ABSTRACT
Mouse embryonic stem cells (mESCs) lack of G1 checkpoint despite that irradiation (IR) activates ATM/ATR-mediated DDR signaling pathway. The IR-induced p53 localizes in the nuclei and up-regulates p21/Waf1 transcription but that does not lead to accumulation of p21/Waf1 protein. The negative control of the p21Waf1 expression appears to occur at 2 levels of regulation. First, both p21/Waf1 gene transcription and the p21/Waf1 protein content increase in mESCs treated with histone-deacetylase inhibitors, implying its epigenetic regulation. Second, proteasome inhibitors cause the p21/Waf1 accumulation, indicating that the protein is a subject of proteasome-dependent degradation in ESСs. Then, the dynamics of IR-induced p21Waf1 protein show its accumulation at long-term time points (3 and 5 days) that coincides with an increase in the proportion of G1-phase cells, down-regulation of Oct4 and Nanog pluripotent gene transcription and activation of endoderm-specific genes sox17 and afp. In addition, nutlin-dependent stabilization of p53 in mESC was also accompanied by the accumulation of p21/Waf1 as well as restoration of G1 checkpoint and an onset of differentiation. Thus, the lack of functional p21/Waf1 is indispensable for maintaining self-renewal and pluripotency of mESCs.
Introduction
Embryonic stem cells (ESCs) are characterized by a high rate of self-renewal and are capable of differentiating into all cell types of the adult organism. Stringent control of the genome integrity in ESCs is provided by a variety of mechanisms that (i) limit the frequency of mutations and (ii) provide detection and repair of DNA defects.Citation1-3 Besides, ESCs have very effective antioxidant mechanisms for removing the alien compounds and lowering the level of ROS as compared to differentiated derivative cells.Citation4-5
Paradoxically, one of the well-studied DNA damage responses appears to be lacking in ESCs. Thus, ESCs do not undergo G1/S cell cycle arrest and demonstrate only a temporal G2/M delay following DNA damaging and stress factors.Citation6-8 Since G1 checkpoint is mediated by the canonical p53-p21/Waf1 pathway, the mechanisms of this pathway regulation in ESCs require detailed study.
Studies of p53 in ESCs are numerous, but most of them are focused on human ESCs (hESCs), while the role of p53 in mouse ESCs (mESCs) remains less understood and more controversial. Earlier work by Aladjem et al.Citation9 showed that mESCs lack of G1 checkpoint due to cytoplasmic localization of p53 and its reduced ability to translocate into the nucleus after DNA damage. But later it has been shown that despite its cytoplasmic localization p53 is capable of activating after irradiation of mESCs.Citation10 Furthermore, according to some data, a basal level of p53 in mESCs can be even higher than in somatic cells due to elevated stability of its mRNA and low expression of negative regulator micro-RNAs 125a and 125b.Citation11 Intriguingly, p53 seems to play an important role in regulation of development-associated genes, in particular p53 can induce differentiation of mESCs after DNA damage by a direct suppression of Nanog.Citation12
Low expression of p21/Waf1 may be a reason for the lack of G1 arrest in mESCs.Citation6,13-15 Previously, we found that in IR-exposed embryonal carcinoma cell (ECC) line F9 p21/Waf1 protein is degraded via proteasome-dependent mechanisms, even though DNA damage activates p21/Waf1 gene transcription.Citation16 Similar data have been later obtained for a particular mESCs line CGR8 and low p21/Waf1 expression has been confirmed for hESCs line H9.Citation6,15
Further detailing the issue, here we checked 2 mechanisms, which might be involved in the negative control of p21/Waf1 expression. First, transcription of p21/Waf1 gene and accumulation of p21/Waf1 protein was found to increase after treatment of mESCs with histone deacetylase inhibitor sodium butyrate (NaBut) suggesting the epigenetic mechanism of p21/Waf1 gene regulation at the level of chromatin structure. Second, mESCs treated with proteasome inhibitors revealed accumulation of p21/Waf1, thus the protein might be a target for proteasome-mediated degradation. Unexpectedly, the dynamics of p21/Waf1 expression at 3 and 5 d after irradiation showed p21/Waf1 accumulation starting from day 3. An increase of the p21/Waf1 content was accompanied by increased proportion of G1-phase cells, downregulation of oct3/4 and nanog pluripotent gene transcription and transcriptional activation of endoderm-specific genes sox17 and afp. From the other hand, stabilization of p53 caused by a small molecule nutlin, which inhibits MDM2-mediated degradation p53, also restored G1 checkpoint control and induced differentiation. Altogether, these data demonstrate that the agents used to promote the accumulation of p21/Waf1 do suppress proliferation, restore G1 checkpoint and direct mESCs to differentiate. Thus, the lack of G1-checkpoint in mESCs caused by lack of p21/Waf1 expression is indispensable for maintenance of self-renewal and pluripotency of mESCs.
Results
p53-p21Waf1 pathway is compromised in mESCs
DNA damage-induced G1/S cell cycle arrest is mediated by activation of the p53-p21/Waf1 signaling pathway. Using western blot analysis with antibodies against p53-Ser15, we found that irradiation (6 Gy) induced the phosphorylation of p53 with a maximum at 2 h post-treatment (). According to immunoflurescence data, the phosphorylated p53 is localized in the nuclei of mESCs (). Immunofluorescence study for p53-Ser15 distribution within the cover glass fields revealed that the intensity of staining is noticeably variable in the irradiated cells. To confirm the variability of nuclear staining, we performed a cytometric assessment of p53-Ser15 distribution in irradiated mESCs population by using a bivariate distribution (p53-Ser15 vs DNA content) according to the previously described protocolCitation17 (Fig. S1). Although all nuclei demonstrate p53-Ser15 staining, however, the amount of bound antibody varies within the stretched clouds of both G1 and G2 phase cells (IR, 2 h).
Figure 1. Gamma-irradiation induces p53 Ser15 phosphorylation and nuclear accumulation. (A) Immunofluorescent staining of mESCs with antibody to phospho-p53 (Ser15) 2 h post-irradiation (6 Gy) (green). Nuclei were stained with To-Pro3 (blue). Scale bar 10 μM. (B) Western blotting analysis of protein extracts from non-irradiated and irradiated mESCs. Blots were stained 2, 4 and 8 h post-irradiation (6 Gy) using antibody to phospho-p53 (Ser15), acetyl-p53 (Lys 379) and total p53. (C) A luciferase assay of control and irradiated mESCs after transient transfection with luciferase reporter plasmid PG13-Luc. At time points 4 and 24 h post-irradiation cells were subjected to luciferase activity assays. Pifithrin-α, an inhibitor p53-transcriptional activity (10 μM), was added to the 4 h reaction as a negative control. Error bars correspond to SEM calculated for 3 replicates. (D) Western blotting analysis of control, transfected with luciferase reporter plasmid PG13-Luc and irradiated with 3 Gy (4 h post-irradiation) mESCs. Irradiated cells were taken as control. Blots were stained using antibody to phospho-histone H2AX (Ser139) (γH2AX), phospho-p53 (Ser15) and total p53.
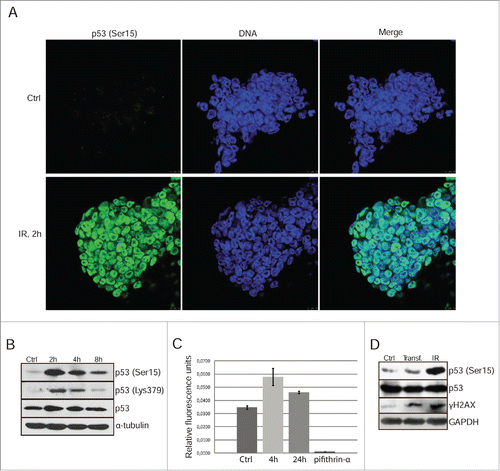
P53-Lys379 acetylation was known to augment the p53 DNA-binding as well as its trans-activating potential.Citation18 Western blot analysis shows that mESCs exposed to irradiation accumulate Lys379-acetylated p53 (). To see whether p53 transactivating potential is becoming increased after irradiation of mESCs, a p53-luciferase reporter plasmid pG13-Luc containing 13 copies of p53 consensus binding site has been used. Transfected mESCs were subjected to X-rays (6Gy) and collected 4 h and 24 h post-irradiation for luciferase assay (). In consistence with immunofluorescence data, irradiated mESCs show enhanced p53-driven activity of pG13-Luc reporter (). Interestingly, non-irradiated cells possess a basal p53 activity that can be suppressed by addition of pifithrin-α, a small molecule inhibitor of p53 transcriptional activity. To verify whether the transient transfection itself could activate DNA damage response and thereby stimulate p53 phosphorylation, we monitored the p53-Ser15 and γH2AX in the transfected mESCs (). According to these data the transfection of pG13-Luc plasmid increased amount of γH2AX, albeit to a lesser extent than irradiation. At the same time, the transfection did not cause p53-Ser15 phosphorylation. Thus, the observed basal level of pG13-Luc activity in non-irradiated mESCs seems not to be associated with transfection procedure.
As for transcription of p53 target genes such as p21/Waf1, RT-PCR analysis revealed a robust accumulation of p21/Waf1 RNA transcripts by 6 h post-irradiation at dosage 6 Gy (, left panel). However, in contrast to NIH3T3 cells, p21/Waf1 protein was not detected at a noticeable level in irradiated mESCs (, right panel). To check possible post-translational degradation of p21/Waf1 protein, 2 proteasome inhibitors with different mechanisms of action have been used - lactacystin (Lc) and MG132. Both inhibitors caused accumulation of p21/Waf1 protein in NIH3T3 cells (data not shown) as well as in mESCs (, left panel). Essentially, this increased level of p21/Waf1 in mES seems to be sufficient for a noticeable accumulation of G1 phase cells with concomitant reducing the proportion of S-phase engaged cells (, right panel). Thus, directed proteasome-dependent degradation of p21/Waf1 may contribute to its down-regulation and eventually to a compromised function of G1 checkpoint in mESCs.
Figure 2. Negative control of p21Waf1 expression occurs at 2 levels of regulation: epigenetic modulation of gene transcription and proteasome-mediated protein degradation. (A) RT-PCR analysis of p21/Waf1 gene transcripts 6 h post-irradiation of mESCs. Gapdh was used as an internal control (left panel). Western blotting analysis of protein extracts from control and irradiated mESCs (8 and 20 h post-irradiation) using antibodies against p21/Waf1 protein. NIH3T3 cells were taken as control (right panel). (B) mESCs were treated with lactacystin (10 μM) and MG132 (10 μM) for 4 h, then extracted proteins were subjected to immunoblotting with antibody to p21/Waf1 protein (left panel). FACS analysis of cell cycle distribution of mESCs treated with lactacystin (Lc) (10 μM) for 20 h (right panel). (C) FACS analysis of cell cycle distribution of untreated mESCs and treated with NaBut (4 mM for 24 h) (left 2 panels). RT-PCR assay for p21/Waf1 transcription in mESCs treated with NaBut (4 mM); gapdh was used as an internal control (middle panel). Western blotting analysis of lysates from untreated (Ctrl) and NaBut-treated mESCs (4 mM for 24 h) using antibodies against p21/Waf1 (right panel). (D) Western blotting analysis of protein extracts from control (Ctrl), treated with NaBut (4 mM, 24 h), and irradiated (2 h, 6 Gy) cells (left panel). Blots were stained using antibody to phospho-p53 (Ser15) and total p53. In the middle panel, RT-PCR analysis of mRNA transcripts of oct3/4, nanog, sox2, afp, and gata6 genes in mESCs treated with 4 mM NaBut for 24 h; gapdh was used as an internal control (middle panel). In the right panel, mESCs treated with 4 mM NaBut for 24 h were analyzed by western blot for Oct3/4, Nanog and Sox2 proteins (right panel).
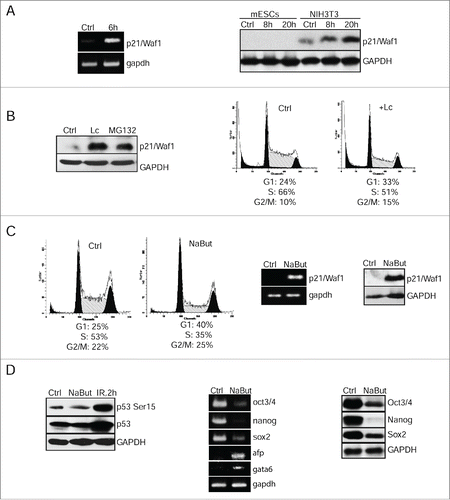
To study an issue of whether p21/Waf1 gene transcription is under negative epigenetic control in mESCs, we used a histone-deacetylase inhibitor, sodium butyrate (NaBut). There are available data that p21/Waf1 gene promoter contains a HDAC-response element operated through Sp1/Sp3 binding site; the Sp1/Sp3 binding site recruits HDAC 1 and 2 activity to repress promoter transcription. Correspondingly, inhibition of histone deacetylase activity at the promoter by HDAC inhibitors leads to histone hyperacetylation and activation of p21Waf1 gene transcription.Citation19 First, NaBut treatment for 20 h caused a significant accumulation of mESCs in G1 phase accompanied by a decline of S-phase cells and a slight increase of cells in G2/M phase (, left panels). Second, NaBut induced both p21/Waf1 gene transcription and protein accumulation that correlated well with the establishment of G1 cell cycle arrest (, middle and right panels). Thus NaBut-accumulated p21/Waf1 does not appear to undergo a proteasome-dependent degradation suggesting that this mechanism might be compromised in HDACI-treated mESCs. We next checked whether p53 activation is necessary for NaBut-induced p21/Waf1 expression in mESCs. As shown in (left panel), p53-Ser15 did not accumulate in NaBut-treated mESCs. Moreover, a p53 inhibitor pifithrin-α alone has little effect on cell cycle parameters of undifferentiated mESCs and is unable to abrogate accumulation of mESCs in G1 phase in NaBut-treated mESCs (Fig. S2). Thus, these results indicate that p53 is not directly involved, if at all, in NaBut-induced G1 checkpoint.
RT-PCR and western blot analysis showed that the NaBut-induced G1 arrest is accompanied by downregulation of oct3/4, nanog and sox2 gene expression both on the level of transcription and protein synthesis as well (, middle and right panels). In turn, such markers of differentiation as gata6 and afp were shown to up-regulate in NaBut-treated mESCs (, middle panels).
Collectively, the negative control of the p21/Waf1 expression occurs on 2 levels of regulation: the epigenetic gene regulation at the level of chromatin structure and a proteasome-mediated degradation.
The long-term consequences of IR-induced p53 activation in mESCs
Accumulating data support an idea that p53 is capable of functioning in mESCs in response to DNA damage. Nevertheless, the p53 activation does not necessarily lead to implementation of the G1 cell cycle checkpoint and seems to play an important role in other cellular processes triggered by genotoxic stresses, for example, apoptotic cell death. The p53 function as a trigger of apoptosis in mESCs was previously documented.Citation20,21 Recently, it has been shown that p53 plays a significant role during retinoic acid-mediated differentiation of hESCs.Citation22 In particular, p53 can drive differentiation of mESCs by direct repression of nanog transcription after DNA damage.Citation12 Thus, p53 protein may have specific functions in opposing self-renewal versus cell death in mESCs. In order to assess outcomes of the p53 activation during DNA damage response of mESCs, we studied long-term consequences of IR-induced p53 activation.
We show that after irradiation with 6 Gy mESCs do not undergo G1 arrest (Fig. S3 A).We carried out caspase-3 activity assay to demonstrate the elevated caspase-3 activity (Fig. S3, B) that correlates with death of a half of cells after 1 day post-irradiation at dose 6 Gy according to MTT assay (Fig. S3, C). To investigate long-term effects of irradiation, a dosage 3 Gy was used as it had less genotoxic stress effect.
We checked p53 activation and subsequent p21/Waf1 protein accumulation at the long time points 1 and 3 d after irradiation with 3Gy. As shown in (left panel), p53-Ser15 phosphorylation is still high on day 1 post-irradiation and then fluctuates on days 3 and 5 that may be accounted for by p53 pulses mediated through p53-Mdm2 and ATM-p53-Wip1 negative feedback loops.Citation23 However, according to qRT-PCR analysis, p21/Waf1 gene transcription increases ∼2 fold already on the first day after irradiation and additionally increased on the 3d and 5th days (, middle panel). Importantly, although the p21/Waf1 mRNA level is clearly increased in a period from 1 to 5 days, the p21/Waf1 protein is barely detected on day 1 and can be seen only at day 3 (, right panel). There is a correlation between an increase of the p21/Waf1 protein content and accumulation of mESCs at G1 phase on the 3rd day post-irradiation as demonstrated by flow cytometry (, compare day 1 and day 3). Thus, over the time after irradiation, the functional p53-p21/Waf1 checkpoint is recovered in the survived mESCs thereby slowing-down their proliferation.
Figure 3. Restoration of G1 checkpoint due to accumulation of p21/Waf1 in irradiated mESCs is accompanied by loss of pluripotency. (A) Western blotting analysis of protein extracts from unirradiated and irradiated mESCs. Blots were stained at 1 and 3 d 3 Gy post-irradiation using antibody to phospho-p53 (Ser15) and total p53 (left panel). The mRNA levels of p21/Waf1 gene was determined by qRT-PCR and standardized by the mRNA levels of gapdh. Data are presented as mean ±SEM, n = 3 (middle panel). Cell lysates from mESCs in the indicated time points were analyzed by Western blotting using antibodies against p21/Waf1 protein (right panel). (B) FACS analysis of cell cycle distribution of unirradiated mESCs and irradiated with 3 Gy. Cells were harvested at 1 and 3 d after irradiation. (C) The mRNA levels of oct3/4, nanog and sox2 genes were determined by qRT-PCR and standardized by the mRNA levels of gapdh. Data are presented as mean ±SEM, n = 3. (D) Western blotting analysis of protein extracts from non-irradiated and irradiated mESCs. Blots were stained 1, 3, 5 d post-irradiation (3 Gy) using antibody to Oct3/4, Nanog and Sox2. A relative densitometry analysis of the Oct3/4, Nanog, Sox2 protein expression (normalized to α-tubulin) was performed using Gel-Pro Analyzer software.
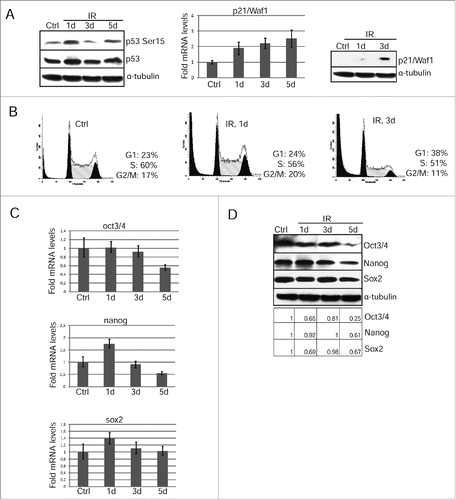
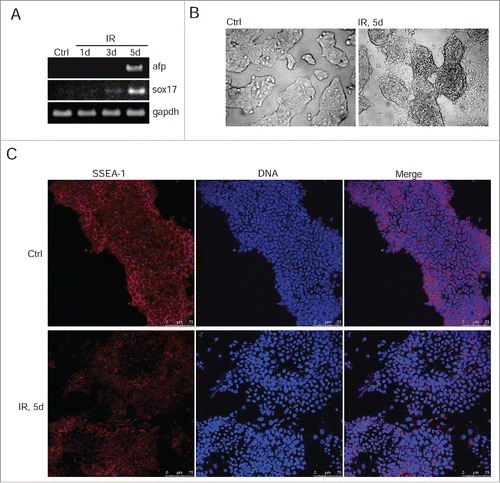
To test whether the irradiation-induced p53 activation does affect the steady-state levels of oct3/4, nanog and sox2 gene expression, we used qRT-PCR and Western blotting in cells on 1, 3, and 5 d after irradiation. For better demonstration, we quantified the band intensities of western blots obtained for oct3/4, nanog and Sox2. The transcription of oct3/4 and nanog genes decreases by ∼2 fold that correlates with lowering the protein levels by day 5 post-irradiation (, left panel). The level of sox2 protein fluctuates in the days following irradiation, however, these changes are not so great that one could state about the actual decrease in its level (). Moreover, the amount of mRNA transcripts for sox2 gene does not change reliably that confirms a conclusion made from an analysis of the Sox2 protein content. (, D). Further analysis of mESCs survived after irradiation showed that down-regulation of oct3/4 and nanog () correlates with induction of expression of endoderm markers sox17 and afp implying an onset of differentiation (). Consistently, the morphology of mESCs colonies changes at day 5 after irradiation. If control undifferentiated IOUD2 colonies are the rounded and are organized as dense masses of cells with clear boundaries, the colonies of irradiated mESCs are characterized by the appearance of separated and flattened cells (). To assess progression of irradiation-induced differentiation in mESCs, we used antibodies to surface marker SSEA-1, which are specific for undifferentiated mESCs. According to immunoflurescence data, the appearance of cells with a flattened cellular morphology is accompanied by a loss of SSEA-1 staining ().
Figure 4. IR-induced mESCs activate expression of differentiation markers. (A) RT-PCR analysis of afp and sox17 genes transcription in non-irradiated mESCs and 1, 3, 5 d post-irradiation (3 Gy). gapdh was used as an internal control. (B) Representative images of colonies of non-irradiated and irradiated (3 Gy) mESCs. Cells were examined at 5 d after irradiation using light microscope. (C) Immunofluorescent staining of mESCs with antibody to a surface marker SSEA-1 5 d post-irradiation (3 Gy) (red). Nuclei were stained with DAPI (blue). Scale bar 75 μM.
Nutlin, an antagonist of MDM2-mediated p53 degradation, restores p53-p21/Waf1-dependent G1 checkpoint in mESCs
To confirm that differentiation induced in survived post-irradiated mESCs depend on the p53 activation, we used MDM2-targeting pharmacological agent nutlin-3. MDM2 ubiquitin ligase is a crucial negative regulator of p53 stability thereby suppressing p53 activity. Nutlin occupies the p53-binding pocket of MDM2 in a way that remarkably mimics the molecular interactions of the crucial amino acid residues from p53.Citation24 Of other nutlins, the nutlin-3 has been shown to inhibit the p53–MDM2 interaction with a higher degree of specificity leading to p53 stabilization and eventually to activation of the p53 pathway. According to our data, nutlin treatment causes p53-Ser15 phosphorylation and accumulation of transcriptionally active p53 protein that induces transcription of its target-gene p21/Waf1 (, left and middle panels). Nutlin increases the p21/Waf1 mRNA level already on the first day of treatment (∼4 fold) that is higher than after irradiation (∼2 fold). In turn, p21/Waf1 protein accumulates by 1 day of nutlin treatment, while in irradiated mESCs p21/Waf1 expression is detected only at day 3 after IR (, right panel). A progressive increase in the number of cells residing in G1 phase and a reduction in a proportion of cells transiting in S phase after nutlin treatment were evidenced by flow cytometry and resulted in G1/S arrest on the 3d day ().
Figure 5. Nutlin-dependent p53 activation is accompanied by loss of mESCs pluripotency. (A) Lysates prepared from mESCs treated with nutlin (10 μM) for 1 and 3 d were analyzed by Western blotting with antibodies specific to p53-Ser15 and total p53 (left panel). The mRNA level of p21/Waf1 gene wase determined by qRT-PCR and standardized by the mRNA levels of gapdh. Data are presented as mean ±SEM, n=3 (middle panel). Western blot analysis of protein extracts from untreated and treated with nutlin mESCs using antibodies against p21/Waf1 protein at the same time point (right panel). (B) Cell cycle parameters of mESCs untreated and treated with nutlin (10 μM) for 1 and 3 d. (C) RNA transcripts from mESCs treated with nutlin (10 μM) for 1, 3 and 5 d were subjected to qRT-PCR assay using primers specific for mouse oct3/4, nanog and sox2. Results of qRT-PCR are displayed as the mean ± SEM, n=3. (D) Lysates prepared from mESCs treated with nutlin (10 μM) for 1, 3 and 5 d were analyzed by Western blotting with antibodies specific for Oct3/4, Nanog and Sox2. A relative densitometry analysis of the Oct3/4, Nanog, Sox2 protein expression (normalized to α-tubulin) was performed using Gel-Pro Analyzer software.
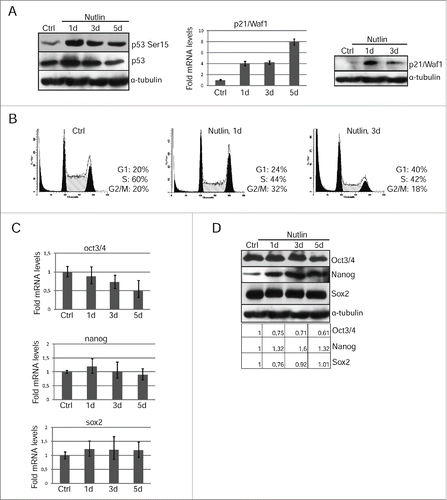
Activation of p53 in nutlin-treated mESCs leads to a two-fold downregulation of oct3/4 mRNA, while there was no a reliable decrease in the amount of nanog and sox2 mRNA transcripts (). At the protein level, there is a decrease only oct3/4 expression on the day 5 of nutlin treatment that in contrast to nanog and Sox2 levels, which have not changed (). A plausible explanation of this phenomenon is that nutlin reduces degradation not only p53, but also other transcription factors, such as nanog and Sox2. For comparison, IR-induced activation of p53 caused down-regulation of pluripotency markers oct3/4 and nanog in mESCs ().
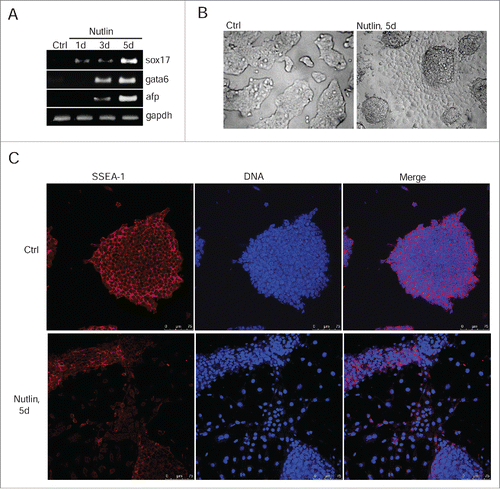
Nevertheless, the reduced expression of oct3/4 in nutlin-treated cells was sufficient to activate transcription of endoderm markers afp and sox17 as well as a mesoderm marker gata6 (). In contrast to irradiated mESCs, the expression of differentiation-associated genes takes place within 1 day of nutlin treatment evidencing for a more rapid induction of differentiation. Nutlin treatment causes a change in morphology of colonies of mESCs in a manner similar to irradiated cells (). Also, nutlin-mediated differentiation of mESCs includes downregulation of ESCs-specific surface marker SSEA-1 ().
Figure 6. Nutlin-dependent activation of p53 induces differentiation of mESCs. (A) RT-PCR analysis of sox17, gata6 and afp RNA transcripts in mESCs untreated and treated with nutlin (10 μM) for 1, 3 and 5 days, gapdh was used as an internal control. (B) Representative images of mESCs colonies untreated and treated with nutlin (10 μM) for 5 d. Cells were examined using light microscope. (C) Immunofluorescent staining of mESCs with antibody to surface-specific marker SSEA-1 (red) after 5 d of nutlin treatment (10 μM). Nuclei were stained with DAPI (blue). Scale bar 75 μM.
Discussion
Execution of G1 checkpoint depends to a larger extent on the functionality of the p53-p21/Waf1 pathway. DNA damage induces p53 phosphorylation on Ser15 as well as acetylation on Lys379, thereby enhancing its trans-activating potential (). Since p53 activated in response to DNA damage, of interest was to check whether p21/Waf1 had been also up-regulated in genotoxically stressed mESCs (). Our data show that despite of an increase of p21/Waf1 mRNA transcripts, no or little p21/Waf1 protein can be detected in irradiated mESCs (, right panel). To clear this point in more details, 2 proteasome inhibitors with different mechanisms and specificity were used. As lactacystin (Lc) and MG132 caused accumulation of p21/Waf1 protein in undamaged mESCs (), one may suggest that expression p21/Waf1 in mESCs is under stringent control at the level of protein degradation. The negative regulation of p21/Waf1 expression appears to be one of the mechanisms involved in maintenance of pluripotency and self-renewal of mESCs. We complemented this finding by showing that p53-independent up-regulation of p21/Waf1 induced by HDAC inhibitor NaBut restored G1 checkpoint and slowed down proliferation of mESCs (). The importance of p21/Waf1 in implementation of G1 checkpoint during RA-mediated differentiation of mESCs was shown by siRNA depletion of p21/Waf1 mRNA.Citation22 Significantly, NaBut induced both p21/Waf1 gene transcription and p21/Waf1 protein accumulation (). It is known that p21/Waf1 gene transcription is under direct epigenetic regulation due to recruitment of HDAC 1 and HDAC 2 activity at the Sp1/Sp3 site of the promoter. The Sp1/Sp3 binding site recruits HDAC 1 and 2 activity to repress promoter transcription. Correspondingly, inhibition of histone deacetylase activity at the promoter by HDAC inhibitors leads to histone hyperacetylation and activation of p21Waf1 gene transcription.Citation19 But there is no data showing the mechanisms of p21/Waf1 stabilization at the protein level in HDACI-treated cells. Available literature data evidence that micro-RNAs can also implement negative control of p21/Waf1 mRNA in mouse and human ESCs by a group of specific micro-RNA, suggesting the expression of p21/Waf1 protein in ESCs might be post-transcriptionally regulated during DDR.Citation25,26 One may suggest that NaBut as a potent modulator of gene activity due to stimulation of acetylation of various chromatin-associated proteins might affect the expression of micro-RNA responsible for negative regulation of p21/Waf1 mRNA thereby allowing its translation and protein accumulation. Thus, p53 activation does not lead to an accumulation of p21/Waf1 protein as its expression is under negative control at the levels of epigenetic gene transcription and protein degradation.
ESCs are highly sensitive to genotoxic insults and rapidly undergo apoptosis to a greater extent than somatic cells.Citation27–29 We detected a significant level of caspase-3 activity in mESCs at 1 and 3 d after irradiation suggesting apoptosis as one of the p53-dependent mechanisms maintaining genome stability of ESCs (Fig. S3 B-E).Citation20,21 However, the role of p53 is not limited by induction of apoptosis. Recent studies have identified involvement of p53 in suppression of pluripotency genes in RA-mediated differentiation of ESCs.Citation22,30 In addition, p53 signaling provides a barrier to reprogramming somatic cells into induced pluripotent stem cells (IPSCs) evidently due to triggering G1 checkpoint and cellular senescence programs.Citation31 However, p53-dependent mechanisms involved in commitment to differentiation and reprogramming are not clearly understood. To unravel p53 functions in modulation of pluripotency and self-renewal, we compared effects produced by agents that activate p53 by different mechanisms (irradiation and nutlin treatment) on pluripotency and renewal of mESCs. According to these results, a primary response of mESCs to irradiation and nutlin is the caspase-3 activation and p53-dependent apoptosis (Fig. S3D, E). It appears that the p53-mediated apoptotic program is dominant over differentiation in mESCs. At least a half of the mESCs population that survived under these stress conditions slows down proliferation due to accumulation of p21/Waf1 protein and restoring the G1 checkpoint (). We estimated heterogeneity of cell population by examining p53 expression in individual cells by using a bivariate distribution flow cytometry (p53-Ser15 vs DNA content). It has been shown that the binding of antibody to p53-Ser15 varies likely due to different response of individual cells accounted for by cellular context (Fig. S1).
The observed changes in cell cycle parameters of IR-survived mESCs are accompanied by a decrease of pluripotency and induction of early differentiation. In irradiated mESCs the p53 activation leads to down-regulation of oct3/4 and nanog and upregulation of differentiation markers sox17 and afp (). Interestingly, nutlin-dependent activation of p53 suppresses only oct3/4 expression transcription but it is sufficient to accelerate differentiation: transcription of sox17 and afp genes has been already observed on day 1 of nutlin treatment ( and ). The observed differences in the expression of p53 targets and pluripotent genes in mESCs cells treated with IR and nutlin appear to be determined by peculiarities of post-translation degradation of the proteins. Available data evidence that nutlin can activate p21Waf1 expression via p53-dependent and p53-independent pathways. For example, Maimets et al showed that nutlin did not cause p53-Ser15 phosphorylation in human ESCs still stabilizing p53 and up-regulating p21Waf1.Citation32 Probably, nutlin can reduce p21Waf1 degradation.
Nevertheless, p53 can function as trans-activator of its target-genes, because high level of p21/Waf1 transcription was observed both in irradiated and nutlin-treated mESCs (, left panel). Although a previous study reported that p53 directly suppresses nanog transcription in mESCs upon DNA damage, a recent system analyses reveal that binding of p53 at promoter region significantly correlates with gene activation but not with repression.Citation31 In consistence with this, p53-mediated repression can occur through interfering with distal enhancer activity within the oct3/4 and nanog genes.Citation33 Thus, p53 appears to use many different ways to repress developmental gene transcription.
Together, literature data and our results suggest that despite the compromised p53-p21/Waf1-dependent G1 checkpoint in mESCs, p53 protein is active and induces transcription of its downstream gene p21/Waf1. However, expression of p21/Waf1 protein is negatively regulated by a mechanism of post-translational proteasomal degradation. In this regard, it is interesting that p21/Waf1 mRNA is effectively translated into the respective p21/Waf1 protein in HDACI-treated mESCs indicating the epigenetic regulation of the gene. Direct p53-independent activation of p21/Waf expression by NaBut treatment is associated with elongation of G1 phase of mESCs cell cycle. Similarly, the accumulation of p21/Waf1 protein is observed in differentiating ESCs and precedes the recovery of G1 checkpoint. Regardless of the agent used to increase the content of p21/Waf1, there follows suppression of proliferation, restoration of G1 checkpoint control and the start of mESC differentiation. Our data evidence that the lack of p21Waf1 expression is the vital event and it contributes to the maintenance of mESCs pluripotent state. Current studies with the iPS cells produced from somatic cells show that a reduced expression of p53 facilitates somatic cell reprogramming to the iPSC as evidenced by using p53-null cells or siRNA to p53.Citation34 Downregulation of p21Waf1, which is an inhibitor of the cyclin-Cdk activity, also increases the efficiency of reprogramming. Thus, to facilitate somatic cell reprogramming, the function of p53-p21Waf1 pathway have to be down-regulated by anywise, indicating that this pathway is antagonistic to pluripotent state and self-renewal.Citation35 Up-regulation of p21Waf1 either by a p53-dependent pathway or p53-independently contributes to a transition from pluripotent state to differentiation.
Materials and methods
Cell culture
Mouse embryonic stem cells IOUD2 were maintained on tissue culture dishes (Corning) coated with 0.2% porcine gelatin (Sigma) in a Dulbecco's modified Eagle's medium DMEM/F12 (1:1) (Gibco) supplemented with 0.1 mM 2-mercaptoethanol (Sigma), 10% fetal bovine serum (HyClone), and 1000 units/ml of recombinant murine LIF at 370C in atmosphere of 5% CO2. Recombinant mLIF was expressed using an expression vector pGEX-2Т-hLIF in E. coli and was further purified on a Glutathione Sepharose column. NIH3T3 cells were maintained in DMEM medium (Gibco) with 10% fetal calf serum (HyClone) and 0.1 mg/mL gentamycin (Biolot) at 37°C and 5% CO2.
Lactacystin (Proteasome Inhibitor IV), MG132 (Carbobenzoxy-L-leucyl-leucinal, Proteasome Inhibitor XI), nutlin-3 (MDM2 Inhibitor IV) and pifithrin-α purchased from Calbiochem were added to the medium in final concentration of 10 μM. Medium was changed daily. Sodium butyrate (NaBut) was used at concentration 4 mM (Sigma).
Cells were irradiated with 3 Gy and 6 Gy using X-ray generator RAP-150/300 (dose rate about 0.5 Gy/min; 300 kV; 15 mA). Immediately after irradiation cells were returned to the incubator for recovery until the appropriate time point.
FACS analysis of cell cycle distribution
For cytometric analysis of DNA content, cells were harvested, washed with PBS, and incubated for 30 min at room temperature in PBS containing 0.01% of saponin (Sigma). Cells were washed twice with PBS and incubated with 100 μg/ml RNase A and propidium iodide for 15 min at 370C. Samples were analyzed by using a flow cytometer Coulter Epics XL (Beckman Coulter) FACscan. Cell cycle phase distribution analysis was performed with MODFIT LT 3.0 software (Verity Software House). Cytometric assessment of p53 phosphorylation was carried out according to the previously described protocol.Citation17 Briefly, cell suspension were washed with PBS and fixed with ice-cold 1% methanol-free formaldehyde solution in PBS. Then cells were pelleted, washed twice with PBS and were stored in ice-cold 70% ethanol at −20°C for at least 12 hours. Cells were incubated overnight at 4ºC with anti-phospho-p53 (Ser15) monoclonal antibody (#9284, Cell Signaling), washed and incubated with secondary antibodies AlexaFlour-488-conjugated goat anti-rabbit F(ab′)2 fragment (Invitrogen) for 1 h at RT, then treated with RNAse (1 mg/ml, 30 min, RT) and 5 μl of 1 mg/ml of PI was added.
MTT-test for cell viability
Evaluation of cell viability was performed by using colorimetric MTT-assay (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma). The amount of formazan product correlates with the quantity of live cells. Cells were seeded in 24-well plates and were irradiated by 6 Gy. After 24 h post-irradiation MTT (0.5 mg/ml dissolved in PBS) was added, and cells were incubated for 1.5 h at 370C in 5% CO2. The resulting formazan precipitate was dissolved in DMSO (Sigma), gently pipetted and dispensed into 96-well plate. The absorbance was measured at 570 nm wavelength using Multiskan EX (Thermo Electron). Each experiment was repeated 6 times followed by calculation of the standard error of the mean.
Immunofluorescence
Cells were seeded and grown on gelatine-coated coverslips, rinsed with cold PBS (phosphate-buffered saline), fixed in 4% paraformaldehyde for 15 min at RT, and permeabilized with 0.25% Triton X-100 for 20 min. After three washes cells were incubated in blocking solution (5% BSA from Sigma in PBS) and then incubated with primary antibodies against p53 (Ser15) (Cell Signaling #9284) or SSEA-1 (Invitrogen #41–1200) at 40C overnight. After washing samples were incubated for 1 h with secondary antibodies AlexaFlour-488/568-conjugated goat anti-rabbit/mouse F(ab′)2 fragment (Invitrogen). Primary and secondary antibodies were dissolved in PBS with 5% BSA and 0.1% Tween-20 (Sigma). Nuclei were stained by 5 min incubation with To-Pro3 (Invitrogen) or DAPI. Images were analyzed with the confocal microscope (Leica).
Protein lysates preparation and Western blotting
For immunoblotting, cell lysates were obtained by incubating cells in RIPA buffer containing PBS solution, 1% Igepal, 0.5% sodium deoxycholate, 0.1% SDS (Sigma), protease and phosphatase inhibitors (cocktail Complete, Roche), 5 mM EGTA, 10 mM β-glycerophosphate). Equal amounts of protein extracts were run on polyacrylamide gel electrophoresis, transferred to PVDF-FL membranes (Millipore) and blotted with antibodies against H2AX (Ser139) (#9718), p53-Ser15 (#9284), p53-Lys379 (#2524), p53 total (#2524), Sox2 (#4900), GAPDH (#2118) from Cell Signaling; oct3/4 (#sc-5279) and nanog (#sc-376915) from Santa Cruz; p21/Waf1 (Invitrogen #AHZ0422), α-tubulin (Sigma #T5168) according to manufacturer's recommendations. HRP-conjugated goat anti-rabbit and rabbit anti-mouse antibodies (Pierce) were used as secondary antibodies. Proteins on membranes were visualized by means of ECL (Amersham). The relative band intensity was quantified using the Gel-pro Analyzer® software.
qRT-PCR and RT-PCR
For qRT-PCR and RT-PCR total cellular RNA was isolated using TRIzol® (Invitrogen) according to a manufacturer's protocol. Reverse transcription was performed with 2 μg RNA, using random hexaprimers (Promega) and M-MuLV Revertase (RevertAid, Fermentas). qPCR was performed using the Real-Time PCR Reagent kit with SYBR Green dye and the reference dye ROX (Syntol), on the 7500 Real-Time PCR System (Applied Biosystems). The reaction parameters were according to the manufacturer's instructions (5 min 95°C, then 60°C 50 s and 95°C 15 s repeated in 45 cycles). The following gene-specific primers were used: oct-3/4-F, CAAGTTGGCGTGGAGACT; oct-3/4-R, TTCATGTCCTGGGACTCCTC; nanog-F, GATGCAAGAACTCTCCTCCA; nanog-R, CAATGGATGCTGGGATACTC; sox2-F, -ACATGAACGGCTGGAGCAACG, sox2-R, CATGTAGGTCTGCGAGCTGGTC; p21/Waf1-F, CCATGAGCGCATCGCAATC; p21/Waf1-R CCTGGTGATGTCCGACCTG; gapdh-F, TGTGTCCGTCG-TGGATCTGA; gapdh-R, TTGCTGTTGAAGTCGCAGGAG. The data were normalized to gapdh mRNA levels. SEM was calculated for 3 technical replicates within the experiment. PCR was performed using Taq polymerase (Fermentas) according to manufacturer's instructions. The following gene-specific primers were used: p21/Waf1-F, AAAGCCTCCTCATCCCG-TGTT C; p21/Waf1-R, GTCACCCTGCCCAACC-TTAGAG; afp-F, TGGTTACACGAGGAAAGCCC; afp-R, AATGTCGG-CCATTCCCTCAC; gata6-F, TCACCATCACCCGACCTACT; gata6-R, GATGAAGGCACGCGCT-TCTG; sox17-F, GCTTT-AAATGGGAGGGAGGGT; sox17-R, CTGGAGGTGCTGCTCATTGTA. Primers for oct3/4, nanog, sox2, gapdh gene were the same as for qPCR.
Dual luciferase reporter assay
Cells grown in 24-well plates (Costar) were transfected with 0.8 μg of p53-responsive reporter plasmid PG13-luc using Lipofectamine® 2000 (Invitrogen) as recommended by the manufacturer. Expression of Renilla was used as an internal control value to which expression of the experimental firefly luciferase reporter gene was normalized. Twenty-four h after transfection mESCs were irradiated with 6 Gy, and then 4 and 24 h post-irradiation were subjected to luciferase activity assays using the Dual-Luciferase Reporter Assay System (Promega). Analysis of luciferase activity was done on luminometer TD-20/20 (Turner Designs). Each experiment was repeated at least 3 times. Diagrams represent means ± SEM.
Caspase-3 activity assay
Fluorescent caspase substrate (Ac-DEVD-AMC, Biomol) was used to measure caspase-3 activity in vitro. Cells were washed with PBS and lysed in ice-cold buffer containing 50 mM HEPES pH 7.4, 5 mM CHAPS, 5 mM DTT, 0.5% NP-40 for 30 min. After centrifugation protein concentration was measured and 100 μg of proteins from each sample were used for the assay. Samples were incubated for 1 h at 37°C in the following buffer: 20 mM Hepes pH 7.4, 0.1% CHAPS, 5 mM DTT, 2 mM EDTA, 40 mM Ac-DEVD-AMC (Sigma). The emission fluorescence at 460 nm produced by cleaved Ac-DEVD-AMC was measured after excitation at 360 nm using fluorimeter GloMax®-Multi Jr.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary_figures_1-3.pptx
Download MS Power Point (137.5 KB)Funding
This work was supported by Russian Science Foundation grant #14–50–00068 (VAP and TVP), a grant from the St. Petersburg State University, contract #1.38.247.2014 (IIS) and the Program "Molecular and Cell Biology" (Russian Academy of Sciences) (BBG).
References
- Stambrook PJ, Tichy ED. Preservation of genomic integrity in mouse embryonic stem cells. Adv Exp Med Biol 2010; 695:v59-75; http://dx.doi.org/10.1007/978-1-4419-7037-4_5.
- Momcilovic O, Choi S, Varum S, Bakkenist C, Schatten G, Navara C. Ionizing radiation induces ataxia telangiectasia mutated-dependent checkpoint signaling and G(2) but not G(1) cell cycle arrest in pluripotent human embryonic stem cells. Stem Cells 2009; 27:1822–35; PMID:19544417; http://dx.doi.org/10.1002/stem.123.
- Suvorova II, Katolikova NV, Pospelov VA. New insights into cell cycle regulation and DNA damage response in embryonic stem cells. Int Rev Cell Mol Biol 2012; 299:161–98; PMID:22959303; http://dx.doi.org/10.1016/B978-0-12-394310-1.00004-7.
- Saretzki G, Armstrong L, Leake A, Lako M, von Zglinicki T. Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells. Stem Cells 2004; 22:962–71; PMID:15536187; http://dx.doi.org/10.1634/stemcells.22-6-962.
- Saretzki G, Walter T, Atkinson S, Passos JF, Bareth B, Keith WN, Stewart R, Hoare S, Stojkovic M, Armstrong L, von Zglinicki T, Lako M. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells 2008; 26:455–64; PMID:18055443; http://dx.doi.org/10.1634/stemcells.2007-0628.
- Malashicheva AB, Kisliakova TV, Savatier P, Pospelov VA. Embryonal stem cells do not undergo cell cycle arrest upon exposure to damaging factors. Tsitologiia 2002; 44:643–8; PMID:12455372.
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol 2006; 209:883–93; PMID:16972248; http://dx.doi.org/10.1002/jcp.20776.
- Chuykin IA, Lianguzova MS, Pospelova TV, Pospelov VA. Activation of DNA damage response signaling in mouse embryonic stem cells. Cell Cycle 2008; 7:2922–8; PMID:18787397; http://dx.doi.org/10.4161/cc.7.18.6699.
- Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol 1998; 8:145–55; PMID:9443911; http://dx.doi.org/10.1016/S0960-9822(98)70061-2.
- Solozobova V, Rolletschek A, Blattner C. Nuclear accumulation and activation of p53 in embryonic stem cells after DNA damage. BMC Cell Biol 2009; 10:46; PMID:19534768; http://dx.doi.org/10.1186/1471-2121-10-46.
- Solozobova V, Blattner C. Regulation of p53 in embryonic stem cells. Exp Cell Res 2010; 316:2434-46; PMID:20542030; http://dx.doi.org/10.1016/j.yexcr.2010.06.006.
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing nanog expression. Nat Cell Biol 2005; 7:165–71; PMID:15619621; http://dx.doi.org/10.1038/ncb1211.
- Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene 2002; 21:8320–33; PMID:12447695; http://dx.doi.org/10.1038/sj.onc.1206015.
- White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev 2005; 1:131–8; PMID:17142847; http://dx.doi.org/10.1385/SCR:1:2:131.
- Barta T, Vinarsky V, Holubcova Z, Dolezalová D, Verner J, Pospísilová S, Dvorák P, Hampl A. Human embryonic stem cells are capable of executing G1/S checkpoint activation. Stem Cells 2010; 28:1143–52; PMID:20518019.
- Malashicheva AB, Kislyakova TV, Aksenov ND, Osipov KA, Pospelov VA. F9 embryonal carcinoma cells fail to stop at G1/S boundary of the cell cycle after gamma-irradiation due to p21WAF1/CIP1 degradation. Oncogene 2000; 19:3858–65; PMID:10951579; http://dx.doi.org/10.1038/sj.onc.1203736.
- Huang X, Darzynkiewicz Z. Cytometric assessment of histone H2AX phosphorylation: a reporter of DNA damage. Methods Mol Biol 2006; 314:73–80; PMID:16673875; http://dx.doi.org/10.1385/1-59259-973-7:073.
- Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of 53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci 2004; 101:2259–64; PMID:14982997; http://dx.doi.org/10.1073/pnas.0308762101.
- Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr 2003; 133:2485S–93S; PMID:12840228.
- Sabapathy K, Klemm M, Jaenisch R, Wagner EF. Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J 1997; 16:6217–29; PMID:9321401; http://dx.doi.org/10.1093/emboj/16.20.6217.
- Corbet SW, Clarke AR, Gledhill S, Wyllie AH. P53-dependent and -independent links between DNA-damage, apoptosis and mutation frequency in ES cells. Oncogene 1999; 18:1537–44; PMID:10102623; http://dx.doi.org/10.1038/sj.onc.1202436.
- Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, Kyba M, Barton MC. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol 2012; 10:e1001268; PMID:22389628; http://dx.doi.org/10.1371/journal.pbio.1001268.
- Zhang XP, Liu F, Wang W. Two-phase dynamics of p53 in the DNA damage response. Proc Natl Acad Sci USA 2011. 108:8990–5; PMID:21576488; http://dx.doi.org/10.1073/pnas.1100600108.
- Shangary S, Wang S. "Small-Molecule Inhibitors of the MDM2-p53 Protein-Protein Interaction to Reactivate p53 Function: A Novel Approach for Cancer Therapy." Annu Rev Pharmacol Toxicol 2008; 49:223–41; http://dx.doi.org/10.1146/annurev.pharmtox.48.113006.094723.
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat Commun 2012; 3:923.
- Dolezalova D, Mraz M, Barta T, Plevova K, Vinarsky V, Holubcova Z, Jaros J, Dvorak P, Pospisilova S, Hampl A. MicroRNAs regulate p21(Waf1/Cip1) protein expression and the DNA damage response in human embryonic stem cells. Stem Cells 2012; 30:1362–72; PMID:22511267; http://dx.doi.org/10.1002/stem.1108.
- Grandela C, Pera MF, Grimmond SM, Kolle G, Wolvetang EJ. p53 is required for etoposide-induced apoptosis of human embryonic stem cells. Stem Cell Res 2007; 1:116–28; PMID:19383392; http://dx.doi.org/10.1016/j.scr.2007.10.003.
- Qin H, Yu T, Qing T, Liu Y, Zhao Y, Cai J, Li J, Song Z, Qu X, Zhou P, et al. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem 2007; 282:5842–52; PMID:17179143; http://dx.doi.org/10.1074/jbc.M610464200.
- Filion TM, Qiao M, Ghule PN, Mandeville M, van Wijnen AJ, Stein JL, Lian JB, Altieri DC, Stein GS. Survival responses of human embryonic stem cells to DNA damage. J Cell Physiol 2009; 220:586–92; PMID:19373864; http://dx.doi.org/10.1002/jcp.21735.
- Akdemir KC, Jain AK, Allton K, Aronow B, Xu X, Cooney AJ, Li W, Barton MC. Genome-wide profiling reveals stimulus-specific functions of p53 during differentiation and DNA damage of human embryonic stem cells. Nucleic Acids Res 2014; 42:205–23; PMID:24078252; http://dx.doi.org/10.1093/nar/gkt866.
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663–76; PMID:16904174; http://dx.doi.org/10.1016/j.cell.2006.07.024.
- Maimets T, Neganova I, Armstrong L, Lako M. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene 2008; 27:5277–87; PMID:18521083; http://dx.doi.org/10.1038/onc.2008.166.
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell 2012; 46:30–42; PMID:22387025; http://dx.doi.org/10.1016/j.molcel.2012.01.020.
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009; 460 (7259):1132–5; PMID:19668191; http://dx.doi.org/10.1038/nature08235.
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 2009; 460:1149–55; PMID:19668189; http://dx.doi.org/10.1038/nature08287.
