Accurate transmission of genetic information across generations is a fundamental trait of all living organisms. The cell overcomes significant hurdles during this process, including the presence of damaged DNA and of naturally occurring structures that blocks the progression of DNA replication. Equally there is the so called ‘end replication problem’, which stems from the inability of de novo synthesis by DNA polymerases.
Several pathways have evolved to promote the completion of DNA replication in the presence of DNA damage. These pathways are collectively known as DNA damage tolerance (DDT) and involve either direct bypass of the DNA lesion using a set of specialized translesion DNA polymerases, or they involve creation of new primer-templates (by strand-invasion) on the newly synthesized sister chromatid using mechanisms involving several homologous recombination (HR) factors. Importantly, canonical HR also promotes DNA damage tolerance later in the cell-cycle, but it is actively repressed during DNA replication, as its use may lead to potentially deleterious chromosomal rearrangements. This HR repression is dependent on post-translational modification of proliferating cell nuclear antigen (PCNA) by SUMOylation.Citation1 Two different factors involved in regulation of HR recognize and act on SUMO-PCNA: Srs2 and Elg1.
Srs2 suppresses recombination by utilizing 2 separate mechanisms. One mechanism involves dismantling Rad51 nucleoprotein filaments (structures required for the strand invasion step) and is dependent on targeting of Srs2 to replication forks by SUMO-PCNA. A recently discovered second mechanism involves limiting the extent of recombination-associated DNA synthesis by disruption of the Polδ/SUMO-PCNA complex.Citation2
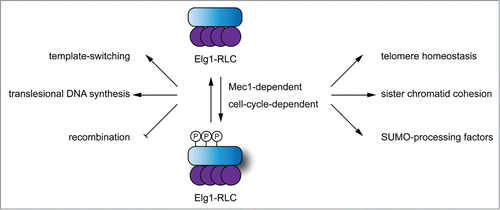
Elg1 shares homology with Rfc1, which is subunit of the canonical complex that loads PCNA on DNA. Along with other Rfc1 homologues, namely Ctf18 and Rad24, they interact with smaller subunits of the RFC complex (Rfc2–5) forming 3 different alternative Rfc1-like complexes (RLC). But in contrast to Rfc1, Elg1 mechanistically removes SUMO-PCNA from chromatin. This differential activity is based on the higher affinity of Elg1 for SUMO-PCNA and its mode of action remains to be defined. Elg1 has been linked with a number of DNA metabolic processes, including regulation of homologous recombination, promotion of template switching at the expense of the translesion DNA synthesis, telomere maintenance, sister chromatid cohesion, proper checkpoint activation, etc (Fig. 1). Additionally, Elg1 interacts with SUMO-processing factors, namely the Slx5–8 SUMO-dependent E3 ubiquitin ligase, known to target SUMOylated proteins for degradation. The many roles of Elg1 in these conceptually different processes thus raised the question of whether and how the Elg1 function is regulated (for a full description of Elg1 function see a recent, comprehensive reviewCitation3).
Work emanating from the lab of Martin Kupiec has been crucial in characterizing the function of Elg1 in various aspects of maintenance of genome stability. In the paper that appears in this issue of Cell cycle,Citation4 Kupiec and co-workers tackle the important question of regulation of Elg1 activity. They found that Elg1 is phosphorylated on serine 112 upon exposure to DNA damage in a Mec1-dependent manner. Furthermore, they found 2 other residues, serine 6 and 8 to become phosphorylated as well, presumably in a cell-cycle stage-dependent manner (Fig. 1). However, Elg1 phosphorylation is not required for resistance to DNA damage by MMS, indicating that other factors, including Srs2, are able to substitute for the Elg1 in MMS resistance. Importantly, by analyzing phenotypes of phospho-deficient and phospho-mimicking mutants in all possible combinations, they found that impairment of Elg1s phosphorylation partially suppresses the MMS sensitivity of Δrad5, pointing to the role of Elg1 in the template switching branch of the DDT and possibly also HR. Furthermore, a proper phosphorylation dynamic of Elg1 is absolutely required for its role in telomere homeostasis. The role of Elg1 in telomere maintenance is independent of recombination and possibly also of Srs2 and/or SUMO-PCNA as well.Citation5 The future work should address whether the Elg1 function in telomere maintenance may thus rely on its interaction with other SUMO-modified proteins and/or the SUMO-processing pathway, which is known to be important for proper telomere homeostasis.
The present work raises several important questions, concerning the role of Elg1 phosphorylation. In the future it will be of interest to see if the phosphorylation of Elg1 affects its ability to remove (SUMO)-PCNA from chromatin, or affects interaction with its multiple interacting partners and regulation of their stability, or whether Elg1 phosphorylation regulates its localization into different genomic loci, e.g., telomeres. In summary, work presented in Shkedy et al.,Citation4 opens up new avenues of research on Elg1 and its role in genome stability.
References
- Karras GI, et al. Mol Cell 2013; 49:536-46; PMID:23260657; http://dx.doi.org/10.1016/j.molcel.2012.11.016
- Burkovics P, et al. EMBO J 2013; 32:742-55; PMID:23395907; http://dx.doi.org/10.1038/emboj.2013.9
- Gazy I, et al. Mutat Res/Rev Mutat Res 2015; 763 IS:267-79; PMID:25795125; http://dx.doi.org/10.1016/j.mrrev.2014.11.007
- Shkedy D, et al. Cell Cycle 2015; PMID:26177013; http://dx.doi.org/10.1080/15384101.2015.1068475
- Smolikov S, et al. Proc Natl Acad Sci USA 2004; 101:1656-61; PMID:14745004; http://dx.doi.org/10.1073/pnas.0307796100