Abstract
Ubiquitination plays a key and complex role in the regulation of c-Myc stability, transactivation, and oncogenic activity. c-Myc is ubiquitinated by a number of ubiquitin ligases (E3s), such as SCFFbw7 and SCFSkp2. Depending on the E3s, ubiquitination can either positively or negatively regulate c-Myc levels and activity. Meanwhile, c-Myc ubiquitination can be reversed by deubiquitination. An early study showed that USP28 deubiquitinates c-Myc via interacting with Fbw7α whereas a recent study reveals that USP37 deubiquitinates c-Myc independently of Fbw7 and c-Myc phosphorylation. Consequently, both USP28 and USP37 stabilize c-Myc and enhance its activity. We recently found the nucleolar USP36 as a novel c-Myc deubiquitinase that controls the end-point of c-Myc degradation pathway in the nucleolus. Here we briefly review the current understanding of ubiquitination and deubiquitination regulation of c-Myc and further discuss the USP36-c-Myc regulatory pathway.
Introduction
As a master transcription regulator, c-Myc (Myc, thereafter) controls the expression of most, if not all, actively transcribed genes in the human genomeCitation1-4 and coordinates many cellular processes, including promoting cell proliferation and growth by enhancing ribosomal biogenesis/protein translation, nucleotide biosynthesis, metabolism, RNA processing and DNA replication, inducing apoptosis, senescence, differentiation, and angiogenesis, as well as regulating stem cell renewal.Citation5-7 Although cells cannot live without Myc, aberrant overexpression of Myc contributes to the development of malignancies in human and in mouse models.Citation6,8 Thus tightly regulated levels of Myc are essential for normal cell growth and proliferation. Among many mechanisms contributing to the deregulated overexpression of Myc, including transcription, gene amplification, and chromosome translocation, increasing studies have shown that Myc protein stabilization due to impaired Myc degradation pathway plays a key role in cancers.Citation9-13
The Myc protein contains a N-terminal unstructured transactivation domain (TAD) essential for Myc transactivation activity, which includes the conserved Myc box I (MBI) and MBII. The MBI contains 2 critical residues, Threonine 58 (T58) and Serine 62 (S62), whose dynamic phosphorylation controls Myc protein stability in response to cell growth signals.Citation5,6,9,10 The MBII is critical for recruiting key Myc transactivation co-activators such as TRAAP, GCN5, TIP48, TIP49, TIP60, CBP/p300, BAF53 as well as Skp2.Citation5,14,15 The central region of Myc contains the MBIII and MBIV followed by a PEST (rich in proline, glutamic acid, threonine, and proline residues) sequence and a nuclear localization signal (NLS), respectively. Located at the C-terminal region is a basic helix-loop-helix leucine zipper (bHLH-LZ) domain that dimerizes with the Max protein and binds to the E-box elements in target gene promoters.Citation5,6,14,15
Myc is a short-lived protein with a half-life less than 30 minutes in non-transformed cells.Citation10,16,17 The fast turnover of Myc protein is tightly controlled by the ubiquitin-proteasome system. Growth signal-controlled Myc turnover is largely through a phosphorylation-ubiquitination cascade. Phosphorylation of Myc at S62, triggered by the Ras-induced Raf-MEK-ERK kinase cascade and/or CDKs in response to growth signals, increases Myc stability. Phosphorylation of T58 occurs sequentially and is mediated by GSK3β, which is held in check by PI(3)K/Akt signaling, also in response to growth factor and receptor tyrosine kinase signaling.Citation10,12,18 When Ras signaling subsides, GSK3β becomes activated and starts to phosphorylate Myc at T58.Citation12,19-22 This phosphorylation facilitates the recruitment of a prolyl isomerase, Pin1, to catalyze the cis to trans isomerization at proline (P) 63 of Myc, leading to the subsequent conformational change that allows the PP2A phosphatase to dephosphorylate Myc at S62.Citation23 Finally, phosphorylated T58 serves as a dock to recruit the T58 phosphorylation-dependent ubiquitin ligase (E3) complex SCFFbw7 to mediate Myc ubiquitination and degradation through the proteasome system.Citation24–27
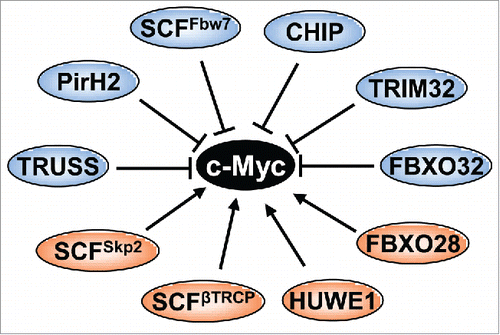
Likewise, there are many other ubiquitin E3s that mediate Myc ubiquitination, resulting in either activation or inhibition of Myc activity and either degradation or stabilization of Myc protein. Moreover, Myc ubiquitination can be reversed by deubiquitinating enzymes (DUBs). In this perspective, we will briefly review current progress toward understanding the ubiquitination-deubiquitination regulation of Myc stability and activity and discuss further our rec-ent finding of the novel nucleolar Myc deubiquitinating enzyme USP36 that positively regulates Myc levels and activity.
Ubiquitination of Myc: Diverse roles in Regulating Myc Stability and Activity
As mentioned above, the SCFFbw7 E3 ligase mediates Myc ubiquitination and degradation upon growth signal deprivation. There are 3 Fbw7 isoforms located in distinct subcellular compartments: Fbw7β and Fbw7α are in the cytoplasm and the nucleoplasm, respectively, whereas Fbw7γ is in the nucleolus.Citation27 When overexpressed, all the 3 isoforms can mediate K48-linked Myc ubiquitination and degradation.Citation26 The T58 phosphorylation-dependent Myc degradation by the SCFFbw7 complex is critical for growth signal-controlled Myc turnover. T58 mutation results in stabilization and enhanced oncogenic activity of Myc, and T58 is one of the “hot spots” for mutation in a subset of Burkitt's lymphomas.Citation22,28-30 Mice harboring the MycT58A mutant develop cancer at a significantly higher penetrance and reduced latency than mice with the wild type c-Myc transgene.Citation31,32 Consistent with the role of Fbw7 in negatively regulating Myc, mutations and deletions of FBW7 are found in multiple human cancers.Citation33 Thus, T58 phosphorylation-dependent ubiquitination by SCFFbw7 clearly plays a key role in regulating Myc turnover and oncogenic activity. Yet, over the past decade, additional ubiquitin E3s have been identified as important Myc regulators ().
SCFSkp2
The SCFSkp2 ubiquitin E3 ubiquitinates Myc and targets Myc for proteasomal degradation.Citation34–36 Surprisingly, this degradation is associated with the increased activity of Myc and overexpression of Skp2 promotes Myc-induced cell cycle progression.Citation34,36 Skp2 binds to the MBII and bHLH-LZ domains of MycCitation34,36 and acts as a co-activator of Myc. Although it remains unknown why Skp2 mediates both the proteasomal degradation and the transactivational activity of Myc, several pieces of evidence support a transcription-coupled proteasomal degradation mechanism. Myc recruits Skp2 and a number of proteasome subunits to Myc target gene promoters such as the cyclin D2 promoter,Citation34,35 suggesting that the 26S proteasome may degrade Myc at its target gene promoters once it finishes its role in activating transcription. Recently, it has been shown that the ubiquitination of Myc at the 6 lysines in the TAD is critical for the canonical transcriptional activity and transformation activity of Myc.Citation37 ARF inhibits the Myc-Skp2 interaction and Skp2-mediated Myc ubiquitination, leading to Myc stabilization and the switch from an oncogenic protein to an apoptotic inducer.Citation37 Supporting the transcription-coupled proteasomal degradation of Myc, we recently found that PIN1 promotes the turnover of Myc at target gene promoters while also increasing its transactivation activity.Citation38 Yet, other studies argued that Skp2 promotes Myc activity independently of the SCF complex, reporting that instead, it cooperates with Myc to recruit the Miz1 transcription factor and p300 coactivator to the target RhoA gene promoter, leading to the increase of RhoA expression and cell invasion and migration.Citation39 Similarly, overexpression of a Skp2 mutant that is unable to associate with the SCF complex also restores Myc activity that is down-regulated by the ERK-dependent reduction of Skp2.Citation40 Nevertheless, Skp2 promotes Myc's transactivation and transformation activity, while it can also ubiquitinate and degrades Myc.
HUWE1/ARF-BP1/HectH9
HUWE1 (also called ARF-BP1 and HectH9), a member of the HECT-domain containing ubiquitin E3s, is another Myc ubiquitin E3 that enhances Myc activity.Citation41 HUWE1 binds to Myc at the TAD domain and mediates K63-linked Myc ubiquitination at the lysines near the NLS.Citation41 This ubiquitination enhances the transcriptional activity of Myc by recruiting p300, without inducing Myc proteasomal degradation. Miz1 competes with Myc to interact with HUWE1 and inhibits HUWE1-mediated Myc ubiquitination. Recently it has been shown that small molecule inhibitors targeting HUWE1 inhibit the activity of Myc in colorectal cancer cells via stabilizing a Miz1-Myc repressive complex that suppresses Myc target genes.Citation42
SCFFBXO28
Similar to HUWE1, SCFFBXO28 also mediates non-proteolytic Myc ubiquitination.Citation43 During cell cycle progression to S phase, CDK1/2-mediated phosphorylation of FBXO28 at Ser 344 activates its ubiquitin E3 activity to polyubiquitinate Myc without inducing proteolytic Myc degradation. FBXO28 binds to Myc at the MBII and possible motifs upstream of the HLH motif. This Myc ubiquitination by SCFFBXO28 stimulates Myc activity also by recruiting p300 to target gene promoters,Citation43 which requires the ubiquitination of several lysines within the 294–367 region of Myc known to be involved in ubiquitin-mediated Myc-p300 interaction.Citation41 Consequently, FBXO28 promotes Myc-driven tumorigenesis and high expression of FBXO28 (as well as high Ser 344 phosphorylation) are associated with poor patient outcomes. Thus, SCFFBXO28-mediated ubiquitination of Myc appears to play an important role in cell cycle regulation of Myc activity, transmitting CDK activity to Myc activity as cells progress to S phase. However, the detailed type of polyubiquitination and why this ubiquitination does not target Myc for degradation remains unknown. How this E3 interplays with other E3s in the cell cycle progression also warrants further investigation.
SCF-TRCP
The fourth Myc E3 that does not mediate Myc degradation is the SCFβ-TRCP. Degradation of Myc by SCFFbw7 has been implicated in controlling Myc stability in G1 and early S phase of the cell cycle.Citation12 Popov et al. recently found that SCFβ-TRCP plays a role in Myc ubiquitination in the subsequent G2 cell cycle phases. This ubiquitination antagonizes SCFFbw7 to differentially ubiquitinate the N-terminus of Myc and stabilizes Myc.Citation44 While SCFFbw7 induces K48-linked ubiquitination, SCFβ-TRCP mediates K48/K63 heterotypic polyubiquitination of Myc and is required for Myc-dependent acceleration of cell cycle progression during S and G2 phases. This ubiquitination also requires the phosphodegron (a phospho-recognition sequence for β-TRCP binding at amino acids 278–283). Therefore, SCFFbw7 and SCFβ-TRCP E3s alternatively act on Myc during cell cycle progression. Consistently, PI3k/mTOR kinase inhibitors inhibit phosphorylation of β-TRCP, resulting in its proteasomal degradation in breast cancer cells, and this correlates with a reduction in Myc.Citation45
TRPC4AP/TRUSS
TRPC4AP (transient receptor potential cation channel, subfamily C, member 4-associated protein), also called TRUSS (tumor necrosis factor receptor-associated ubiquitous scaffolding and signaling protein), was identified as a novel Myc-interacting E3.Citation46 TRUSS interacts with both c-Myc and N-Myc at their C-terminal bHLH-LZ domain and promotes Myc ubiquitination. Further studies showed that TRUSS complex contains DDB1 (damage-specific DNA-binding protein 1), CUL4A (Cullin 4A), DDA1, and the small RING finger protein RBX1 (which is the receptor for a DDB1-CUL4 E3 ligase complex) and mediates the interaction of MYC with the DDB1-CUL4 E3 ligase for selective Myc degradation through the proteasome.Citation46 Consistently, TRUSS suppresses Myc transactivation and transformation activity and is downregulated in a number of cancers.Citation46 Interestingly, it was recently shown that TRUSS itself is ubiquitinated by Skp2 and degraded through the proteasome.Citation47 TRUSS is primarily expressed during the G1 phase of cell cycle and its level starts to decline with the expression of the S-phase specific E3 ligase Skp2. Overexpression of Skp2 correlates with the reduction of TRUSS in human cancers, providing support that Myc ubiquitin E3s coordinately regulate Myc and can inter-regulate each other during the cell cycle.
TRIM32
Schwamborn et alCitation48 has shown that TRIM32 is an Ring-finger ubiquitin ligase for Myc that controls Myc protein stability. The RING finger mutant TRIM32C24A failed to ubiquitinate and degrade Myc. TRIM32-mediated Myc degradation is essential for TRIM32 to induce neuronal differentiation. However, whether and how TRIM32 plays a role in tumorigenesis by regulating Myc remains unclear.
Pirh2
Pirh2 is originally shown to be a p53 ubiquitin ligase. To analyze the in vivo function of Pirh2, Hakem et alCitation49 generated Pirh2 deletion mice. Consistent with the increased levels of p53, increased p53-dependent apoptosis was observed in the mice in response to DNA damage. They also found that Myc levels were increased in these mice. Subsequent study revealed that Pirh2 associated with Myc through 2 contacts: the C-terminal Ring-finger containing domains of Pirh2 interacts with the Myc MBII whereas the N-terminal domain of Pirh2 interacts with the bHLH-LZ domain of Myc.Citation49 Downregulation of Pirh2 is associated with poor outcome of a number of human cancers including breast, ovarian, and lung cancers.Citation49 Genetic deletion of Pirh2 in mice also results in plasma cell hyperplasia associated with the increased Myc, γ-globulinemia, kidney failure, premature death, tumorigenesis, and shortened lifespan.Citation49 Approximately 25% of Pirh2-/- and 17% of Pirh2+/- sick mice also developed solid tumors including sarcomas, liver, testes, mammary and lung tumors, indicating that Pirh2 is a tumor suppressor in vivo.
CHIP
CHIP (carboxyl terminus of Hsc70-interacting protein) is a chaperone-associated U-box-containing E3 ligase that links a chaperone to the 26S proteasome by ubiquitinating chaperone substrates that then direct the chaperone toward the proteasome.Citation50 Overexpression of CHIP could accelerate the turnover rate of Myc protein, whereas knockdown of CHIP by RNAi stabilizes Myc.Citation51 CHIP interacts with and ubiquitinates Myc and suppresses Myc activity. Interestingly, the association between CHIP and Myc is dependent on the chaperone, particularly Hsp70. Further study needs to clarify the interaction between Myc and CHIP and whether CHIP regulates Myc levels under physiological conditions.Citation51 Overall, the activity of CHIP to inhibit Myc is consistent with its putative tumor suppressor function.
SCFFBXO32
Recently, it has been shown that SCFFBXO32 promotes Myc ubiquitination through K48-linked ubiquitin chains and targets it for proteasome degradation, independently of its phosphorylation at either T58 or S62.Citation52 FBXO32 interacts with the MBII, MBIV and PEST motifs. The ubiquitination site is mapped at K326 on Myc. Unlike FBXO28, FBXO32 suppresses Myc transactivation activity and inhibits cell growth. Moreover, FBXO32 itself is a Myc target gene, forming a negative regulatory loop.
Deubiquitination of Myc: When and Where?
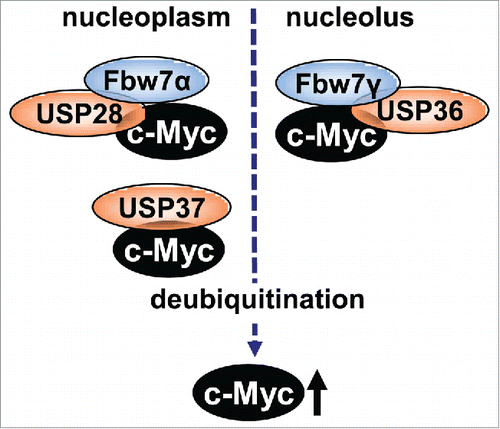
Adding to the complexity of the ubiquitination regulation of Myc, deubiquitination regulation has emerged as another important mechanism for the control of Myc. Similar to many other reversible posttranslational modifications, ubiquitination of Myc can be reversed by deubiquitination catalyzed by DUBs. The human genome encodes approximately 95 DUBs that are classified into 5 families.Citation53,54 Ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor associated proteases (OTUs), Machado-Joseph disease (or Josephin domain) proteins (MJDs), and JAB1/MPN/MOV34 proteins (JAMMs). The USP family member USP28 is the first DUB discovered that deubiquitinates and stabilizes Myc.Citation55 Knockdown of USP28 reduces the levels and activity of Myc. Interestingly, USP28 binds to Myc through Fbw7α in the nucleoplasm, but not Fbw7γ in the nucleolus, and reverses Fbw7α-, but not Fbw7γ-, mediated Myc ubiquitination and degradation,Citation55 suggesting that it deubiquitinates and stabilizes Myc in the nucleus, but not in the nucleolus. Consistently, USP28 overexpression is found in colon and breast cancers and USP28-mediated stabilization of Myc is essential for the cancer cell proliferation observed in the study.Citation55 In response to DNA damage, USP28 dissociates from Fbw7α, allowing Myc to be degraded by Fbw7α, providing a mechanism for Myc degradation upon DNA damage.Citation56
Interestingly, a recent study has increased the complexity of this USP28-Fbw7α-Myc axis. Schulein-Volk et alCitation57 demonstrated that USP28 has a dose-dependent and dual regulation of Fbw7 function. An elegant USP28 knockout mouse study revealed that USP28 primarily controls the stability of Fbw7 by antagonizing its auto-ubiquitination and degradation. Thus, while heterozygous deletion of USP28 can still maintain stable Fbw7 to ubiquitinate and degrade its substrates, complete deletion of USP28 triggers Fbw7 auto-destruction, resulting in the stabilization of the Fbw7 substrates such as Myc and cyclin E.Citation57 Thus both overexpression of USP28 and its complete deletion stabilize Myc and promote cell transformation.Citation57 Further complexity was revealed in an intestine study, where it was shown that deletion of USP28 rescued the cytotoxic effects of Fbw7-deficiency in primary fibroblasts and meliorated the hyperproliferation and the impaired goblet and Paneth cell differentiation in mice with intestine-specific deletion of Fbw7 (Fbw7ΔIEC).Citation58 This correlates with correcting the aberrant accumulation of Fbw7 substrates such as Myc, c-Jun and NICD1. These results suggest that USP28 can function independent of a direct interaction with Fbw7. Using peptide binding assays, it was shown that USP28 binds to the same Myc phosphodegron recognized by Fbw7 in MBI; yet USP28 binds to the non-phosphorylated form whereas Fbw7 binds to the phosphorylated form of this Myc degron.Citation58 These studies demonstrate a complex regulation of Myc by the USP28-Fbw7 interplay, suggesting an important role in the dynamic and finely tuned regulation of Myc activity necessary for normal cellular function.
Recently, USP37 was found to be another DUB that deubiquitinates and stabilizes Myc.Citation59 Interestingly, USP37 regulation of Myc does not require either Fbw7 or the phosphorylation of Myc at T58. Instead, it directly interacts with Myc at the MBIII region. Functionally, USP37 promotes cell proliferation and metabolism by stabilizing Myc and it is also overexpressed in tested human lung cancers.Citation59 Whether the activity of USP37 coordinates with that of USP28 requires further study.
Both USP28 and USP37 regulate Myc stability in the nucleoplasm. Studies indicate that Myc is mainly degraded in the nucleolus and proteasome inhibition leads to accumulation of Myc in the nucleolus.Citation27,60,61 Importantly, Myc-mediated cell growth and tumorigenesis is well tied with its role in enhancing ribosome biogenesis in the nucleolus.Citation62,63 Thus, the nucleolus certainly plays a key role in regulating Myc levels and oncogenic activity and it is important to understand whether and how Myc can be deubiquitinated in the nucleolus. Toward this goal, we recently discovered the nucleolar USP36 as a novel Myc DUB and a positive Myc regulator in the nucleolus.Citation64
USP36 is a Novel Nucleolar DUB for Myc
USP36 was initially identified as a DUB overexpressed in ovarian cancers.Citation65 Later, it was found that USP36 is mainly located in the nucleolus and regulates the stability of nucleolar proteins nucleophosmin and fibrillarian, both of which are involved in rRNA processing and ribosomal biogenesis.Citation66,67 Knockdown of USP36 impaired rRNA synthesis, processing and ribosomal biogenesis in the nucleolus.Citation67 Further supporting this role, a yeast homolog ubp10 controls the stability of the largest subunit of RNA polymerase I and is essential for RNA Pol I activity and yeast cell growth; and Ubp10 deletion-induced yeast growth retardation can be rescued by human USP36.Citation68 Like many nucleolar proteins, USP36 likely shuttles between the nucleolus and the nucleoplasm and a small fraction of USP36 is located in the nucleoplasm,Citation64 indicating that USP36 may have additional functions in the nucleoplasm. Further, it has been shown that the Drosophila USP36 homolog (dUSP36) regulates autophagyCitation69 and the NFκB-associated inflammation signaling by targeting the IMD proteins,Citation70 suggesting that USP36 executes multiple functions in cells.
We found that USP36 directly deubiquitinates and stabilizes Myc, and this can occur in the nucleolus. USP36 directly binds to Myc, and it interacts specifically with Fbw7γ in the nucleolus, but not Fbw7α in the nucleoplasm, and it forms a tertiary complex with Myc and Fbw7γ.Citation64 Consistently, ablation of USP36 significantly reduced the levels of Myc and inhibited cell proliferation. Its overexpression was also seen in many human cancers such as breast and lung cancers, implying the oncogenic nature of USP36.Citation64 As Myc is a master regulator of ribosomal biogenesis, our results further support the role of USP36 in ribosomal biogenesis and demonstrate USP36 as a critical Myc regulator.Citation64 Yet, many questions still remain.
Although USP36 regulates Myc in the nucleolus, it increases the steady state levels of Myc in both the nucleoplasm and the nucleolus,Citation64 suggesting that the deubiquitinated nucleolar Myc can shuttle back to the nucleoplasm. This is consistent with the ability of USP36 to enhance Myc transactivation activity of Pol II-driven transcription. These data do not completely exclude the possibility that USP36 might also directly deubiquitinate Myc in the nucleoplasm, as a small fraction of USP36 is localized in the nucleoplasm and it can directly interact with Myc without the need of Fbw7. Also our ubiquitination assays were conducted in cells treated with proteasome inhibitor, which results in the accumulation of the ubiquitinated Myc in the nucleolus.Citation27,60,61 The nucleolar-localization defective mutant of USP36 could solve this issue. Most likely, at normal homeostatic conditions, USP36 mainly controls Myc ubiquitination and turnover in the nucleolus. It will be interesting to determine whether USP36 localization and its regulation of Myc changes under stress conditions and whether post-translational modifications can regulate the cellular localization and function of USP36.
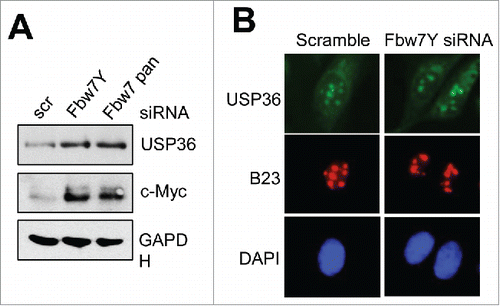
It is also important to further elucidate the interplay between USP36 and Fbw7. We show that USP36 interacts specifically with Fbw7γ and thus the detailed interaction needs to be analyzed. Why it only interacts with Fbw7γ, but not Fbw7α, will be interesting to determine. Do USP36 and Fbw7γ mutually regulate each other, given that an ubiquitin E3 often complexes with a corresponding DUB to dynamically regulate a substrate? We indeed observed that siRNA-mediated knockdown of Fbw7γ or all Fbw7 isoforms increases the levels of USP36 () without affecting its nucleolar localization (), suggesting that Fbw7 may ubiquitinate and destabilize USP36. Conversely, it is interesting to test whether USP36 could suppress Fbw7γ autoubiquitination and stabilize it, similar to the role of USP28 on Fbw7.Citation57,58 It is also important to differentiate the role of USP36 as a DUB to directly deubiquitinate Myc or as an Fbw7γ inhibitor to suppress Fbw7γ-mediated Myc ubiquitination, or a combined effect of both.
Figure 2. Fbw7γ regulates the levels of USP36, but not its nucleolar localization. Hela cells transfected with scrambled, Fbw7γ specific siRNA or pan-siRNA targeting all Fbw7 isoforms were subjected to immunoblot to detect the indicated proteins (A) and IF staining with anti-USP36 (green) or B23 (red) antibodies (B).
Another important question is whether the USP36- Fbw7γ axis interplays with the USP28- Fbw7α axis to regulate Myc. Both USP28 and USP36 deubiquitinate Myc and they interact with Fbw7α and Fbw7γ, respectively. Interestingly, USP36 can inhibit Myc degradation mediated by either Fbw7α or Fbw7γ, suggesting that Fbw7α ubiquitination of Myc in the nucleoplasm collaboratively contributes to Fbw7γ-mediated Myc ubiquitination in the nucleolus, either by adding priming ubiquitins or by facilitating translocation of Myc to the nucleolus, which would be antagonized by USP28.Citation71 Indeed, we observed that overexpression of USP36 and USP28 cooperatively further increase Myc levels compared to individual overexpression of USP36 or USP28 (). Thus, it is important in future studies to examine whether the USP28-Fbw7α and USP36-Fbw7γ axis regulate Myc ubiquitination sequentially at the respective cell compartments and how they may interplay to dynamically control Myc ubiquitination and degradation ().
Figure 3. USP36 and USP28 collaboratively enhance Myc levels. 293 cells transfected with V5-USP36, Flag-USP28, or both plasmids were assayed by immunoblot using anti-Myc, anti-V5 and anti-Flag antibodies as indicated.
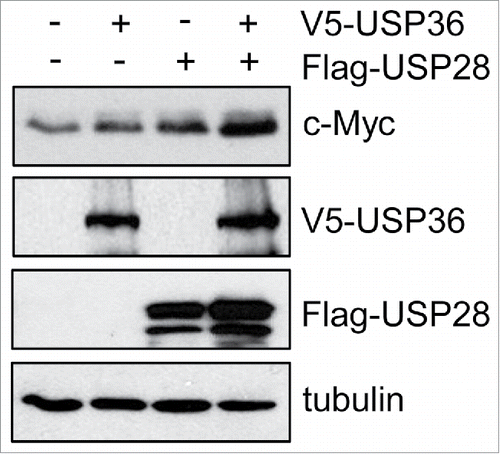
Figure 4. A diagram showing Fbw7 ubiquitinating and coordinating DUBs known to stabilize Myc. USP28 and USP37 act in the nucleoplasm whereas USP36 deubiquitinates Myc in the nucleolus.
Finally, we show that USP36 itself is a Myc target gene, suggesting that USP36 forms a positive feedback regulatory loop with Myc and plays an important role in rapid Myc stabilization upon growth stimulation. Indeed, we show that knockdown of USP36 drastically inhibited Myc induction following serum stimulation. Intriguingly, following its peak induction at 2 hours after serum stimulation, Myc levels decrease, while USP36 remains at relatively high levels for up to 12 hours,Citation64 due to the nature of its longer half-life (data not shown). This indicates that USP36 may lose its activity to target Myc before its level declines, although the underlying mechanism is not known. It will be interesting to examine whether posttranslational modifications of USP36 may be involved. Also, USP36 levels are likely regulated through cell cycle progression and/or USP36 regulation of Myc may be regulated by the cell cycle. Nevertheless, USP36 induction by Myc and in turn stabilization of Myc by USP36 suggest an elegant feed-forward regulatory loop that ensures a rapid control of Myc levels and activity following growth stimulation or in response to other extrinsic or intrinsic cues.
Conclusions
Emerging evidence indicates that deubiquitination regulation of Myc is equally important for the control of Myc protein stability and activity as ubiquitination regulation. Whether other DUBs besides the above mentioned USPs could down-regulate Myc activity with or without affecting Myc stability similar to some of the ubiquitin E3s is an interesting question to be explored. Does USP36 co-regulate Myc with ubiquitin E3s other than Fbw7 during alternate cell cycle phases or under different physiological conditions? Does it co-regulate Myc with DUBs other than USP28? It has been shown that SCFβ-TRCP antagonizes SCFFbw7 to ubiquitinate Myc at the N-terminus. Does USP36 deubiquitinate Myc that is ubiquitinated by SCFβ-TRCP or SCFSkp2 in the nucleus? Finally, it is important to study the USP36 regulation of Myc using in vivo models to further understand its role in regulating Myc, nucleolar dynamics and ribosomal biogenesis. Myc is a master regulator of ribosome biogenesis; Myc-overexpressing cells require or are adapted to the elevated ribosomal biogenesis/translation activity for uncontrolled cell proliferation and growth (oncogenic addiction). As USP36 is mainly localized in the nucleolusCitation64,66 and plays a role in ribosome biogenesis,Citation66,68 knockdown or inhibition of USP36 may severely attenuate ribosomal biogenesis (by directly inhibiting Myc and other nucleolar targetsCitation66,68 involved in ribosomal biogenesis), leading to a synthetic lethality in Myc-addicted cells. It will be interesting to investigate whether USP36 regulates other nucleolar proteins involved in rRNA biogenesis and whether it has a nucleolar scaffold protein function independent of its DUB activity. Nevertheless, our data showing that USP36 stabilizes Myc and its levels correlate with the levels of Myc in tumors suggest that targeting USP36 could be a means to develop novel cancer therapeutics.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by NIH/NCI grants R01 CA186241 to M-S. D. and R. S. and R01 CA160474 to M-S. D., and R01 CA 100855 to R. S., as well as a grant from the Medical Research Foundation (MRF) of Oregon to X.-X. S.
References
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012; 151:56-67; PMID:23021215; http://dx.doi.org/10.1016/j.cell.2012.08.026
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012; 151:68-79; PMID:23021216; http://dx.doi.org/10.1016/j.cell.2012.08.033
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014; 511: 488-92; PMID:25043028; http://dx.doi.org/10.1038/nature13537
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 2014; 511:483-7; PMID:25043018; http://dx.doi.org/10.1038/nature13473
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 2005; 6:635-45; PMID:16064138; http://dx.doi.org/doi:10.1038/nrm1703
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008; 8:976-90; PMID:19029958; http://dx.doi.org/10.1038/nrc2231
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 2010; 10: 301-9; PMID:19029958; http://dx.doi.org/10.1038/nrc2231
- Nesbit CE, Tersak JM, aProchownik EV. MYC oncogenes and human neoplastic disease. Oncogene 1999; 18:3004-16; PMID:20332779; http://dx.doi.org/10.1038/nrc2819
- Farrell AS, Sears RC. MYC degradation. Cold Spring Harb Perspect Med 2014; 4: pii: a014365; PMID:24591536; http://dx.doi.org/10.1101/cshperspect.a014365
- Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol 2006; 16:288-302; PMID:16938463; http://dx.doi.org/10.1016/j.semcancer.2006.08.004; PMID:16938463
- Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell 1999; 3:169-79; PMID:10078200; http://dx.doi.org/10.1016/S1097-2765(00)80308-1
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 2000;14:2501-14; PMID:11018017; http://dx.doi.org/10.1101/gad.836800
- Sears RC. The life cycle of C-myc: from synthesis to degradation. Cell cycle 2004; 3:1133-7; PMID:15467447; http://dx.doi.org/10.4161/cc.3.9.1145
- Conacci-Sorrell M, McFerrin L, Eisenman, RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med 2014; 4, a014357; PMID:24384812; http://dx.doi.org/10.1101/cshperspect.a014357
- Tu WB, Helander S, Pilstal R, Hickman KA, Lourenco C, Jurisica I, Raught B, Wallner B, Sunnerhagen M, Penn LZ. Myc and its interactors take shape. Biochim Biophys Acta 2015; 1849:469-83; PMID:24933113; http://dx.doi.org/10.1016/j.bbagrm.2014.06.002
- Ciechanover A, DiGiuseppe JA, Bercovich B, Orian A, Richter JD, Schwartz AL, Brodeur GM. Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proc Natl Acad Sci U S A 1991; 88:139-43; PMID:1846034
- Flinn EM, Busch CM, Wright AP. myc boxes, which are conserved in myc family proteins, are signals for protein degradation via the proteasome. Mol Cell Biol 1998; 18:5961-69; PMID:9742113
- Lutterbach B, Hann SR. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol 1994; 14:5510-22; PMID:8035827; http://dx.doi.org/10.1128/MCB.14.8.5510
- Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem 2003; 278:51606-12; PMID:14563837; http://dx.doi.org/10.1074/jbc.M310722200
- Chang DW, Claassen GF, Hann SR, Cole MD. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol Cell Biol 2000; 20:4309-19; PMID:10825194; http://dx.doi.org/10.1128/MCB.20.12.4309-4319.2000
- Lutterbach B, aHann SR. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol 1994; 14:5510-22; PMID:8035827; http://dx.doi.org/10.1128/MCB.14.8.5510
- Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv Cancer Res 2002; 84:81-154; PMID:11885563; http://dx.doi.org/10.1016/S0065-230X(02)84004-0
- Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol 2004; 6:308-18; PMID:15048125; http://dx.doi.org/10.1038/ncb1110
- Moberg KH, Mukherjee A, Veraksa A, Artavanis-Tsakonas S, Hariharan IK. The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr Biol 2004; 14:965-74; PMID:15182669; http://dx.doi.org/10.1016/j.cub.2004.04.040
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 2004; 23:2116-25; PMID:15103331; http://dx.doi.org/10.1038/sj.emboj.7600217
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman B E. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A 2004; 101:9085-90; PMID:15150404; http://dx.doi.org/10.1073/pnas.0402770101
- Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol 2004; 14:965-74; PMID:15498494; http://dx.doi.org/10.1016/j.cub.2004.09.083
- Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 2000; 95:2104-10; PMID:10706881
- Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol. 2000; 20:2423-35; PMID:10713166; http://dx.doi.org/10.1128/MCB.20.7.2423-2435.2000
- Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J 1999; 18, 717-26; PMID:9927431
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 2005; 436:807-11; PMID:16094360; http://dx.doi.org/10.1038/nature03845
- Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, Sears RC. Phosphorylation regulates c-Myc's oncogenic activity in the mammary gland. Cancer Res 2011; 71:925-36; PMID:21266350; http://dx.doi.org/10.1158/0008-5472.CAN-10-1032
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 2008; 8:83-93; PMID:18094723; http://dx.doi.org/10.1038/nrc2290
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell 2003; 11:1189-200; PMID:12769844; http://dx.doi.org/10.1016/S1097-2765(03)00193-X
- von der Lehr N, Johansson S, Larsson LG. Implication of the ubiquitin/proteasome system in Myc-regulated transcription. Cell Cycle 2003; 2:403-7; PMID:12963825; http://dx.doi.org/10.4161/cc.2.5.484
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell 2003; 11:1177-88; PMID:12769843; http://dx.doi.org/10.1016/S1097-2765(03)00173-4
- Zhang Q, Spears E, Boone DN, Li Z, Gregory MA, Hann SR. Domain-specific c-Myc ubiquitylation controls c-Myc transcriptional and apoptotic activity. Proc Natl Acad Sci U S A 2013; 110:978-83; PMID:23277542; http://dx.doi.org/10.1073/pnas.1208334110
- Farrell AS, Pelz C, Wang X, Daniel CJ, Wang Z, Su Y, Janghorban M, Zhang X, Morgan C, Impey S, et al. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol Cell Biol 2013; 33:2930-49; PMID:23716601; http://dx.doi.org/10.1128/MCB.01455-12
- Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, Wu J, Nakayama KI, Kang HY, Huang HY, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat Cell Biol 2010;12:457-67; PMID:20383141; http://dx.doi.org/10.1038/ncb2047
- Li X, Bian Y, Takizawa Y, Hashimoto T, Ikoma T, Tanaka J, Kitamura N, Inagaki Y, Komada M, Tanaka T. ERK-dependent downregulation of Skp2 reduces Myc activity with HGF, leading to inhibition of cell proliferation through a decrease in Id1 expression. Mol Cancer Res 2013; 11:1437-47; PMID:24177224; http://dx.doi.org/10.1158/1541-7786.MCR-12-0718
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 2005; 123:409-21; PMID:16269333; http://dx.doi.org/10.1016/j.cell.2005.08.016
- Peter S, Bultinck J, Myant K, Jaenicke LA, Walz S, Muller J, Gmachl M, Treu M, Boehmelt G, Ade CP, et al. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol Med 2014; 6:1525-41; PMID:25253726; http://dx.doi.org/10.15252/emmm.201403927
- Cepeda D, Ng HF, Sharifi HR, Mahmoudi S, Cerrato VS, Fredlund E, Magnusson K, Nilsson H, Malyukova A, Rantala J, et al. CDK-mediated activation of the SCF(FBXO) (28) ubiquitin ligase promotes MYC-driven transcription and tumourigenesis and predicts poor survival in breast cancer. EMBO Mol Med 2013; 5:999-1018; PMID:23776131; http://dx.doi.org/10.1002/emmm.201202341
- Popov N, Schulein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(β-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol 2010; 12:973-81; PMID:20852628; http://dx.doi.org/10.1038/ncb2104
- Yi YW, Kang HJ, Bae EJ, Oh S, Seong YS, Bae I. β-TrCP1 degradation is a novel action mechanism of PI3K/mTOR inhibitors in triple-negative breast cancer cells. Exp Mol Med 2015; 47, e143; PMID:25721419; http://dx.doi.org/10.1038/emm.2014.127
- Choi SH, Wright JB, Gerber SA, Cole MD. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev 2010; 24:1236-41; PMID:20551172; http://dx.doi.org/10.1101/gad.1920310
- Jamal A, Swarnalatha M, Sultana S, Joshi P, Panda SK, Kumar V. The G1 phase E3 ubiquitin ligase TRUSS that gets deregulated in human cancers is a novel substrate of the S-phase E3 ubiquitin ligase Skp2. Cell Cycle, 2015; 14:2688-700; PMID:26038816; http://dx.doi.org/10.1080/15384101.2015.1056946
- Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 2009; 136:913-25; PMID:19269368; http://dx.doi.org/10.1016/j.cell.2008.12.024
- Hakem A, Bohgaki M, Lemmers B, Tai E, Salmena L, Matysiak-Zablocki E, Jung YS, Karaskova J, Kaustov L, Duan S, et al. Role of Pirh2 in mediating the regulation of p53 and c-Myc. PLoS genetics 2011; 7:e1002360; PMID:22125490; http://dx.doi.org/10.1371/journal.pgen.1002360
- Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol 1999; 19:4535-45; PMID:10330192
- Paul I, Ahmed SF, Bhowmik A, Deb S, Ghosh MK. The ubiquitin ligase CHIP regulates c-Myc stability and transcriptional activity. Oncogene 2013; 32:1284-95; PMID:22543587; http://dx.doi.org/10.1038/onc.2012.144
- Mei Z, Zhang D, Hu B, Wang J, Shen X, a Xiao W. FBXO32 Targets c-Myc for Proteasomal Degradation and Inhibits c-Myc Activity. J Biol Chem 2015; 290:16202-14; PMID:25944903; http://dx.doi.org/10.1074/jbc.M115.645978
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 2009; 10:550-63; PMID:19626045; http://dx.doi.org/10.1038/nrm2731
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005; 123:773-86; PMID:16325574; http://dx.doi.org/10.1016/j.cell.2005.11.007
- Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol 2007; 9:765-74; PMID:17558397; http://dx.doi.org/10.1038/ncb1601
- Popov N, Herold S, Llamazares M, Schulein C, Eilers M. Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell cycle 2007; 6:2327-31; PMID:17873522; http://dx.doi.org/10.1016/j.celrep.2014.09.057
- Schulein-Volk C, Wolf E, Zhu J, Xu W, Taranets L, Hellmann A, Janicke LA, Diefenbacher ME, Behrens A, Eilers M, et al. Dual regulation of Fbw7 function and oncogenic transformation by Usp28. Cell Rep 2014; 9:1099-109; PMID:25437563; http://dx.doi.org/10.1016/j.celrep.2014.09.057
- Diefenbacher ME, Chakraborty A, Blake SM, Mitter R, Popov N, Eilers M, Behrens A. Usp28 counteracts Fbw7 in intestinal homeostasis and cancer. Cancer Res 2015; 75:1181-6; PMID:25716680; http://dx.doi.org/10.1158/0008-5472.CAN-14-1726
- Pan J, Deng Q, Jiang C, Wang X, Niu T, Li H, Chen T, Jin J, Pan W, Cai X, et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene 2015; 34:3957-67; PMID:25284584; http://dx.doi.org/10.1038/onc.2014.327
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 2005; 7:303-10; PMID:15723053; http://dx.doi.org/10.1038/ncb1225
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 2005; 7:311-8; PMID:15723054; http://dx.doi.org/10.1038/ncb1224
- Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem 2008; 105:670-7; PMID:18773413; http://dx.doi.org/10.1002/jcb.21895
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 2010; 10:301-9; PMID:20332779; http://dx.doi.org/10.1038/nrc2819
- Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci U S A 2015; 112:3734-9; PMID:25775507; http://dx.doi.org/10.1073/pnas.1411713112
- Li J, Olson LM, Zhang Z, Li L, Bidder M, Nguyen L, Pfeifer J, Rader JS. Differential display identifies overexpression of the USP36 gene, encoding a deubiquitinating enzyme, in ovarian cancer. Int J Med Sci 2008; 5:133-42; PMID:18566677; http://dx.doi.org/10.7150/ijms.5.133
- Endo A, Kitamura N, Komada M. Nucleophosmin/B23 regulates ubiquitin dynamics in nucleoli by recruiting deubiquitylating enzyme USP36. J Biol Chem 2009; 284:27918-23; PMID:19679658; http://dx.doi.org/10.1074/jbc.M109.037218
- Endo A, Matsumoto M, Inada T, Yamamoto A, Nakayama KI, Kitamura N, Komada M. Nucleolar structure and function are regulated by the deubiquitylating enzyme USP36. J Cell Sci 2009; 122:678-86; PMID:19208757; http://dx.doi.org/10.1242/jcs.044461
- Richardson LA, Reed BJ, Charette JM, Freed EF, Fredrickson EK, Locke MN, Baserga SJ, Gardner RG. A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Rep 2012; 2:372-85; PMID:22902402; http://dx.doi.org/10.1016/j.celrep.2012.07.009
- Taillebourg E, Gregoire I, Viargues P, Jacomin AC, Thevenon D, Faure M, Fauvarque MO. The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy 2012; 8:767-79; PMID:22622177; http://dx.doi.org/10.4161/auto.19381
- Thevenon D, Engel E, Avet-Rochex A, Gottar M, Bergeret E, Tricoire H, Benaud C, Baudier J, Taillebourg E, Fauvarque MO. The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe 2009; 6:309-20; PMID:19837371; http://dx.doi.org/10.1016/j.chom.2009.09.007
- Amati B, Sanchez-Arevalo Lobo VJ. MYC degradation: deubiquitinating enzymes enter the dance. Nat Cell Biol 2007; 9:729-31; PMID:17603505; http://dx.doi.org/10.1038/ncb0707-729