2014; 16:550-60; PMID:24814515; http://dx.doi.org/10.1038/ncb2959
- Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, Gerlich DW. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 2009; 136:473-84; PMID:19203582; http://dx.doi.org/10.1016/j.cell.2008.12.020
- Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science 2012; 336:220-5; PMID:22422861; http://dx.doi.org/10.1126/science.1217180
- Mackay DR, Makise M, Ullman KS. Defects in nuclear pore assembly lead to activation of an Aurora B-mediated abscission checkpoint. J Cell Biol 2010; 191:923-31; PMID:21098116; http://dx.doi.org/10.1083/jcb.201007124
- Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol 2009; 21:796-805; PMID:19836940; http://dx.doi.org/10.1016/j.ceb.2009.09.008
- Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal 2011; 4:rs5; PMID:21712546; http://dx.doi.org/10.1126/scisignal.2001497
- Norden C, Mendoza M, Dobbelaere J, Kotwaliwale CV, Biggins S, Barral Y. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell 2006; 125:85-98; PMID:16615892; http://dx.doi.org/10.1016/j.cell.2006.01.045
- Mendoza M, Norden C, Durrer K, Rauter H, Uhlmann F, Barral Y. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat Cell Biol 2009; 11:477-83; PMID:19270692; http://dx.doi.org/10.1038/ncb1855
- Capalbo L, Montembault E, Takeda T, Bassi ZI, Glover DM, D'Avino PP. The chromosomal passenger complex controls the function of endosomal sorting complex required for transport-III Snf7 proteins during cytokinesis. Open Biol 2012; 2:120070; PMID:22724069; http://dx.doi.org/10.1098/rsob.120070
- Chen CT, Hehnly H, Doxsey SJ. Orchestrating vesicle transport, ESCRTs and kinase surveillance during abscission. Nat Rev Mol Cell Biol 2012; 13:483-8; PMID:22781903; http://dx.doi.org/10.1038/nrm3395
- Fadri-Moskwik M, Weiderhold KN, Deeraksa A, Chuang C, Pan J, Lin SH, Yu-Lee LY. Aurora B is regulated by acetylation/deacetylation during mitosis in prostate cancer cells. FASEB J 2012; 26:4057-67; PMID:22751009; http://dx.doi.org/10.1096/fj.12-206656
- You J, Li Q, Wu C, Kim J, Ottinger M, Howley PM. Regulation of aurora B expression by the bromodomain protein Brd4. Mol Cell Biol 2009; 29:5094-103; PMID:19596781; http://dx.doi.org/10.1128/MCB.00299-09
- Bailey JK, Fields AT, Cheng K, Lee A, Wagenaar E, Lagrois R, Schmidt B, Xia B, Ma D. WD repeat-containing protein 5 (WDR5) localizes to the midbody and regulates abscission. J Biol Chem 2015; 290:8987-9001; PMID:25666610; http://dx.doi.org/10.1074/jbc.M114.623611
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006; 442:96-9; PMID:16728974; http://dx.doi.org/10.1038/nature05140
- Ochoa CD, Alexeyev M, Pastukh V, Balczon R, Stevens T. Pseudomonas aeruginosa exotoxin Y is a promiscuous cyclase that increases endothelial tau phosphorylation and permeability. J Biol Chem 2012; 287:25407-18; PMID:22637478; http://dx.doi.org/10.1074/jbc.M111.301440
