Abstract
The Anaphase Promoting Complex/Cyclosome (APC/C) ubiquitin ligase activated by its G1 specific adaptor protein Cdh1 is a major regulator of the cell cycle. The APC/CCdh1 mediates degradation of dozens of proteins, however, the kinetics and requirements for their degradation are largely unknown. We demonstrate that overexpression of the constitutive active CDH1m11 mutant that is not inhibited by phosphorylation results in mitotic exit in the absence of the FEAR and MEN pathways, and DNA re-replication in the absence of Cdc7 activity. This mode of mitotic exit also reveals additional requirements for APC/CCdh1 substrate degradation, which for some substrates such as Pds1 or Clb5 is dephosphorylation, but for others such as Cdc5 is phosphorylation.
Introduction
The anaphase promoting complex/cyclosome (APC/C) is a multi-subunit ubiquitin-E3 ligase that targets multiple proteins for proteasomal degradation and is required for initiation of anaphase by the degradation of securin (Pds1).Citation1-5 During the mitotic cell cycle the APC/C is sequentially activated by 2 substrate specific activators - Cdc20, which is active from anaphase to telophase, and Cdh1, which is active from telophase until the onset of S-phase. During meiosis the APC/C is further activated by Ama1.Citation6 Cdh1 activity is inhibited by phosphorylation by Cdc5 and Cdc28-Clb, which is reversed by dephosphorylation by Cdc14.Citation7-19 The mechanisms of mitotic exit have been thoroughly investigated and reviewed in depth.Citation20-22 Briefly, the securin Pds1 inhibits the separase Esp1, which cleaves cohesins causing anaphase initiation and sister chromatid separation. Esp1 activity and destruction of Clb5, activate the FEAR (Cdc Fourteen Early Release) network causing early release of Cdc14 from the nucleolus into the nucleus, Movement of the nucleus into the bud activates Tem1 and the MEN (Mitosis Exit Network) cascade of Cdc15 and Dbf2, which allows full dissociation of Cdc14 from Net1 and its release into the cytoplasm. The MEN pathway and Cdc14 activity are required for cytokinesis.Citation23 Following degradation of Cdc5 by APC/CCdh1, Cdc14 is re-sequestered into the nucleolus.Citation24 The reduction in Cdc28-Clb activity by the APC/C and by synthesis and activation of the Cdc28 inhibitor Sic1, which are both promoted by the Cdc14 phosphatase, drive the cells into G1.Citation19,25,26 The identity of the APC/C substrates whose destruction is essential for mitotic exit is disputed - a D-box deleted Clb2 (clb2Δdb) causes arrest in telophaseCitation16 whereas APC/CCdc20 is dispensable in cells lacking Pds1 and Clb5. If Sic1 is additionally overexpressed, the APC/C as a whole becomes dispensable.Citation11,13
The effects of both native Cdh1 and non-phosphorylatable, constitutively active CDH1m4, m9 or m11, overexpression have been studied in depth.Citation18,27,28 Ectopic overexpression of Cdh1 in nocodazole arrested cells results in degradation of Clb2, re-budding, and arrest in G2 with replicated DNA.Citation18,28 We have found that these processes occur with a bypass of the MEN and FEAR pathways, and consequently the roles of these pathways in the regulation of APC/CCdh1 substrates were found to involve both phosphorylation and dephosphorylation of substrates.
Results
CDH1m11 expression enables mitotic exit with bypass of the MEN and FEAR pathways
Previous studies have noted that cells overexpressing CDH1m11 form buds and replicate their DNA before arresting in G2 with extremely elongated buds.Citation18,28 We used stabilized substrates described previouslyCitation29 to determine which APC/C substrate degradation is required for mitotic exit when CDH1m11 is overexpressed. Whereas wild-type cells clearly show budding 90 minutes after induction of CDH1m11 expression, cells expressing KEN-box deleted Clb2 do not bud () nor do they express the G1 genes Sic1 or Cln2 (). In addition, Cdc5 is stabilized, suggesting Clb2 degradation to be a prerequisite for Cdc5 degradation as well as for exit from mitosis ().
Figure 1. CDH1m11 expression enables mitotic exit with bypass of the MEN and FEAR pathways. (A) Cells were arrested with nocodazole for 3 hours and CDH1m11 expression induced by addition of 4% galactose. Cells were stained with calcofluor white to visualize cell walls and septa. (B) Diploid cells were arrested with nocodazole and galactose was added to induce CDH1m11 expression. Samples were taken at indicated times and processed for immunoblotting. (C) Cells were arrested with nocodazole at 37°C as indicated and galactose was added to induce CDH1m11 expression. (D) Cells were arrested with nocodazole for 2.5 hours and CDH1m11 expression induced by addition of 4% galactose. Cells were stained with calcofluor white to visualize cell walls and septa. In all strains, rebudding occurred. (E) Cells expressing Cdc20 under the control of the MET25 promoter were grown overnight in SC-Ura-Met 2% raffinose media. 80mg/liter methionine was added to repress Cdc20 expression. After 5 hours, cells were transferred to glass bottom Petri dishes and immobilized using Concavalin A. Galactose was added to induce CDH1m11 expression. Images of Myo1-GFP were taken at indicated times. Although rebudding occurs, Myo1-GFP remains at the bud neck.
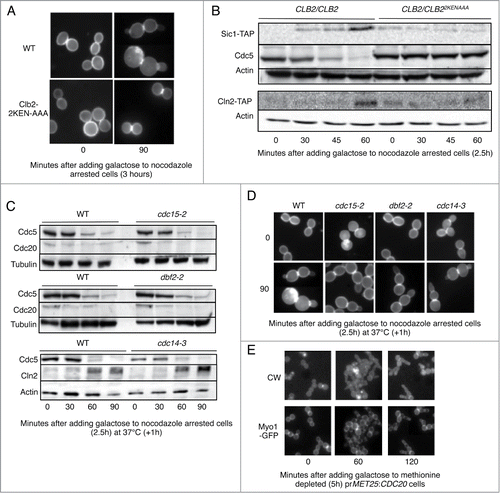
Exit from mitosis as a result of Esp1 overexpression is MEN dependent.Citation14 Using temperature sensitive MEN mutants, we examined whether Cdc15, Dbf2 or Cdc14 are required for mitotic exit when CDH1m11 is overexpressed. shows that the activity of none of these proteins was required for degradation of Cdc5, and that Cln2 accumulated normally in Cdc14-3 cells at 37°C. shows that both wild-type and MENts cells bud without cell separation. This implies that mitotic exit induced by overexpression of CDH1m11 does not require the MEN pathway.
Prior studies involving CDH1m11 have not distinguished between lack of cell separation as opposed to lack of cytokinesis.Citation18,28 For example, ace2Δ cells undergo cytokinesis but do not separate.Citation30,31 We stained chitin with calcofluor white as a determinant of cell separation and compared it to Myo1-GFP localization to determine whether cytokinesis occurs in cells upon induction of CDH1m11 expression. As Myo1-GFP failed to localize properly in the presence of nocodazole we used cells expressing a sole copy of CDC20 from a MET25 promoter. These cells were arrested in metaphase by adding methionine. Upon induction of CDH1m11 expression, cells bud but Myo1-GFP remains at the bud neck, indicating lack of cytokinesis (). An even more direct indication that cytokinesis did not take place is the fact that the nucleus (RFP-PUS1) can easily move back and forth through the bud neck ( and Supp. Movie 1). This figure further shows that while Myo1 remains on the original bud neck, it also subsequently accumulates on the new bud necks that form on the elongated bud caused by the expression of CDH1m11.
Figure 2. Expression of CDH1m11 results in mitotic exit without Cdc14 release or cytokinesis. (A) prMET25:CDC20 prGAL:CDH1m11 cells bearing plasmids for RFP-Pus1 (nuclear marker) and Myo1-GFP (contractile ring) were arrested for 5 hours with methionine in raffinose containing medium and galactose was added to induce CDH1m11 expression. The images are stills from Movie S1. (B) Cdc14-GFP cells were arrested with nocodazole for 3 hours, washed and released into drug-free medium with galactose. The images are stills from Movie S2. (C) Cdc14-GFP prGAL:CDH1m11 cells were arrested with nocodazole for 3 hours. Galactose was added to induce CDH1m11 expression. The images are stills from Movie S3.
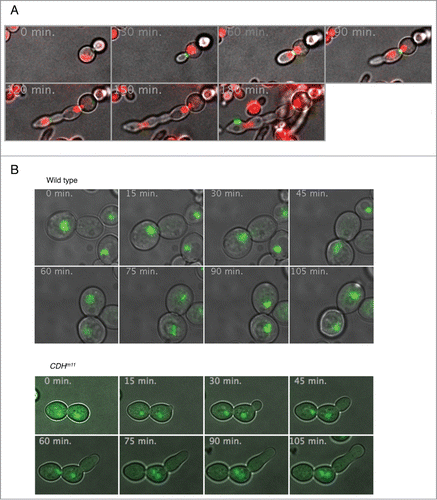
Cdc14-GFP localization upon CDH1m11 overexpression was examined in cells arrested with nocodazole. ( and Supp. Movies 2 and 3). (top) and supplemental movie 2 show that, as previously reported,Citation19,24,32,33 wild type Cdc14-GFP is released from the nucleolus during mitosis (min. 30 to 60) and re-sequestered to it in G1 (min. 75 and 90). In contrast, in cells expressing CDH1m11, Cdc14-GFP remained in the nucleolus throughout mitosis ( bottom and Supp. Movie 3). Taken together these observations show that the mode of mitotic exit upon CDH1m11 overexpression is MEN and Cdc14 independent, but does require at least partial Clb2 degradation.
Expression of CDH1m11 overrides the requirement for Cdc7 for DNA replication
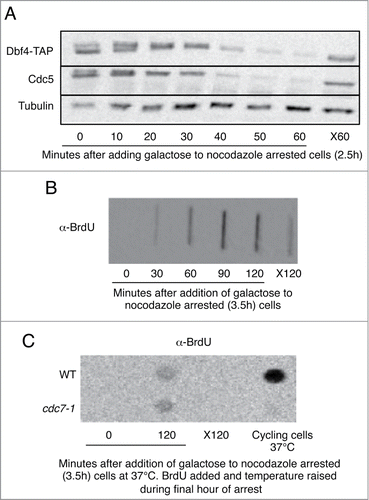
Activity of Cdc7 in complex with its cyclin Dbf4 is required for DNA replication.Citation34,35 Dbf4 has been reported to be an APC/CCdh1 substrate,Citation36 and is degraded upon overexpression of CDH1m11 in nocodazole arrested cells (). It is thus surprising that CDH1m11 expressing cells replicate their DNA as determined by BrdU incorporationCitation37 (). As Cdc5 is also absent it cannot be a suppressor of Cdc7-Dbf4.Citation38 It is possible that a small, but sufficient amount of Dbf4 evades degradation. To eliminate Cdc7 activity we utilized a cdc7-1 strain, which does not incorporate BrdU at the restrictive temperature ( – far right column). Strikingly, expression of CDH1m11 for 2 hours led to incorporation of similar amounts of BrdU in both wild-type and cdc7-1 cells (). This suggests that Cdc7-Dbf4 is not required for DNA replication when CDH1m11 is expressed.
Figure 3. Expression of CDH1m11 overrides the requirement for Cdc7 for DNA replication. (A) Cells were arrested with nocodazole for 2.5 hours and then BrdU was added for an additional hour. Galactose was added to induce CDH1m11 expression. Samples were taken for processing at the indicated times. One sample was left for the duration of the experiment without galactose to determine the incorporation of BrdU into non-replicating cells (X120). (B) Cells were arrested with nocodazole for 2.5 hours and galactose was added to induce CDH1m11 expression. Samples were taken at indicated times and processed for immunoblotting. One sample was left without galactose treatment for the duration of the experiment as a control against prolonged exposure to nocodazole (X60). (C) Cells were arrested with nocodazole for 2.5 hours and then BrdU was added for an additional hour, during which the temperature was raised slowly to 37°C. Galactose was added to induce CDH1m11 expression. Samples were taken for processing at indicated times. One sample was left for the duration of the experiment without galactose to determine the incorporation of BrdU into non-replicating cells (X120). Another sample was grown at 37°C for the duration of the experiment without nocodazole to test the efficacy of the cdc7-1 mutation.
Dephosphorylation is required for Pds1 and Clb5 degradation
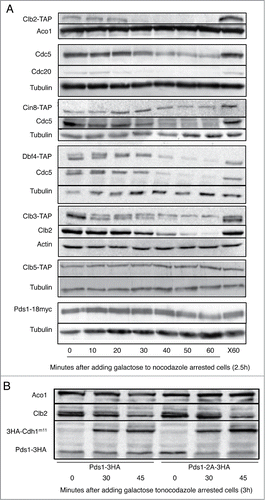
We utilized CDH1m11 expression to induce exit from mitosis as a means to examine the effects of bypass of the FEAR and MEN pathways on APC/C substrate degradation kinetics. Close examination of the timing of the disappearance of known APC/C targets upon induction of CDH1m11 expression reveals 3 broad classes of substrates (Clb2, Clb3 and Cdc20) are degraded within 30 minutes, the degradation of Cdc5, Cin8 and Dbf4 is delayed, and Clb5 and Pds1 are, as previously reported,Citation26,28 stable (). Pds1 degradation by Cdc20 is prevented by phosphorylation by Cdc28-Clb2 and enabled by dephosphorylation by Cdc14. The targets of this dephosphorylation are the T27 and S71 residues of Pds1 and a Pds1T27A,S71A mutant is targeted by the APC/C in the absence of Cdc14 activity,Citation39 shows that these mutations also convert Pds1 from being stable upon ectopic CDH1m11 to being a substrate of APC/CCdh1-m11.
Figure 4. Kinetics of APC/C substrate degradation. (A) Cells were arrested with nocodazole for 3 hours and galactose was added to induce CDH1m11 expression. Samples were taken at indicated times and processed for immunoblotting. One sample was left without galactose treatment for the duration of the experiment as a control against prolonged exposure to nocodazole (X60). (B) Cells were arrested with nocodazole for 3 hours and galactose added to induce CDH1m11 expression. Samples were taken at indicated times and processed for immunoblotting.
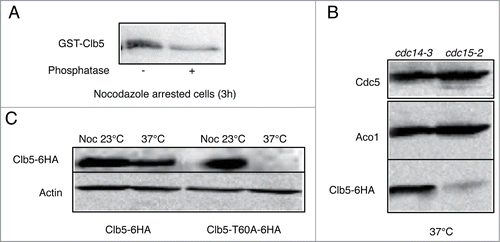
We hypothesized that a similar mechanism might apply to Clb5. shows that Clb5 in nocodazole arrested cells is phosphorylated. We arrested cells expressing endogenous wild type Cdh1 in telophase by using either cdc14-3 or cdc15-2 temperature sensitive mutant. Both mutants arrest at the restrictive temperature in telophase, but in the latter FEAR has been active so that at least nuclear targets of Cdc14 can be expected to have been dephosphorylated. Indeed, Clb5 is stable in cdc14-3 cells but is absent in cdc15-2 cells (), showing Clb5 stability to be regulated by Cdc14. A T60A mutation restores Clb5 degradation to cdc14-3 cells arrested in telophase (), suggesting that this residue is dephosphorylated by Cdc14 in order for Clb5 degradation to occur.
Figure 5. Degradation of Clb5 requires its dephosphorylation. (A) Cells were arrested in metaphase with nocodazole for 2.5h, and GST6H-Clb5 expressed from a GAL promoter for an additional hour. Cells were lysed and GST6H-Clb5 pulled down using glutathione beads. Half of the extract was treated with λ-phosphatase for one hour. (B) cdc14-3 and cdc15-2 cells expressing Clb5-6HA were arrested in telophase by elevation of the temperature to 37°C for 3 hours. (C) cdc14-3 cells expressing either wild-type Clb5-6HA or Clb5T60A-6HA were either arrested at metaphase with nocodazole for 3 hours at 23°C, or in telophase by elevation of the temperature to 37°C for 3 hours.
Phosphorylation of Cdc5 is required for its degradation
Degradation of Cdc5 upon CDH1m11 expression is preceded by Cdc20 and Clb5 degradation (), and is dependent upon at least a partial loss of Clb2 (). The first 70 amino acids of Cdc5 are required for its degradation.Citation12 We examined alanine and glutamate mutations of all of the reported phosphorylation sitesCitation40 within this region (S2, S18, T23, T29, T42, T70). The only mutation that stabilizes Cdc5 under a rapamycin arrest in G1 is Cdc5T29A ( andCitation29). Cdc5T29A is also stable upon expression of CDH1m11 (), which indicates that phosphorylation of Cdc5 at this site is a second pre-requisite for its degradation in addition to Clb2 degradation. Such a mechanism of phosphorylation promoting APC/C substrate ubiquitinylation has been previously identified for human Mcl1.Citation41,42 Mutation of K26A similarly prevented Cdc5 degradation in G1 or upon CDH1m11 expression () suggesting a minimal kinase consensus sequence of KxxT. This kinase would need to be active during telophase, because if Cdc5 were to be phosphorylated early, its degradation would not be delayed with respect to Clb2 or Cdc20, but independently from Cdc14.
Figure 6. Phosphorylation of Cdc5 is required for its degradation. (A) Cells expressing Cdc5 (and mutants) from a CEN plasmid were arrested with nocodazole for 3 hours and galactose was added to induce CDH1m11 expression. Samples were taken at indicated times and processed for immunoblotting. One sample was left without galactose treatment for the duration of the experiment as a control against prolonged exposure to nocodazole (X60). Another sample was treated with rapamycin (and was not treated with nocodazole) for the duration of the experiment to provide a control for G1 arrest. Both Cdc5T29A and Cdc5K26A are stable in rapamycin. (B) Cells were arrested with nocodazole for 2.5h and released into rapamycin containing media with 1M sorbitol, with and without staurosporine (40 μg/ml) for 1 hour. (C) Cells containing a plasmid with prGAL:GST6H-Cdc5 were arrested in rapamycin for 2.5 hours, and pulsed with galactose to express GST-Cdc5 or 1 additional hour during which the temperature was slowly raised to 37°C. The media was switched to media containing rapamycin (1μg/ml), glucose (4%) and cycloheximide (200 μg/ml) to cease GST-Cdc5 production. During the chase, samples were withdrawn at the indicated time points.
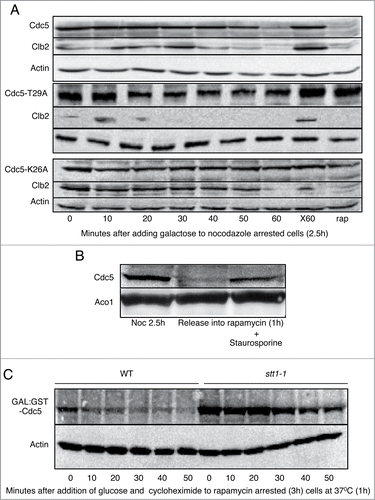
One such kinase that is activated during telophase is PKC1.Citation43,44 Cells expressing endogenous wild type Cdh1 were arrested in metaphase, and released into either rapamycin with or without the PKC inhibitor staurosporine.Citation45 Staurosporine impedes Cdc5 degradation (). To confirm that Pkc1 activity is required for Cdc5 degradation we conducted pulse-chase experiments using pkc1-ts (stt1-1)Citation46 cells arrested in G1 with rapamycin. Whereas GST-Cdc5 is rapidly degraded upon cessation of translation during G1 in wild-type cells, GST-Cdc5 was markedly stabilized in stt1-1 cells (). Together, this shows that Pkc1 is required for Cdc5 degradation, likely due to phosphorylation of T29.
Discussion
Sister chromatid separation during anaphase and cell division in telophase are 2 of the most crucial cell cycle transitions. Under normal conditions these 2 events are tightly coupled by the activity of the APC/C. This coupling is accomplished by the sequential activation of the APC/C by Cdc20 and Cdh1, which mediate the degradation of multiple mitotic proteins. This sequential APC/C activation is regulated by various phosphorylation events by kinases that are themselves degradation substrates of the APC/C, mainly by the Clb cyclins and by Cdc5. Of particular relevance for the present study is the substantial phosphorylation of Cdh1 that prevents its premature activation. Under physiological conditions Cdc20 initiates the cascade that leads to the dephosphorylation of Cdh1 and is thus essential for its activation. We used here the CDH1m11 mutant to uncouple anaphase and cytokinesis in order to study the respective roles of various components of this pathway. CDH1m11 is no longer inhibited by phosphorylation and is thus constitutively active. Cdc20 was inhibited by nocodazole and degraded by CDH1m11 during these experiments (). This enabled a detailed analysis of the respective roles of Cdc20 and Cdh1. In spite of being free from any inhibition and of being over expressed, CDH1m11 still maintained its exquisite substrate specificity as shown here and by usCitation47 and othersCitation39,48 for all the substrates we tested.
Cells expressing CDH1m11 failed to degrade Pds1 and Clb5 and to activate the pivotal Cdc14 phosphatase (). They did bypass the MEN and FEAR pathways and entered G1 without undergoing cytokinesis and cell separation ( Supp. Mov 1-3). In contrast a single copy of Clb2 with a mutant KEN box, resistant to Cdh1 specific degradation, was sufficient to prevent CDH1m11 mediated entry into G1 (). These observations reconcile 2 previous conflicting observations. The first was that Pds1 and Clb5 are the only essential substrates of the APC/C in mitosis.Citation11 The second was that degradation of Clb2 by the APC/C is essential for mitotic exit.Citation16 Here we show that both views are correct but for different reasons - Pds1 and Clb5 must be degraded so that the sister chromatids can be separated and the MEN and FEAR pathway be activated, while Clb2 must be degraded so that cells can enter G1. Comparing regular release from metaphase arrest with forcing mitotic exit by overexpression of CDH1m11 could thus in future be used as a tool to further study Cdc14 activity and function.
Pds1 degradation by the APC/CCdc20 depends on the dephosphorylation of residues T27 and S71 of Pds1 by Cdc14.Citation39,49 We suspected that the stability of Pds1 upon CDH1m11 overexpression was due to the lack of Cdc14 activity in these cells. Indeed Pds1T27A,S71A was degraded upon CDH1m11 overexpression in metaphase arrested cells (). Removal of the Cdc28-Clb phosphorylation sites of Pds1 thus converted Pds1 to an APC/CCdh1 substrate. Clb5 is also stable upon CDH1m11 overexpression in metaphase arrested cells, and is stable in cdc14-3 but not cdc15-2 cells arrested in telophase. Applying a similar rationale to that of Pds1 degradation, we found that mutation of residue T60 of Clb5 to an alanine suffices to permit Clb5 degradation in telophase arrested cdc14-3 cells (). It therefore seems that Clb5 degradation is regulated, like that of Pds1, by dephosphorylation by Cdc14. These observations further suggest that other substrates considered APC/CCdc20 or APC/CCdh1 “specific” could in fact be targeted for degradation by both activators, and that their apparent specificity depends on their cell cycle specific phosphorylation status.
In contrast to the degradation of Clb5 and Pds1, which are inhibited by phosphorylation, degradation of Cdc5 requires phosphorylation on T29.Citation47 We have further characterized here this motif to include K26 (−3 position), which is a sequence common to many kinases (). We speculated that Pkc1, which is activated during the end of mitosis, independently from Cdc14, could be the regulating kinase (). Interestingly Pkc1 is locally activated at the bud neck by Cdc5 activity on Tus3.Citation44 Cdc5 activity thus sets up the conditions for its own degradation with the appropriate timing, which is a recurrent pattern of regulation in the APC/C pathway.
Metaphase or α-factor arrested cells ectopically expressing CDH1 or CDH1m11 replicate their DNA before their terminal early G2-like arrest.Citation18,28 The kinase Cdc7 and its cyclin Dbf4 are essential for DNA replication.Citation34 However Dbf4 is an APC/CCdh1 substrate, both in cycling cellsCitation36 and upon ectopic expression of CDH1m11 in metaphase arrested cells. We found that Cdc7 activity is not required for DNA replication as determined directly by incorporation of BrdU when CDH1m11 is ectopically expressed in metaphase cells (). It is possible that the increase in Clb5 levels, which is not degraded under these conditions, compensates for lack of Cdc7-Dbf4 activity, and/or that targets of Cdc7-Dbf4 are dephosphorylated by Cdc14.
Methods
Yeast strains
Yeast transformations were performed by the LiOAc method.Citation50 Cassettes containing GALL:3HACdh1m11 were kind gifts from W. Zachariae and F. Cross and integrated at the URA3 (StuI) and TRP1 (EcoRV) loci respectively. prMET25:Cdc20 (TRP1) was a gift from F. Cross. Clb22KEN-AAA was generated by excision of part of CLB2 by replacement with URA3 and replacement with a fragment from plasmid FC733 (prCLB5-clb22KEN-AAA). The resultant strain (GALL:Cdh1m11 (TRP1) Clb22KEN-AAA in W303α) was mated with relevant TAP strains from the TAP collection (BY4741 background). MEN pathway mutants were kind gifts from A. Amon and S. Piatti. Strains derived from CVy63a (BrdUinc LEU2 bar1Δ)Citation37 were used for all BrdU experiments. This strain was mated with cdc7-1Citation35 to generate cdc7-1 BrdUinc (LEU2) GALL:Cdh1m11 (TRP1). Pds13HA and Pds1-2A3HA (T27A, S71A) were kind gifts from D. Morgan.Citation39 A plasmid containing prCLB5:Clb5-6HACitation51 was integrated at the Clb5 promoter by cutting with BstXI. Plasmids containing Cdc5 and Cdc5T29A were kind gifts from D. Kellogg.Citation40 pEG(KT) plasmids (2μ URA3) containing GAL:GST6H-Cdc5 and GAL:GST6H-Clb5 were kind gifts from B. Andrews.Citation52 Mutagenesis was performed with the Stratagene Quikchange kit. STT1/stt1-1Citation46 cells were a kind gift from D. Pellman. The GFP-Myo1 (pRS316 CEN URA3) plasmidCitation53 was a gift from E. Bi. PUS1-RFP (pRS315 CEN LEU2) was a gift from M. Hochstrasser.
Experimental
For CDH1m11 overexpression experiments, cells growing with raffinose (2%) as their carbon source were arrested in metaphase using nocodazole (10 μg/ml) (Sigma) for 2.5 hours, methionine addition (200 mg/l) or temperature elevation to 37°C depending upon the strain used for 2.5 hours. Cdh1m11 expression was induced with warm 4% galactose. For APC/C substrate degradation experiments, a sample was treated with rapamycin (1 μg/ml) for 2.5 hours as a control for G1 arrest. Another sample was left without galactose for the duration of the experiment, as a control for prolonged exposure to nocodazole. For DNA labeling experiments BrdU (300 μg/ml) was added for an additional hour before addition of galactose, and if necessary the temperature was raised to 37°C during this period (i.e. for cdc7-1 experiments). Samples were processed as previously described.Citation37 For experiments involving staurosporine (40 μg/ml) or stt1-1 cells, cells were grown in the presence of 1M sorbitol. Cells were subsequently arrested with nocodazole (10 μg/ml) or rapamycin (1 μg/ml) (LC labs) for 2.5h and the temperature of the media was raised to 37°C for 1 hour if required. For nocodazole release experiments, cells were arrested with 10 μg/ml nocodazole for 2.5 hours, washed 3 times and released into media containing rapamycin (1 μg/ml) where indicated. For IP of GST-Clb5, GST6H-Clb5 was expressed from a GAL promoter for one hour in nocodazole arrested cells (2.5 h before addition of 4% galactose). Cells were lysed in the presence of yeast protease inhibitors (100X) and yeast phosphatase inhibitor cocktails 1 and 2 (100X, Sigma). Half of the extract was treated with λ-phosphatase for one hour. GST-6H-Clb5 was pulled-down using glutathione beads and extracted into sample buffer. For pulse chase experiments with GAL:GST6H-Cdc5, proteins were expressed during this hour in rapamycin (1 μg/ml) arrested cells (2.5 h previously), and then the media were switched to media containing rapamycin (1 μg/ml), glucose (4%) and cycloheximide (200 μg/ml).
For microscopy experiments, cells were immobilized using Concavalin A on glass bottom petri-dishes (Mattek) and filmed using a Roper CCD Camera mounted on an Olympus IX70 microscope with a 60x oil objective. 1mg/ml Calcoflour white (Sigma) followed by 3 washes was used to stain the chitin of cells fixed with formaldehyde.
Immunological procedures and antibodies
Cells were harvested and killed using 20% TCA, shredded with glass beads and extracted into 2X sample buffer. 10% acrylamide gels were used for SDS-PAGE. Antibodies used were rabbit anti-TAP 1/1000 (Open Biosystems), goat anti-Cln2 1/200 (Santa Cruz), mouse anti-GST 1/200 (Santa Cruz), mouse anti-BrdU (1/1000, Becton Dickinson), mouse-anti-myc 1/1000 (9E10), mouse anti-HA 1/1000 (12CA5), rabbit-anti-Cdc5 1/200 (Santa-Cruz, y300), rabbit anti-Clb2 1/500 (a kind gift from A. Amon), rabbit-anti-βactin 1/500 (Epitomics), mouse anti-tubulin 1/5000 (Sigma B512) and rabbit-anti-Aco1 1/20000 (a kind gift from O. Pines). Secondary antibodies were from Jackson Laboratories.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
1078036_supplemental_movies__3_.zip
Download Zip (6.7 MB)Acknowledgments
Many heartfelt thanks to A. Amon, B. Andrews, O. Aparicio, F. Cross, D. Pellman, E. Bi, M. Hochstrasser, D. Kellogg, H. Masumoto, D. Morgan, S. Piatti, D. Stuart and W. Zachariae for strains, plasmids and antibodies. We also thank M. Lavy, Y. Lavy, R. Sclafani, O. Feine, J. Sajman, and S. Creat, for many stimulating discussions.
Funding
This research was funded by grant BSF 2007288 from the US-Israel Binational Science Foundation, and a grant from the Israel Cancer Research Fund. K.J.S.L was a recipient of an integration fellowship from the Ministry of Adsorption.
References
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 1995; 81:269-78; PMID:7736579; http://dx.doi.org/10.1016/0092-8674(95)90337-2
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 1995; 81:279-88; PMID:7736580; http://dx.doi.org/10.1016/0092-8674(95)90338-0
- Lamb JR, Michaud WA, Sikorski RS, Hieter PA. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. Embo J 1994; 13:4321-8; PMID:7925276
- Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science 1996; 274:1201-4; PMID:8895471; http://dx.doi.org/10.1126/science.274.5290.1201
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev 1996; 10:3081-93; PMID:8985178; http://dx.doi.org/10.1101/gad.10.24.3081
- Cooper KF, Mallory MJ, Egeland DB, Jarnik M, Strich R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci U S A 2000; 97:14548-53; PMID:11114178; http://dx.doi.org/10.1073/pnas.250351297
- Huang JN, Park I, Ellingson E, Littlepage LE, Pellman D. Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J Cell Biol 2001; 154:85-94; PMID:11448992; http://dx.doi.org/10.1083/jcb.200102007
- Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci 1997; 110(Pt 13):1523-31; PMID:9224769
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol 1999; 9:227-36; PMID:10074450; http://dx.doi.org/10.1016/S0960-9822(99)80111-0
- Rudner AD, Murray AW. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol 2000; 149:1377-90; PMID:10871279; http://dx.doi.org/10.1083/jcb.149.7.1377
- Shirayama M, Toth A, Galova M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 1999; 402:203-7; PMID:10647015; http://dx.doi.org/10.1038/46080
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. Embo J 1998; 17:1336-49; PMID:9482731; http://dx.doi.org/10.1093/emboj/17.5.1336
- Thornton BR, Toczyski DP. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat Cell Biol 2003; 5:1090-4; PMID:14634663; http://dx.doi.org/10.1038/ncb1066
- Tinker-Kulberg RL, Morgan DO. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev 1999; 13:1936-49; PMID:10444592; http://dx.doi.org/10.1101/gad.13.15.1936
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 1997; 278:460-3; PMID:9334304; http://dx.doi.org/10.1126/science.278.5337.460
- Wasch R, Cross FR. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 2002; 418:556-62; PMID:12152084; http://dx.doi.org/10.1038/nature00856
- Yeong FM, Lim HH, Wang Y, Surana U. Early expressed Clb proteins allow accumulation of mitotic cyclin by inactivating proteolytic machinery during S phase. Mol Cell Biol 2001; 21:5071-81; PMID:11438663; http://dx.doi.org/10.1128/MCB.21.15.5071-5081.2001
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 1998; 282:1721-4; PMID:9831566; http://dx.doi.org/10.1126/science.282.5394.1721
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell 1998; 2:709-18; PMID:9885559; http://dx.doi.org/10.1016/S1097-2765(00)80286-5
- Li M, Zhang P. The function of APC/CCdh1 in cell cycle and beyond. Cell Div 2009; 4:2; PMID:19152694; http://dx.doi.org/10.1186/1747-1028-4-2
- Simpson-Lavy KJ, Oren YS, Feine O, Sajman J, Listovsky T, Brandeis M. Fifteen years of APC/cyclosome: a short and impressive biography. Biochem Soc Trans 2010; 38:78-82; PMID:20074039; http://dx.doi.org/10.1042/BST0380078
- Dumitrescu TP, Saunders WS. The FEAR Before MEN: networks of mitotic exit. Cell Cycle 2002; 1:304-7; PMID:12461288; http://dx.doi.org/10.4161/cc.1.5.147
- Jimenez J, Cid VJ, Cenamor R, Yuste M, Molero G, Nombela C, Sanchez M. Morphogenesis beyond cytokinetic arrest in Saccharomyces cerevisiae. J Cell Biol 1998; 143:1617-34; PMID:9852155; http://dx.doi.org/10.1083/jcb.143.6.1617
- Visintin C, Tomson BN, Rahal R, Paulson J, Cohen M, Taunton J, Amon A, Visintin R. APC/C-Cdh1-mediated degradation of the Polo kinase Cdc5 promotes the return of Cdc14 into the nucleolus. Genes Dev 2008; 22:79-90; PMID:18172166; http://dx.doi.org/10.1101/gad.1601308
- Toyn JH, Johnson AL, Donovan JD, Toone WM, Johnston LH. The Swi5 transcription factor of Saccharomyces cerevisiae has a role in exit from mitosis through induction of the cdk-inhibitor Sic1 in telophase. Genetics 1997; 145:85-96; PMID:9017392
- Lopez-Aviles S, Kapuy O, Novak B, Uhlmann F. Irreversibility of mitotic exit is the consequence of systems-level feedback. Nature 2009; 459:592-5; PMID:19387440; http://dx.doi.org/10.1038/nature07984
- Robbins JA, Cross FR. Requirements and Reasons for Effective Inhibition of the Anaphase Promoting Complex Activator Cdh1. Mol Biol Cell 2010; 21(6):914-25; PMID:20089834
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 1997; 90:683-93; PMID:9288748; http://dx.doi.org/10.1016/S0092-8674(00)80529-2
- Simpson-Lavy KJ, Sajman J, Zenvirth D, Brandeis M. APC/CCdh1 specific degradation of Hsl1 and Clb2 is required for proper stress responses of S. cerevisiae. Cell Cycle 2009; 8:3003-9; PMID:19713762; http://dx.doi.org/10.4161/cc.8.18.9616
- King L, Butler G. Ace2p, a regulator of CTS1 (chitinase) expression, affects pseudohyphal production in Saccharomyces cerevisiae. Curr Genet 1998; 34:183-91; PMID:9745020; http://dx.doi.org/10.1007/s002940050384
- Ufano S, Pablo ME, Calzada A, del Rey F, Vazquez de Aldana CR. Swm1p subunit of the APC/cyclosome is required for activation of the daughter-specific gene expression program mediated by Ace2p during growth at high temperature in Saccharomyces cerevisiae. J Cell Sci 2004; 117:545-57; PMID:14709718; http://dx.doi.org/10.1242/jcs.00880
- Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet 2004; 38:203-32; PMID:15568976; http://dx.doi.org/10.1146/annurev.genet.38.072902.093051
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 2002; 108:207-20; PMID:11832211; http://dx.doi.org/10.1016/S0092-8674(02)00618-9
- Jackson AL, Pahl PM, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol 1993; 13:2899-908; PMID:8474449
- Ogi H, Wang CZ, Nakai W, Kawasaki Y, Masumoto H. The role of the Saccharomyces cerevisiae Cdc7-Dbf4 complex in the replication checkpoint. Gene 2008; 414:32-40; PMID:18372119; http://dx.doi.org/10.1016/j.gene.2008.02.010
- Ferreira MF, Santocanale C, Drury LS, Diffley JF. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol Cell Biol 2000; 20:242-8; PMID:10594027; http://dx.doi.org/10.1128/MCB.20.1.242-248.2000
- Viggiani CJ, Aparicio OM. New vectors for simplified construction of BrdU-Incorporating strains of Saccharomyces cerevisiae. Yeast 2006; 23:1045-51; PMID:17083135; http://dx.doi.org/10.1002/yea.1406
- Kitada K, Johnson AL, Johnston LH, Sugino A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol 1993; 13:4445-57; PMID:8321244
- Holt LJ, Krutchinsky AN, Morgan DO. Positive feedback sharpens the anaphase switch. Nature 2008; 454:353-7; PMID:18552837; http://dx.doi.org/10.1038/nature07050
- Mortensen EM, Haas W, Gygi M, Gygi SP, Kellogg DR. Cdc28-dependent regulation of the Cdc5/Polo kinase. Curr Biol 2005; 15:2033-7; PMID:16303563; http://dx.doi.org/10.1016/j.cub.2005.10.046
- Biggs JR, Peterson LF, Zhang Y, Kraft AS, Zhang DE. AML1/RUNX1 phosphorylation by cyclin-dependent kinases regulates the degradation of AML1/RUNX1 by the anaphase-promoting complex. Mol Cell Biol 2006; 26:7420-9; PMID:17015473; http://dx.doi.org/10.1128/MCB.00597-06
- Harley ME, Allan LA, Sanderson HS, Clarke PR. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. Embo J 2010; 29(14):2407-20; PMID:20526282
- Andrews PD, Stark MJ. Dynamic, Rho1p-dependent localization of Pkc1p to sites of polarized growth. J Cell Sci 2000; 113(Pt 15):2685-93; PMID:10893184
- Yoshida S, Kono K, Lowery DM, Bartolini S, Yaffe MB, Ohya Y, Pellman D. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science 2006; 313:108-11; PMID:16763112; http://dx.doi.org/10.1126/science.1126747
- Krause SA, Xu H, Gray JV. The synthetic genetic network around PKC1 identifies novel modulators and components of protein kinase C signaling in Saccharomyces cerevisiae. Eukaryot Cell 2008; 7:1880-7; PMID:18806213; http://dx.doi.org/10.1128/EC.00222-08
- Yoshida S, Ikeda E, Uno I, Mitsuzawa H. Characterization of a staurosporine- and temperature-sensitive mutant, stt1, of Saccharomyces cerevisiae: STT1 is allelic to PKC1. Mol Gen Genet 1992; 231:337-44; PMID:1538690; http://dx.doi.org/10.1007/BF00292700
- Simpson-Lavy KJ, Brandeis M. Phosphorylation of Cdc5 regulates its accumulation. Cell Div 2011; 6:23; PMID:22204387; http://dx.doi.org/10.1186/1747-1028-6-23
- Lu D, Hsiao JY, Davey NE, Van Voorhis VA, Foster SA, Tang C, Morgan DO. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J Cell Biol 2014; 207(1):23-39; PMID:25287299
- Lu D, Hsiao JY, Davey NE, Van Voorhis VA, Foster SA, Tang C, Morgan DO. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J Cell Biol 2014; 207:23-39; PMID:25287299; http://dx.doi.org/10.1083/jcb.201402041
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 1999; 15:963-72; PMID:10407276; http://dx.doi.org/10.1002/(SICI)1097-0061(199907)15:10B%3c963::AID-YEA399%3e3.0.CO;2-W
- Raithatha SA, Stuart DT. Meiosis-specific regulation of the Saccharomyces cerevisiae S-phase cyclin CLB5 is dependent on MluI cell cycle box (MCB) elements in its promoter but is independent of MCB-binding factor activity. Genetics 2005; 169:1329-42; PMID:15654101; http://dx.doi.org/10.1534/genetics.104.036103
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 2006; 21:319-30; PMID:16455487; http://dx.doi.org/10.1016/j.molcel.2005.12.011
- Caviston JP, Longtine M, Pringle JR, Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol Biol Cell 2003; 14:4051-66; PMID:14517318; http://dx.doi.org/10.1091/mbc.E03-04-0247
