ABSTRACT
CAPNS1 is essential for the stability and function of ubiquitous CAPN1 and CAPN2. Calpain modulates by proteolytic cleavage many cellular substrates and its activity is often deregulated in cancer cells, therefore calpain inhibition has been proposed as a therapeutical strategy for a number of malignancies. Here we show that CAPNS1 depletion is coupled to impairment of MCF7 and MCF10AT cell lines growth on plate and defective architecture of mammary acini derived from MCF10A cells. In soft agar CAPNS1 depletion leads to cell growth increase in MCF7, and decrease in MCF10AT cells. In both MCF7 and MCF10AT, CAPNS1 depletion leads to the enlargement of the stem cell compartment, as demonstrated by mammosphere formation assays and evaluation of stem cell markers by means of FACS and western blot analysis. Accordingly, activation of calpain by thapsigargin treatment leads to a decrease in the stem cell reservoir. The expansion of the cancer stem cell population in CAPNS1 depleted cells is coupled to a defective shift from symmetric to asymmetric division during mammosphere growth coupled to a decrease in NUMB protein level.
Introduction
CAPNS1 regulatory subunit is essential for the stability and function of ubiquitous calpain-1, CAPN1 and calpain-2, CAPN2.Citation14 Both CAPN1 and CAPN2 are generally overexpressed upon transformation.Citation15 CAPN1 expression is associated with relapse-free survival in breast tumors from patients treated with trastuzumab following adjuvant chemotherapy.Citation16 Conversely, high expression of CAPN2 in basal-like or triple-negative disease was associated with adverse breast cancer-specific survival.Citation17 We reported that CAPNS1 interferes with PP2A-Akt interaction, consequently affecting FoxO3A-dependent cell death in mouse embryonic fibroblasts and human MCF7 cells. We therefore suggested that ubiquitous Calpain inhibition might be exploited as a tool to induce apoptosis in tumors sensitive to FoxO activation.Citation1 In accordance with our findings, CAPN2 regulates proliferation, survival, migration, and tumorigenesis of breast cancer cells in mouse models, via the PP2A-Akt-FoxO-p27Kip1 signaling axis.Citation6 CAPN1 stabilizes USP1 deubiquitinating enzyme during the G1 phase of the cell cycle in human osteosarcoma cells and fibroblasts, indeed calpain inhibits USP1 proteasomal degradation, acting as a break on the CDK5/APC/Ccdh1 axis.Citation2 USP1 belongs to the class of Fanconi anemia proteins that are required for DNA repair of interstrand crosslinks and consequently for genome stability maintenance. FANCD1/BRCA2, another essential protein of the Fanconi anemia repair pathway, also plays a key role in the BRCA repair pathway, a safeguard against breast cancer, suggesting the existence of functional overlapping between the 2 machineries. USP1 stabilizes the inhibitor of differentiation ID2 and consequently preserves cellular stemness in undifferentiated osteosarcomasCitation19 therefore represents suitable targets for differentiation therapy of specific tumors.Citation3 Notably, a recent study reported that the expression of the USP1 target ID2 was directly associated with the Nottingham Prognostic Index, worsening clinical outcome and with worse disease-free and overall survival in human breast cancer.Citation18 In this study we unveiled a multifacet effect of CAPNS1 depletion in breast cancer biology, including a destabilizing effect of NUMB protein, a well known protein switch from symmetric to asymmetric cellular division.Citation13,10,8,12
Results
CAPNS1 depletion in MCF7 and MCF10AT cells perturbs both anchorage dependent and independent growth
In order to investigate the effect of CAPNS1 depletion on anchorage dependent and independent growth of breast cancer cells, colony assays and soft agar assays were performed using both luminal type MCF7 and basal like MCF10AT cell line. CAPNS1 depleted MCF7 and MCF10AT cells and respective wild type controls were seeded on tissue culture plates and 2 weeks later the number of colonies was counted after staining with crystal violet. As it is shown in the representative pictures and in the graphs of A and B, CAPNS1 depletion is coupled to a reduction of cellular growth on tissue culture plates. The effect of calpain on anchorage dependent growth of breast cancer cells confirms a previous study that demonstrated an essential role for this protease in cell adhesion mediated by paxillin cleavage.Citation4
Figure 1. CAPNS1 depletion alters breast cancer cells growth on plate and in soft agar. Control and CAPNS1 depleted MCF7 cells (1A) or MCF10AT (1B) were plated on 6 cm dishes at the indicated concentration. Two weeks later, the cells were stained with crystal violet and images were acquired by Epson scanner. Images of representative experiments are shown. Colonies were counted using image J software; the graph reports the average of 3 independent experiments (p values <0.01). Control and CAPNS1 depleted MCF7 cells (1C) or MCF10AT (1D) were resuspended in 0.3 % agar and the indicated number of cells was plated on 1% agar according to standard procedures. Three weeks later colonies were counted. The graphs report the average number of colonies counted in 3 independent experiments (p values <0.01). Representative pictures are shown next to the graphs.
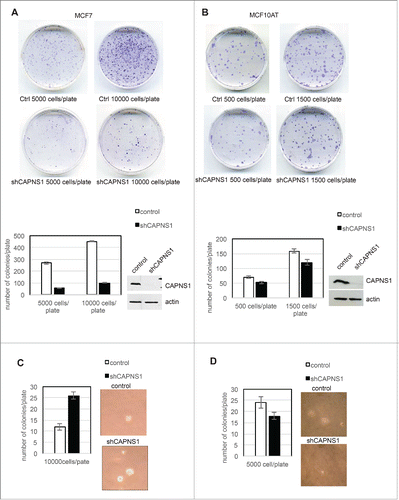
Next, CAPNS1 depleted MCF7 and MCF10AT cells and respective controls were employed for soft agar assays according to standard procedures. Three weeks after cells seeding, pictures were taken and analyzed by Image J software. Typical colonies are shown in C, 1D, and the average number of colonies counted in 3 independent experiments are indicated in the graphs. CAPNS1 depletion enhances the growth of MCF7 in soft agar, while its loss impairs the growth of MCF10AT cells in the same semisolid medium. A similar outcome of CAPNS1 depletion on cell growth both on plates and in soft agar was reported for the highly metastatic AC2M2 mouse mammary cells.Citation6 It is not surprising that interfering with essential enzymes like calpain may produce opposite effects not only in different genetic settings, but also in alternative growing environments.
Calpain modulates mammary acini formation
To assess whether calpain may play a role in the maturation of mammary acini, we performed 3D studies using MCF-10A cells, a widely used model system for understanding epithelial cell biology. When plated in a mixture of collagen and laminin MCF10A cells form 3D structures that resemble acini structures of the human breast. The size of the acini is sharply reduced in CAPNS1 depleted cells as compared to control ones (A, C), although the number of acini is not affected (). Staining with GM130, β-catenin and E-cadherin specific antibodies () revealed that the architecture of acini derived from CAPNS1 depleted cells is perturbed. These data suggest that calpain is required for the correct formation of the basic unit of breast epithelium and may play an important role in the initiation of the differentiation process in this system.
Figure 2. CAPNS1 depletion does not affect mammary acini number, but is involved in mammary acini growth and architecture. (A) Representative bright field pictures of control (left panel) and shCAPNS1 (right panel) MCF10A cells grown in 3D. Scale bar corresponds to 50 μm. (B) Average number of acini counted in 3 independent experiments (p value = 0.2). (C) Percentage of acini of different dimensions as indicated. The graph represent mean values obtained in 3 independent experiments (p values <0.01). (D) Representative immunofluorescence pictures of control (upper panels) and shCAPNS1 (lower panels) MCF10A cells grown in 3D, stained with the indicated antibodies. Scale bar corresponds to 50 μm. (E) Mouse embryonic stem cells were silenced with control siRNA or CAPNS1 specific siRNA and grown in differentiation media. The graph indicates the quantification of mRNA expression of the indicated transcripts by real time PCR analysis (p values <0.01).
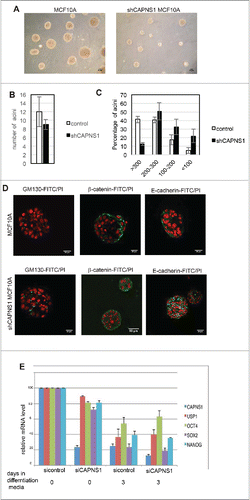
The same type of analysis was performed in MCF10AT cells, the Rasv12 transformed derivative of MCF10A cells. In this system the differences between control and CAPNS1 depleted cells appear less evident, since also control cells have lost the capacity to form acini with well defined cavities, as already reported by others (Supplementary 1A, 1B).
To address the role of CAPNS1 in stem cell maintenance and differentiation in a more general context, we employed mice embryonic stem (ES) cells. CAPNS1 was knocked out by siRNA in mice ES cells prior the induction of differentiation and the kinetics of disappearance of stem cell markers was investigated by real time PCR. CAPNS1 depletion does not alter the kinetics of disappearance of stem cell markers, indicating that its depletion is not sufficient to affect differentiation in this system. However, CAPNS1 mRNA levels clearly decrease upon differentiation, just as the standard stem cell markers, OCT4 and SOX2 and NANOG (). A similar behavior is observed for USP1 that is involved in a calpain-modulated axis in osteosarcoma and MEFs.Citation2
Calpain modulates breast cancer stem cells compartment
Cancer stem cells are considered the fuel of cancer recurrence and therefore, a satisfactory chemotherapy must be effective also against this cancer reservoir. The assay of choice to assess the role of specific genes/pathways in stem cell maintenance/growth in vitro is the mammospheres formation assay. We first applied this methodology to MCF7 cells where CAPNS1 depletion is linked to a decrease of growth in 2D, but an increase of growth in soft agar. In this system CAPNS1 depletion is linked to an increase in mammosphere formation efficiency (), and to an alteration of mammospheres shape that is less spheric respect to control ().
Figure 3. Calpain affects mammosphere forming efficiency. Control and CAPNS1 depleted MCF7 and MCF10AT cells were plated in 96 wells multiwell plates (1000 cells/well) in mammosphere specific medium. After ten days, the mammospheres were collected and disaggregated. Mammospheres derived cells were counted and plated as for the first generation and 7 d later picture were taken. (A) typical MCF7 mammospheres. Scale bar corresponds to 50 μm. (B) reports the average number of second generation MCF7 mammospheres obtained in 3 independent experiments (p values <0.01). (C) typical MCF10AT mammospheres. Scale bar corresponds to 50 μm. (D) reports the average number of second generation MCF10AT mammospheres obtained in 3 independent experiments (p values <0.01). (E) shows the average number of mammospheres counted in 3 independent experiments in control and shCAPNS1 MCF10AT treated or not treated with thapsigargin (p values<005). Briefly, control and CAPNS1 depleted MCF10AT cells were plated on 96 wells multiwells (1000 cells/well) in mammosphere specific medium. After 7 days, the mammospheres were incubated or not with 100 nM thapsigargin for 6 hours. Mammospheres derived cells were counted and plated as for the first generation and 7 d later the number of second generation mammospheres was counted. Control and CAPNS1 depleted MCF10AT cells derived from second generation mammospheres were stained with anti CD44 and anti CD24 antibodies and analyzed by FACS. Adherent cells were analyzed in parallel. The dot plot (F) and the graph (G) are the results of a typical FACS analysis.
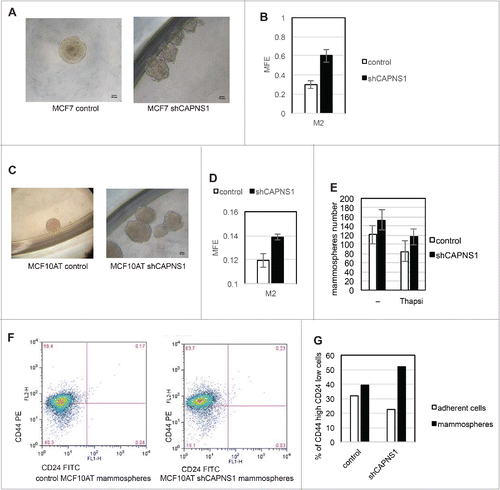
Next, we applied mammosphere assay to MCF10AT cells where calpain depletion is coupled to a decrease of both anchorage dependent and independent growth. The shape of mammospheres derived from CAPNS1 depleted cells is more irregular as compared to the shape of control mammospheres, as it is the case for MCF7 cells (). Also in this system, CAPNS1 depletion is coupled to an increase in mammospheres formation efficiency (). On the basis of these findings we hypothesized that calpain activation could hamper the expansion of the stem cell population. To verify this hypothesis, 100 nM thapsigargin was added for 6 hours to the first generation mammospheres to induce calcium release from the endoplasmic reticulum and activate calpain. Such treatment does not significantly affect cell viability, as monitored by a cell viability assay and FACS analysis performed 48 hours after treatment (Data not shown). Control and CAPNS1 depleted MCF10AT cells were employed for a standard mammosphere formation assay. After counting the mammospheres of the first generation M1, the cells were either treated or not with 100 nM thapsigargin to activate calpain. Six hours later, the mammospheres number was counted again before harvesting and disaggregation. Afterwards, equal amounts of cells were seeded in mammosphere specific medium. The number of mammospheres in the subsequent generation, M2, decreases in thapsigargin treated cells, and the entity of the reduction in control cells is almost twice the reduction occurring in CAPNS1 depleted cells (). Notably, calpain activation by ionomycin has the same effect of thapsigargin on mammosphere forming efficiency, and the calcium chelator BAPTA prevents such effect (data not shown). These results show that transient calpain activation may limit stem cell expansion.
In order to further verify the increase of stem cell population in CAPNS1 depleted cells, parental adherent cells and mammospheres derived cells were collected and analyzed by FACS to evaluate the surface expression of the widely recognized staminality markers CD44 and CD24. Cells derived from mammospheres originated from CAPNS1 depleted cells are clearly more enriched in the CD24 low/CD44 high population as compared to control cells (F, G).
CAPNS1, a novel marker of breast cancer stem cells and mouse embryonic stem cells
CAPNS1 depleted MCF10AT cells and respective controls were sorted by FACS in order to isolate the CD44 high/CD24 low population from each cell line. An aliquot of sorted cells was utilized for nuclear/cytoplasmic fractionation and western blot analysis, and the remaining cells were employed for mammosphere formation assays. As expected, E-cadherin protein level is lower in sorted cells as shown in , indicating that epithelial-mesenchymal transition is taking place in CD44 high/CD24 low MCF10AT cells. Notably, as already observed for osteosarcomas and MEFs, full length USP1 (indicated by an arrow) decreases in CAPNS1 depleted cells. Interestingly, both calpain and USP1 are more abundant in the sorted CD44 high/CD24 low cell population, enriched in cancer stem cells. In particular, CAPN1 is present also in the nuclear compartment and USP1 is present also in the cytoplasmic compartment in the cancer stem cells enriched population. Our findings suggest that calpain and USP1 may be novel markers of breast cancer stem cells.
Figure 4. CAPNS1 depletion is coupled to breast cancer stem cells expansion.CD44+/CD24- population was sorted by FACS analysis from CAPNS1 depleted and control MCF10AT cells. Nuclear and cytoplasmic extracts obtained from sorted cells and analyzed by western blot with the indicated antibodies (A). In parallel, sorted CD44+/CD24- cells were utilized for mammosphere formation assay. (B) reports the average mammosphere forming efficiency for the first (M1) and second generation (M2) (p values <005). (C) reports the average MFE of second generation mammospheres with diameter larger than 250 micrometres (p values <005). (D, E) Control and shCAPNS1 MCF10AT were stained with PHK26 vital dye and after extensive washing were plated in mammosphere forming medium. Pictures were taken after 2 weeks. Scale bar corresponds to 50 μm. The percentage of PKH26 stained area of 4 planes of 4 control and 4 shCAPNS1 mammospheres was measured by Image J and is reported in the graph. (F) CAPNS1 depleted and control adherent MCF10AT cells and cells derived from first and second generation mammospheres derived from the same cells were lysed and analyzed by western blot with the indicated antibodies.
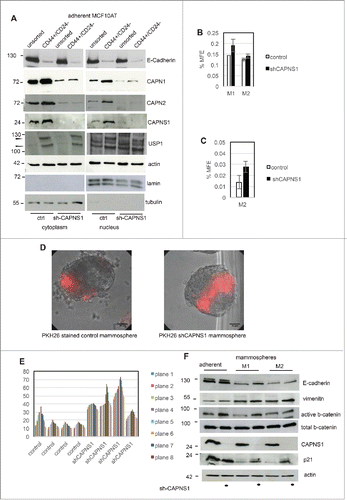
As already observed for unsorted cells, the CD44 high/CD24 low population derived from CAPNS1 depleted cells is characterized by an increase in first generation mammosphere forming efficiency (). This result is in accordance with the higher surface expression of CD44 in CAPNS1 depleted sorted cells, shown in F, G. The number of second generation mammospheres originated from CD44 high/CD24 low MCF10AT cells is unaffected by CAPNS1 depletion possibly due to exhaustion of stem cell potential. Indeed, the size of mammospheres originated from CAPNS1 depleted cells, is significantly larger, as quantified and reported in .
Calpain acts as a switch from symmetric to asymmetric division
On the basis of the data reported above, we hypothesized that calpain may be involved in the switch from symmetric division to asymmetric division at the onset of differentiation of human mammary cells. In order to investigate whether calpain is involved in this switch, we followed the distribution of the vital dye PKH26 during the growth of mammosphere originated from control cells and from CAPNS1 depleted cells. Two representative pictures are shown in and the quantification of the percentage of the surface of the mammospheres that retains PKH26 is shown in graph 4E. Notably, the surface of stained mammospheres, that reflects the number of stained cells, is larger when CAPNS1 is depleted.
As a first approach to understand the molecular basis of the expansion in cancer stem cells coupled to CAPNS1 depletion, the levels of key proteins in cell extracts from adherent cells and mammospheres of the first and second generation were evaluated by western blot. As it is shown in , comparing adherent cells to mammospheres, E-cadherin protein levels decrease, while vimentin becomes more abundant both in control and CAPNS1 depleted cells, as expected when epithelial/mesenchimal transition is taking place. Notably, active beta-catenin, that positively modulates mammary stem cells long-term expansion, is more abundant in CAPNS1 depleted adherent and M1 cells as compared to the control counterparts, in accordance with the increase in cancer stem cells characteristics reported above for CAPNS1 depleted cells. Genotoxic stress was shown to activate p21 and consequently to induce symmetric division in mammary cancer stem cells, leading to the expansion of stem cell compartment.Citation7 Interestingly, p21 is more abundant in mammospheres from CAPNS1 depleted cells, as compared to control cells, further supporting the expansion of the stem cell compartment in CAPNS1 depleted mammospheres.
CAPNS1 reintroduction rescues the wild type phenotype and reduces stem cells expansion
To verify whether CAPNS1 reintroduction could rescue the phenotype observed in CAPNS1 depleted mammospheres, we produced a novel set of MCF10AT cell lines: a control cell line, a CAPNS1 depleted cell line and a CAPNS1 depleted cell line, expressing a CAPNS1 mRNA resistant to CAPNS1 silencing (). Mammosphere formation assay was performed, confirming an increase in mammosphere formation efficiency in CAPNS1 depleted cells. Notably, CAPNS1 reintroduction is coupled to a reduction of mammosphere forming efficiency, further confirming that the expansion of the stem cell compartment observed in CAPNS1 depleted cells is dependent on CAPNS1 ().
Figure 5. CAPNS1 depletion is linked to expansion of PKH26 stained cells that can be rescued upon CAPNS1 reintroduction. (A) CAPNS1 expression in MCF10AT cell lines: control MCF10AT, shCAPNS1 MCF10AT, shCAPNS1 MCF10AT stably expressing siRNA resistant CAPNS1. (B) Average number of mammosphere number obtained in 3 independent experiments with MCF10AT derived cell lines described above (p values <005). (C) Average of PKH26 stained cells in mammospheres obtained in 3 independent experiments (p values <005). (D) Subsequent pictures taken 3, 5, 7, 9 d after seeding of representative mammosphere obtained from the above described cell lines. Scale bar corresponds to 50 μm.
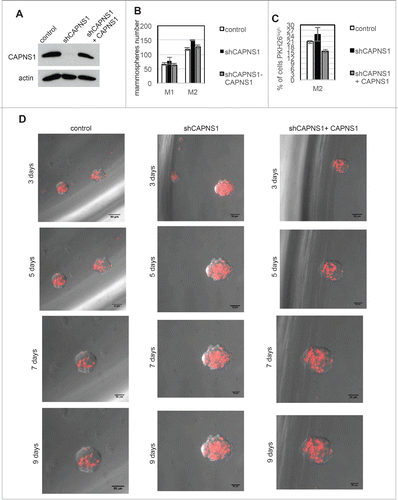
The three cell lines were further employed for mammosphere formation and staining with PKH26. Pictures were taken at subsequent time points, namely 3, 5, 7, 10 d after staining and seeding. Typical pictures are shown in . In order to quantify the number of PKH26 positive cells, FACS analysis was performed 9 days after staining (). These novel sets of experiments further confirm that the number of cells retaining the PKH26 die negatively correlates with CAPNS1 protein level.
NUMB protein level is lower in CAPNS1 depleted cells
Numb is an essential modulator of the shift between symmetric and asymmetric division, therefore we evaluated NUMB protein level in CAPNS1 depleted MCF7, MCF10A and MCF10AT cells, as compared to control cells. NUMB level is lower in CAPNS1 depleted MCF7 cells, while it is apparently unaffected by CAPNS1 depletion in MCF10A and MCF10AT cells (). However, analysis of nuclear and cytoplasmic fractions of MCF10AT cells shows that NUMB protein level is lower in cytoplasms extracted from CAPNS1 depleted as compared to control ones (). The decrease in NUMB protein level coupled to CAPNS1 depletion is sharper in the cytoplasms of CD44 high/CD24 low population ().
Figure 6. CAPNS1 depletion is coupled to a decrease in NUMB protein level.NUMB protein level was compared by western blot in CAPNS1 depleted versus control cells, using total cell lysates of MCF7, MCF10A and MCF10AT cells (A) nuclear and cytoplasmic extracts of MCF10AT cells (B) and cytoplasms from CD44 high/CD24 low sorted MCF10AT cells (C). (D) CAPNS1 specific, USP1 specific or a control siRNA were transfected in the indicated cell lines. 48 hours later, lysates were analyzed by western blot to monitor USP1 and CAPNS1 levels. Actin was used as loading control.
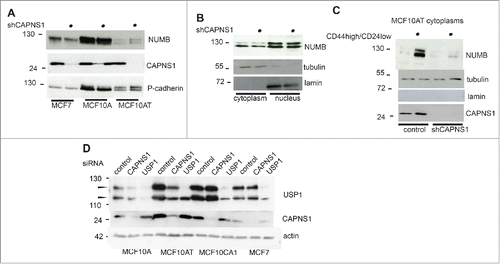
Why the increase in the expansion of the stem cell compartment is sharper in MCF7 cells than in MCF10AT cells? We previously reported that CAPNS1 positively modulates USP1 stability in osteosarcoma and MEFs. Since USP1 inhibition/depletion was shown to induce differentiation in various cellular contexts, we hypothesized that USP1 reduction might buffer the effect due to NUMB reduction.
CAPNS1 was transiently depleted in MCF10A, MCF10AT, MCF10CA1, MCF7 cells and USP1 level was evaluated by western blot. As expected, USP1 level decreases in MCF10A and MCF10AT cells, upon CAPNS1 depletion, but it is unaffected in MCF7 cells.
Discussion
CAPNS1 depletion is coupled to a reduction of growth on plates in both epithelial MCF7 and basal like MCF10AT cancer cells. On the other hand, CAPNS1 depletion has opposite effects on soft agar growth of MCF10AT and MCF7 and suggest that when cancer cells bypass the first environmental barrier and become able to grow in an anchorage independent manner, the overall role of calpain may vary in different genetic settings. The positive effect of CAPNS1 on MCF10AT cells growth in soft agar is in accordance with the previously reported negative role of APC/Ccdh1 in the same experimental system.Citation5 Indeed, calpain negatively modulates APC/Ccdh1Citation2,11 and therefore in CAPNS1 depleted cells, APC/Ccdh1 is more active and growth results impaired.
The negative effect of CAPNS1 depletion on the growth of MCF10AT cells demonstrated by colony and soft agar assays is in accordance with the results obtained by Wai Chi-Ho et al. in ACM2 cells.Citation6 Interestingly, they demonstrate that while CAPN2 knockout reduced tumor growth in a breast tumor engraftment model, lung metastasis was not affected by CAPN2 depletion and the percentage of mice with metastasis in the 2 reported experiments was higher when CAPN2 depleted cells were injected. This result is in agreement with our finding on the expanded cancer stem cell compartment in CAPNS1 depleted MCF7 and MCF10AT cells. Accordingly, we have shown that calpain activation reduces the expansion of this compartment in a calpain dependent manner. Cancer stem cells are resistant to standard therapies that reduce the bulk of the tumor. Thus new therapies specifically targeting this tumor reservoir are warranted.
Altogether previously published data and our findings highlight a role of calpain as a master director of cellular architecture and cell fate decision. In this study we have also demonstrated by mammosphere formation assays, western blot and FACS analysis of breast cancer stem cell markers that calpain depletion increases the amplification of the breast cancer stem cell compartment. The vital dye PKH26 used to stain the individual cancer stem cells is inherited by a larger number of daughter cells when CAPNS1 is depleted, indicating a longer persistence of symmetric division. Accordingly, NUMB protein, a master regulator of the shift between symmetric to asymmetric division that is coupled to differentiation, is reduced in CAPNS1 depleted cells.
Materials and methods
Chemicals and reagents
Thapsigargin and PKH26 were bought from Sigma-Aldrich. Lipofectamine™ RNAiMAX from Invitrogen. USP1 specific siRNA with the following sequence: AACCCUAUGUAUGAAGGAUAU was purchased from Eurofins Mwg/Operon. RNAi oligonucleotides specific for CAPN1, Cdk5 and cdh1 were purchased from Santa Cruz Biotechnology. CAPN2 RNAi and SMARTpool for CAPNS1 silencing were purchased from DHARMACON, Inc., Dallas, TX (USA). AllStars Negative Control siRNA (Qiagen) was used as negative control.
Cell culture and transfections
MCF-7 cells were grown in Eagle’ s Minimum Essential Medium supplemented with 10% fetal calf serum, 1% Penicillin/Streptomycin (Lonza) and Non essential Amino Acid Solution 100X (Sigma). MCF10A and MCF10AT cells were cultured in DMEM:F12 media (1:1) supplemented with 10μg/ml Insulin (Sigma), 20ng/ml EGF (PEPROTECH), 500ng/ml hydrocortisone (Sigma), 5% horse serum, 10U/ml Penicillin, 100μg/ml Streptomycin and 1% HEPES (Gibco). MCF10CA1 were cultured in DMEM:F12 media (1:1) supplemented with 10% foetal calf serum, 1% Penicillin/Streptomycin (Lonza). MDA-MB231 and MDA-MB468 were grown in Dulbecco's modified Eagle's Medium (DMEM) supplemented with 10% foetal calf serum, 1% Penicillin/Streptomycin (Lonza). SKBR3 cells were grown in RPMI supplemented with 10% foetal calf serum, 1% Penicillin/Streptomycin (Lonza). Mouse embryonic stem (ES) cells were cultured on 0.2% gelatin-coated plates in mESC self-renewal medium: Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% knockout serum replacement (Gibco), 1% nonessential amino acids (Gibco), 1 mM sodium pyruvate (Invitrogen), 0.1 mM ß-mercaptoethanol, 1% penicillin/streptomycin (Invitrogen), and 1,000 U/ml mouse leukemia inhibitory factor (LIF). All cells were grown in 5% CO2 at 37°C in a humidified incubator. MCF10A, MCF10AT, MCF10CA1 (obtained from Karmanos Cancer Institute, US), MCF-7, MDA-MB-231, MDA-MB468 and SKBR3 cells at 60–80% confluence, and ES cells at low confluence were transiently oligofected using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions.
Viral constructs
shCAPNS1 was obtained by annealing the following sense oligonucleotide: 5′- GAT CCC CGC CAC AGA ACT CAT GAA CAT TCA AGA GAT GTT CAT GAG TTC TGT GGC TTT TTA - 3′ to the following antisense oligonucleotide: 5′ - AGC TTA AAA AGC CAC AGA ACT CAT GAA CAT CTC TTG AAT GTT CAT GAG TTC TGT GGC GGG G - 3′. The annealed product was inserted by standard procedures in pRetroSuper vector, previously linearized by BglII and HindIII digestion, and checked by sequencing.
Cell infections
293GP packaging cells were transfected with the calcium-phosphate method with pRetroSuper-shCAPNS1 or vector alone, after 72 hours the supernatant was harvested, filtered and added to MCF10AT or MCF-7 cells. The infected cells were selected by the addition of puromycin and after 7 d the expression of CAPNS1 was checked by western blot.
Colony formation assay
Cells were seeded at 1000 cells/ml and 2000 cells/ml for MCF-7 and 100 cells/ml and 300 cells/ml for MCF10AT in a 21 cm2 cell culture dish and incubate at 37°C in a humidified, 5% CO2 atmosphere. After incubation for 15 days, cell culture plates containing colonies were gently washed with PBS twice, fixed with 4% paraformaldehyde for 20 minutes at room temperature and stained with 0.5% Crystal violet. Excess stain was removed by washing repeatedly with PBS and colonies were counted. For quantification, the colonies were analyzed using ImageJ software. Three reduplicate dishes were used from each condition.
Soft-agar assay
To evaluate anchorage-independent growth, 5 × 103 and 1 × 104 MCF-7 cells and MCF10AT were resuspended in 0.3% agarose in growth media. Cells were plated on a solidified bed of 1% agarose in growth media in 21 cm2 plates. Plates were incubated for 3 weeks at 37°C. Bright field images of MCF-7 and MCF10AT colonies were taken using a 10× objective. The number of colonies per field was counted using ImageJ software. The experiments were conducted in triplicate.
Three dimensional culture
3D culture was carried out based on the method as already described by others.Citation9 Briefly, 12 mm round glass coverslips were coated with 80 μl of ice-cold rBM (Cultrex) and incubated at 37°C for 20 minutes to allow rBM to solidify. The coated coverslips were placed in a 24-well plate with the rBM facing up. Single cell suspensions containing 7000 cells/well for MCF10A and 5000 cells/well for MCF10AT, were resuspended in 250μl of complete growth medium with 2% (v/v) of Cultrex and plated on top of the rBM coated chamber slides and incubated at 37°C for 30 minutes to let the cells attach. Then 250μl of the assay medium was added per well and the cells were cultured al 37°C and 5% CO2 for 3 weeks, with the medium being changed every 3 d.
Immunostaining and microscopy
The three-dimensional structures in rMB Cultrex were fixed in 4% paraformaldehyde at room temperature for 20 minutes, permeabilized with 1% Triton X-100 in PBS for 15 minutes and washed with 100 mM glycine in PBS, 3 times at 10 min/wash. After the 3-dimensional structures were blocked with IF (immunofluorescent) blocking buffer (0.1% BSA, 0.2% Triton X-100, 0.05% Tween 20, 7.7 mM NaN3, in PBS) containing 10% normal goat serum for 1h and incubated overnight at 4°C with the following primary antibodies: anti-GM130 (BD Biosciences), anti-E-cadherin (Takara bio) and anti-βcatenin (Santa Cruz). Samples were then washed with IF buffer (0.1% BSA, 0.2% Triton X-100, 0.05% Tween 20, 7.7 mM NaN3, in PBS) 3 times at 20 min/wash and stained with secondary antibody for 1h at room temperature. This was followed by 3 washes with IF buffer at 10 min each. To stain the nuclei, the slides were incubated with RNase 200μg/ml for 15 min at room temperature and then stained with Propidium Iodide 25μg/ml for 5 min at room temperature, washed 3 times with PBS and mounted on glass coverslips. Confocal microscopic images of the acinus structures were captured by the Z-stacking function for serial confocal sectioning (LSM-510 META confocal microscope) and then analyzed by ImageJ software. The experiments were conducted in triplicate.
Mammosphere cultures
To generate mammospheres, confluent monolayers cells were trypsinized, counted and plated as single suspension (1000 cells/well) in a low attachment plates (Corning) and grown in non-adherent culture conditions, as described in Dontu et al, 2003. MCF10AT cells were grown in a serum-free mammary epithelial growth medium (MEBM) (Lonza) supplemented with 2 % of B27 (Invitrogen), 20 ng/mL hEGF (PEPROTECH), 20 ng/mL bFGF (Provitro), and 4 μg/mL heparin (Sigma), 10 ng/ml hydrocortisone (Sigma) and 5 μg/ml Insulin (Sigma) and 1% Penicillin/Streptomycin (Lonza) while MCF-7 cells were grown in a serum-free mammary epithelial growth medium (MEBM) supplemented with B27 (Invitrogen), 20 ng/mL hEGF (PEPROTECH) 5 μg/ml Insulin (Sigma) and 1% Penicillin/ Streptomycin (Lonza). Primary mammospheres after 10 d were collected by gentle centrifugation (800 rpm) and dissociated enzymatically to be re-seeded to obtain secondary mammospheres. The number of mammospheres (≥ 250 μm) per well was then counted and percentages of mammospheres forming efficiencies (% MFE) was calculated as a number of mammospheres divided by the plated cell number and multiplied for a hundred.
PKH26 assay
Primary mammospheres were collected and dissociated enzymatically, resuspendend in PBS and labeled with PKH26 (Sigma) 10−7 M for 5 min. To stop the staining, an equal volume of serum was added to the cells and incubated for 1 minute to allow binding of excess dye. Labeled cells were re-plated (1000cells/well) in ultralow attachment plates in mammospheres culture medium to obtain secondary mammospheres. After 10 days, secondary mammospheres were analyzed by confocal microscopy.
Drug treatments
Cells in 3D culture and primary mammospheres were treated with 100nM thapsigargin. For cells in 3D culture, the drug was added after a week and replaced every 3 days, while for primary mammospheres, 100nM of thapsigargin was added 6h before collection, dissociation, and re-seeding to obtain secondary mammospheres.
Flow cytometric analysis and cell sorting
Single cell suspension of adherent cells or mammospheres was stained with CD24-FITC (BD Biosciences) and CD44-PE (BD Biosciences). Appropriate isotype controls were used to set the threshold for CD44 and CD24 positive cells. Cells were incubated with the appropriate antibodies at room temperature in the dark for 30 minutes, then washed with PBS and finally resuspended in 500μL of PBS. 10,000 cells were analyzed for each condition using a FACSCalibur flow cytometer (BD Biosciences); data were analyzed using FlowJo software (Tree Star, Ashland, USA).
To separate CD44high/CD24low MCF10AT cells, flow cytometric cell sorting was performed on single-cell suspensions previously stained with CD44 and CD24 antibody.
Quantitative Real Time PCR (qRT-PCR) analysis of ES cells
ES cells were transiently transfected using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions, using a control siRNA (AllStars Negative Control siRNA, Qiagen) or a siRNA targeting mouse CAPNS1 (5′ - CAGGCTATATACAAACGCTTT - 3′). 72 hours later, ES cells were re-seeded and subjected to a second round of siRNA silencing, and grown for 72 hours in mESC differentiation medium: DMEM supplemented with 15% ES cell certified serum (Invitrogen), 1% non-essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 0.1 mM ß-mercaptoethanol and 1% penicillin/streptomycin (Lonza).
For quantitative real-time PCR, total RNA from ES cells was purified using QIAzol lysis reagent (Qiagen). 1.5 μg of total RNA was treated with DNase (RQ1, Promega) and subjected to reverse transcription in the presence of random primers (Promega) or oligo dT primers using SuperScriptIII Reverse Transcriptase (Invitrogen) according to the manufacturer's suggestions. Quantitative real-time PCR was performed on StepOnePlus real-time PCR machine (Applied Biosystems), using SYBR Green Universal PCR Master Mix (Applied Biosystems) and the following oligonucleotides targeting mouse genes: CAPNS1 for TTCATCAGCTGCTTGGTCAG, rev AGCCACTCCTGGATGTTCAC; USP1 for CATTTGAAGTGCTTTGCTGCT, rev CTTGGAAAGTCCACCACCAT; Gapdh: for TTCACCACCATGGAGAAGGC, rev CCCTTTTGGCTCCAC; Nanog for TTCTTGCTTACAAGGGTCTGC, rev AGAGGAAGGGCGAGGAGA; Oct4 for CAGGGACACCTTTCCCAGGG, rev TTTAAGAACAAAATGATGAG; Sox2 for TGCTGCCTCTTTAAGACTAGGG, rev TCGGGCTCCAAACTTCTCT.
Quantitative RT (qRT-pCR) was performed in triplicate and relative expression levels were normalized to controls by using the comparative Ct (ΔΔCt) method, using Gapdh mRNA as internal control.
Western blot analysis
Western blotting was performed on cells protein lysates (50 mM Tris-HCl pH 7.5; 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5 mM NaF, 1 mM Sodium orthovanadate, complete protease inhibitor cocktail Sigma-Aldrich). When specified, nuclear and cytoplasmic extracts were prepared using nuclear and cytoplasmic extraction reagents (ThermoScientific) according to manufacturer instructions. Lysates were clarified by centrifugation for 15 min at 4°C and protein concentrations were assessed using Bradford protein assay (BioRad Laboratories). Samples containing equal amounts of protein were boiled in SDS sample buffer, resolved using SDS-PAGE (15-20 μg protein per lane) and transferred to nitrocellulose membranes. The blots were then probed with the following antibodies: anti-β-catenin active (Abcam), anti-E-cadherin (Takara bio), anti-Capns1, anti-Capn2, anti-sβ-catenin, anti-α Actin (Sigma-Aldrich), anti-Capn1, anti-p21, anti-p53, anti-β-catenin, anti-Cdc27, anti-Cdc6 (Santa Cruz), anti-cdh1 (Calbiochem), anti-USP1 (Bethyl Laboratories) and anti-vimentin (BD Bioscences), overnight at 4°C, then washed, incubated with horseradish peroxidase-conjugated secondary antibodies (Sigma), and analyzed by enhanced chemiluminescence
Statistical analysis
Results are expressed as means ± standard deviations of at least 3 independent experiments. Statistical analysis was performed using Student's t test with the level of significance set at P < 0.05.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KCCY_S_1121325.pdf
Download PDF (485.7 KB)Funding
This work was supported by a Project funded under the Cross-Border Cooperation Programme Italy-Slovenia 2007–2013 by the European Regional Development Fund and national funds to F. Demarchi; a FIRB grant CINECA RBAPMLP2W, and CTN01_00177_817708 to C. Schneider.
References
- Bertoli C, Copetti T, Lam EW, Demarchi F, Schneider C. Calpain small-1 modulates Akt/FoxO3A signaling and apoptosis through PP2A. Oncogene 2009; 28:721–33; PMID:19029949; http://dx.doi.org/10.1038/onc.2008.425
- Cataldo F, Peche LY, Klaric E, Brancolini C, Myers MP, Demarchi F, Schneider C. CAPNS1 regulates USP1 stability and maintenance of genome integrity. Mol Cell Biol 2013; 33:2485–96; PMID:23589330; http://dx.doi.org/10.1128/MCB.01406-12
- Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, Maloney DJ, Jadhav A, Simeonov A, Zhuang Z, Selective and cell-active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem Biol 2011; 18:1390–400; PMID:22118673; http://dx.doi.org/10.1016/j.chembiol.2011.08.014
- Cortesio CL, Boateng LR, Piazza TM, Bennin DA, Huttenlocher A. Calpain-mediated proteolysis of paxillin negatively regulates focal adhesion dynamics and cell migration. J Biol Chem 2011; 286:9998–10006; PMID:21270128; http://dx.doi.org/10.1074/jbc.M110.187294
- Fujita T, Liu W, Doihara H, Date H, Wan Y. Dissection of the APCCdh1-Skp2 cascade in breast cancer. Clin Cancer Res: An Off J Am Assoc Cancer Res 2008; 14:1966–75; PMID:18381934; http://dx.doi.org/10.1158/1078-0432.CCR-07-1585
- Ho WC, Pikor L, Gao Y, Elliott BE, Greer PA. Calpain 2 regulates Akt-FoxO-p27(Kip1) protein signaling pathway in mammary carcinoma. J Biol Chem 2012; 287:15458–65; PMID:22427650; http://dx.doi.org/10.1074/jbc.M112.349308
- Insinga A, Cicalese A, Faretta M, Gallo B, Albano L, Ronzoni S, Furia L, Viale A, Pelicci PG. DNA damage in stem cells activates p21, inhibits p53, and induces symmetric self-renewing divisions. Proc Natl Acad Sci U S A 2013; 110:3931–6; PMID:23417300; http://dx.doi.org/10.1073/pnas.1213394110
- Kechad A, Jolicoeur C, Tufford A, Mattar P, Chow RW, Harris WA, Cayouette M. Numb is required for the production of terminal asymmetric cell divisions in the developing mouse retina. J Neurosci: Off J Soc Neurosci 2012; 32:17197–210; PMID:23197712; http://dx.doi.org/10.1523/JNEUROSCI.4127-12.2012
- Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods 2007; 4:359–65; PMID:17396127; http://dx.doi.org/10.1038/nmeth1015
- Lerner RG, Petritsch C. A microRNA-operated switch of asymmetric-to-symmetric cancer stem cell divisions. Nat Cell Biol 2014; 16:212–4; PMID:24576899; http://dx.doi.org/10.1038/ncb2924
- Maestre C, Delgado-Esteban M, Gomez-Sanchez JC, Bolanos JP, Almeida A. Cdk5 phosphorylates Cdh1 and modulates cyclin B1 stability in excitotoxicity. EMBO J 2008; 27:2736–45; PMID:18818692; http://dx.doi.org/10.1038/emboj.2008.195
- Rennstam K, McMichael N, Berglund P, Honeth G, Hegardt C, Ryden L, Luts L, Bendahl PO, Hedenfalk I. Numb protein expression correlates with a basal-like phenotype and cancer stem cell markers in primary breast cancer. Breast Cancer Res Treat 2010; 122:315–24; PMID:19795205; http://dx.doi.org/10.1007/s10549-009-0568-x
- Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development 2002; 129:4843–53; PMID:12361975
- Sorimachi H, Hata S, Ono Y. Expanding members and roles of the calpain superfamily and their genetically modified animals. Exp Anim 2010; 59:549–66; PMID:21030783; http://dx.doi.org/10.1538/expanim.59.549
- Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nat Rev Cancer 2011a; 11:364–74; PMID:21508973; http://dx.doi.org/10.1038/nrc3050
- Storr SJ, Woolston CM, Barros FF, Green AR, Shehata M, Chan SY, Ellis IO, Martin SG. Calpain-1 expression is associated with relapse-free survival in breast cancer patients treated with trastuzumab following adjuvant chemotherapy. Int J Cancer. J Int du Cancer 2011b; 129:1773–80; PMID:21140455; http://dx.doi.org/10.1002/ijc.25832
- Storr SJ, Lee KW, Woolston CM, Safuan S, Green AR, Macmillan RD, Benhasouna A, Parr T, Ellis IO, Martin SG. Calpain system protein expression in basal-like and triple-negative invasive breast cancer. Ann Oncol: Off J Euro Soc Med Oncol/ESMO 2012; 23:2289–96; PMID: 22745213; http://dx.doi.org/10.1093/annonc/mds176
- Wazir U, Jiang WG, Sharma AK, Newbold RF, Mokbel K. The mRNA expression of inhibitors of DNA binding-1 and -2 is associated with advanced tumour stage and adverse clinical outcome in human breast cancer. Anticancer Res 2013; 33:2179–83; PMID:23645773
- Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, Cao TC, Carano RA, Dixit VM. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 2011; 146:918–30; PMID:21925315; http://dx.doi.org/10.1016/j.cell.2011.07.040
