Accurate chromosomal DNA replication is essential for normal development and genome stability. Under conditions of replication stress or increased DNA damage, completion of replication crucially depends on the ability of cells to activate DNA damage tolerance (DDT) pathways, which mediate replication fork reactivation and gap-filling. Two main modes of DDT are present in all eukaryotic organisms. An error-prone DDT mode uses translesion synthesis polymerases capable of replicating across DNA lesions, but also occasionally introduces mutations. The other DDT mode, error-free, is mediated by recombination and involves a switch from the damaged template to a homologous one, usually the sister chromatid (template switching). Damage-bypass by template switching genetically depends on PCNA polyubiquitylation, which is induced by replication stress and replication-associated DNA damage, and on homologous recombination activities.Citation1 However, during replication, various regulatory mechanisms ensure global repression of recombination. The most elucidated mechanism for counteracting replication-associated recombination involves SUMOylated PCNA, a PCNA modification that occurs coincidently with replication.Citation2 This modification acts to recruit the Srs2 anti-recombinase to replication forks and to suppress recombination.Citation3,4 Notably, SUMOylation of PCNA is still present, even increased, in response to DNA damage.Citation2 As DDT is essential to prevent conversion of single-stranded DNA to deleterious double strand breaks, but recombination is globally suppressed during replication, the question arising is if replicating cells use preferentially translesion synthesis-mediated damage-bypass during replication. This does not appear to be the case. Contrary to this, various findings indicate that mutagenesis is suppressed early during replication, while damage-bypass by template switching is instead favored.Citation5,6 However, how cells engage in recombination-mediated DDT by template switching at sites of perturbed replication, while recombination is globally suppressed, remained puzzling.
In a recent study in the lab, we uncovered a SUMO-orchestrated regulatory mechanism, mediated by the conserved SUMO-like domains (SLDs)-containing protein Esc2, that enables recombination-mediated DDT specifically at sites of compromised replication.Citation7 Mechanistically, we found that Esc2 preferentially binds structures arising at stalled and damaged replication forks. In this environment, Esc2 engages via its SLDs in regulatory interactions with SIM-containing replisome-associated proteins. Critical for Esc2 pro-recombination function at such sites of perturbed replication is its counteraction of the anti-recombinase Srs2. The locally reduced Srs2 levels subsequently allow local Rad51 filament formation and template switching-mediated damage-bypass. We uncovered that Esc2 controls Srs2 local levels by promoting its turnover and limiting its chromatin recruitment. Crucial in this process is Esc2 interaction with a conserved SUMO–targeted ubiquitin ligase (STUbL) complex, Slx5/Slx8, and their subsequent joint action in mediating proteasome-dependent Srs2 turnover (). In addition, Esc2 limits de novo Srs2 recruitment at sites of damaged forks by favoring local stable association of the PCNA unloader, Elg1. It is likely that this second process is linked to the Srs2 turnover regulation. As Elg1 and Srs2 bind to and potentially compete for SUMOylated PCNA, lower levels of Srs2 will make space for increased Elg1 binding, accelerating unloading of PCNA and therefore limiting Srs2 recruitment. Thus, we propose that Esc2 acts with STUbLs to initiate a feedback circle of Srs2 down-regulation that enables local engagement of recombination at sites of perturbed replication ().
Figure 1. Model for Esc2 role in facilitating recombination at sites of perturbed replication. Esc2 binds stalled replication forks and interacts via its SUMO-like domains (SLDs) with Srs2 and Slx5/8. This guides Slx5/Slx8-mediated proteasome-dependent degradation of the anti-recombinase Srs2, enabling Rad51 filament formation and recombination-mediated DNA damage tolerance.
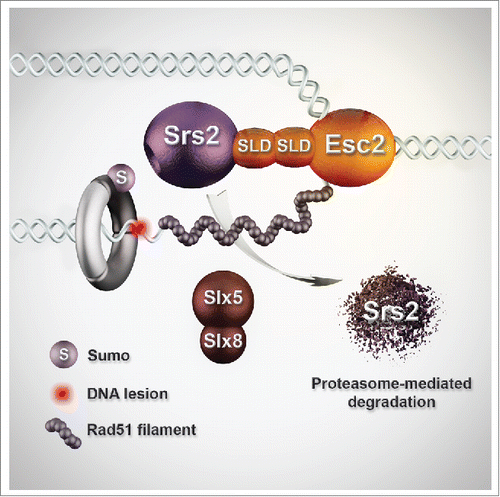
STUbLs are known to mediate proteasome-degradation of SUMOylated substrates. Although Srs2 is SUMOylated and this modification can occur in the absence of Esc2, we identified Esc2 to be a crucial regulator of Srs2 turnover.Citation7 Esc2 role in Srs2 turnover relies on its ability to robustly bind stalled forks and on its SLDs-mediated interaction with the SIMs of Slx5 and Srs2. We thus propose that SLDs of Esc2 act as a platform to recruit Slx5-Slx8 to its substrates at stalled forks, and possibly in other chromosomal contexts in which Esc2 functions are important. In the light of this scenario, STUbL substrates may not need to be SUMOylated in order to be degraded, as Esc2 could recruit Slx5-Slx8 via its SLDs to the substrates. Whether Esc2 is required for the STUbL-mediated turnover of other replisome-associated factors, and if that also serves recombination-mediated DDT by template switching, will remain to be addressed by future studies.
To date, Srs2 regulation was described primarily in the context of SUMOylated PCNA, which functions to recruit Srs2 to chromatin and therefore to inhibit recombination during replication.Citation3,4 Our recent findings identified a mechanism that guides recombination specifically at sites of replication stress, by opposing Srs2 accumulation.Citation7 This newly identified mechanism is also SUMO-orchestrated, involving SIM/SLD interactions between the SIM motifs of Srs2 and STUbL components and the SLDs of Esc2. Our results indicate that the SLD/SIM-coordinated regulatory mechanism acts to locally curb down the anti-recombination effect caused by SUMOylated PCNA, enabling recombination-mediated DDT at sites of replication stress (). Thus, 2 SUMO-mediated pathways act in distinct ways and crosstalk to ensure both global repression of recombination at undamaged chromosomes and local error-free recombination at sites of DNA damage.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Branzei D, et al. Nature 2008; 456:915-20; PMID:19092928; http://dx.doi.org/10.1038/nature07587
- Hoege C, et al. Nature 2002; 419:135-41; PMID:12226657; http://dx.doi.org/10.1038/nature00991
- Papouli E, et al. Mol Cell 2005; 19:123-33; PMID:15989970; http://dx.doi.org/10.1016/j.molcel.2005.06.001
- Pfander B, et al. Nature 2005; 436:428-33; PMID:15931174
- Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat Genet 2009; 41:393-5; PMID:19287383; http://dx.doi.org/10.1038/ng.363
- Gonzalez-Huici V, et al. EMBO J 2014; 33:327-40; PMID:24473148; http://dx.doi.org/10.1002/embj.201387425
- Urulangodi M, et al. Genes Dev 2015; 29:2067-80; PMID:26443850; http://dx.doi.org/10.1101/gad.265629.115
