Meiosis is an important process for sexually reproducing organisms to generate haploid gametes. Meiosis effectively reduces the genome ploidy by half, involving one round of chromosome replication followed by 2 rounds of chromosome segregation in meiosis I and meiosis II. Chromosome segregation in meiosis I differs from that in mitosis and meiosis II, in that homologous chromosomes segregate but sister chromatids remain attached. Precocious separation of sister chromatids in meiosis I underlies the main mechanism contributing to chromosomal disorders such as trisomy diseases.
In order to ensure co-segregation of the sister chromatids in meiosis I, the kinetochores are reorganized at the sister centromeres so that sister kinetochores receive microtubules from the same spindle poles (i.e., monopolar spindle-kinetochore attachment). However, the mechanism contributing to the control of sister kinetochore reorganization has not yet been elucidated in detail. We recently found that a nuclear pore complex (NPC) component, Nup132, is involved in regulating meiotic kinetochore assembly in the fission yeast Schizosaccharomyces pombe.Citation1
S. pombe preferentially reproduces through mitosis but will enter meiosis under conditions of nutrient limitation. The chromosomes are dynamically rearranged as the cells transit from the mitotic cell cycle to meiosis. In mitotic cells, the centromeres stably associate with the spindle pole body (SPB), the yeast microtubule-organizing center, and the telomeres are distant from the SPB. On the other hand, upon entry into meiosis, the telomeres are brought toward the SPB and the centromeres are released.Citation2 Association of the centromeres and the SPB occurs through interactions between the KMN (KNL1/Spc7-Mis12-Nuf2) complex and Csi1.Citation3 Csi1 is an SPB-association protein, whereas the KMN complex constitutes the outer kinetochores at the centromeres and remains assembled throughout the mitotic cell cycles in yeasts. However, in early meiosis, Csi1 and the KMN complex disassemble from the SPB and the centromeres, respectively ().Citation1,4,5 Without interactions between the KMN complex and Csi1, the centromeres can be liberated from the SPB in early meiotic prophase. In late prophase, Csi1 re-associates with the SPB. Concomitantly, the KMN complex reassembles at the centromeres and facilitates centromere recapture at the SPB.
Figure 1. Illustration of meiotic chromosome rearrangements upon kinetochore reorganization. Kinetochore reorganization during meiosis with or without Nup132. The KMN complex disassembles from the centromeres in early meiotic prophase, and reassembles in late meiotic prophase to recapture centromeres to the SPB. In the nup132Δ mutant, the kinetochores are partially assembled in early meiotic prophase. Such preassembled kinetochores may lead to an unfavorable organization for sister chromatid co-segregation in meiosis I.
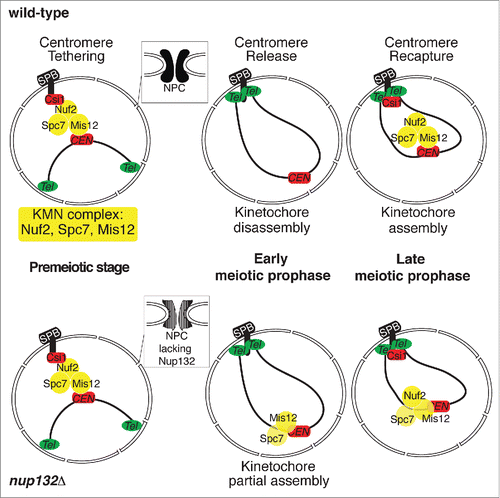
In the nup132Δ mutant, dynamic association of Csi1 to the SPB occurs as normal; however, this mutation impacts assembly of the KMN complex at the centromeres.Citation1 The KMN components Mis12 and Spc7 stay at the centromeres in early meiotic prophase in the nup132Δ mutant, while the KMN complex normally disassembles in the wild-type cells. The centromeres with preassembled Mis12 and Spc7 remain detached from the SPB in early meiotic prophase, possibly because the Nuf2-Ndc80 complex is not incorporated into the KMN complex until late meiotic prophase. The spindle assembly checkpoint (SAC) component Bub1 is also precociously recruited to the prophase centromeres. This preassembled kinetochore subsequently affects the microtubule-kinetochore attachments at the first meiotic division in the nup132Δ mutant. Through live-cell imaging of the behaviors of the centromere along the meiotic spindles, we found that the sister centromeres unsteadily move between the spindle poles in the nup132Δ mutant. In contrast, due to monopolar spindle-kinetochore attachment in meiosis I, the sister centromeres usually steadily move together toward the same spindle pole in the normal condition. These results indicate that the sister kinetochores of the nup132Δ mutant receive microtubules from the opposite spindle poles in meiosis I. Thus, inappropriate microtubule-kinetochore attachment activates the SAC and results in prolongation of meiosis I. Activation of the SAC facilitates correction of the erroneous microtubule-kinetochore attachment, so that the sister chromatids still co-segregate with high fidelity in meiosis I in the nup132Δ mutant. Nonetheless, in the absence of the SAC component Mad2 or Bub1, the prolongation of meiosis I is shortened, and the frequency of precocious sister chromatid segregation is increased in the nup132Δ mutant. Curiously, sister chromatids were found to segregate almost randomly in the nup132Δbub1Δ double mutant. In addition to the SAC, Bub1 also functions in recruiting the cohesin protector Sgo1 to the centromeric region.Citation6 However, the synergistic effects of nup132Δ and bub1Δ in sister chromatid segregation do not simply occur because of the loss of Sgo1, given that simultaneous deletion of mad2 and sgo1 in the nup132Δ mutant does not phenocopy the nup132Δbub1Δ mutant.Citation1 This result suggests that Bub1 has another regulatory role in meiotic kinetochore assembly.
Nup132, the S. pombe homolog of human Nup133, is a component of the Nup107–160 subcomplex. The Nup107-160 subcomplex supports the NPC structure across the nuclear pore. In metazoans, the Nup107-160 subcomplex relocates from the nuclear envelope to the kinetochores upon nuclear envelope breakdown, and has been implicated in kinetochore function in mitosis.Citation7 Therefore, it is likely that Nup132 and its metazoan homolog play a similar and conserved role in meiotic kinetochore assembly. However, unlike the case in metazoans, the fission yeast undergoes mitosis or meiosis I without nuclear envelope breakdown, and the majority of Nup132 stays on the nuclear envelope upon kinetochore assembly. Therefore, future work is required to elucidate precisely how Nup132 regulates kinetochore assembly.
References
- Yang H-J, et al. J Cell Biol 2015; 211:295-308; PMID:26483559; http://dx.doi.org/10.1083/jcb.201501035
- Chikashige Y, et al. Cell 2006; 125:59-69; PMID:16615890; http://dx.doi.org/10.1016/j.cell.2006.01.048
- Hou H, et al. J Cell Biol 2012; 199:735-44; PMID:23166349; http://dx.doi.org/10.1083/jcb.201208001
- Asakawa H, et al. Mol Biol Cell 2005; 16:2325-38; PMID:15728720
- Hayashi A, et al. Mol Biol Cell 2006; 17:5173-84; PMID:17035632
- Kawashima SA, et al. Science 2010; 327:172-7; PMID:19965387; http://dx.doi.org/10.1126/science.1180189
- Zuccolo M, et al. EMBO J 2007; 26:1853-64; PMID:17363900
