The discovery of N6-methyladenosine (m6A) in polyadenylated RNA can be traced back to the 1970s. However, the biological functions of m6A in mammalian mRNA had remained mysterious for decades. This modification was brought back into the spotlight several years ago after the development of m6A-seq and the elucidation of its enzyme system. Transcriptome-wide m6A mapping has revealed asymmetric distribution of mRNA methylation with the majority of m6A sites enriched near the stop codon.Citation1,2 Recognizing the core consensus sequence GGAC, the responsible methyltransferase has been identified as a functional complex consisting of METTL3, METTL14, and WTAP. The discovery of FTO, a demethylase that acts on m6A, demonstrates the reversibility and the dynamic nature of m6A. The YTH domain family proteins, in particular YTHDF2, serve as the major m6A-binding proteins that regulate a diverse array of RNA metabolism, including mRNA splicing and degradation.Citation3
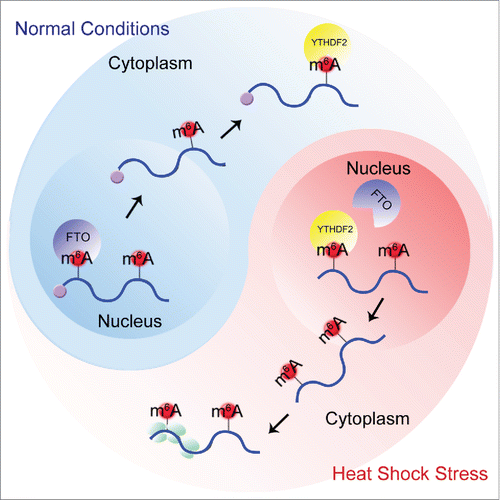
While the distribution pattern of m6A under physiological conditions is well understood, it remains unclear how the m6A landscape might change under different growth conditions. Using m6A-seq, we observed that heat shock stress triggered the m6A modification in the 5' untranslated region (5'UTR) of mRNAs.Citation4 5’UTR methylation had not previously received much attention, presumably due to the relatively low levels of m6A at this site under the normal conditions. Remarkably, elevated m6A levels selectively occurred in stress-induced transcripts like Hsp70 mRNA. In the elucidation of the mechanism underlying stress-induced 5’UTR methylation, we discovered several surprising features of the m6A "reader" YTHDF2 (). First, YTHDF2 is upregulated on both the mRNA and protein levels in response to heat shock stress, in a manner similar to Hsp70 induction. Second, upon heat shock treatment, nearly all of the YTHDF2 proteins translocate from the cytosol into the nucleus. Third, the nuclear YTHDF2 preserves m6A modification in the 5’UTR by competing with the demethylase FTO. As a result, only transcripts induced during stress exhibit elevated 5’UTR methylation.
Figure 1. A proposed model for the stress-inducible feature of m6A in the 5'UTR and its role in cap-independent translation. Under physiological conditions (blue), FTO removes m6A from the 5'UTR in the nucleus. Under heat shock stress (red), YTHDF2 relocates to the nucleus and protects the 5'UTR m6A. The resultant increase of 5'UTR methylation leads to cap-independent translation in the cytoplasm.
Cells often use methylation to distinguish portions of DNA from one another, as is apparent from the differences between methylation in self and foreign DNA, or parental and daughter DNA strands. We reason that 5’UTR methylation acts similarly—it permits the translation machinery to identify stress-induced messages from preexisting mRNAs, thereby achieving selective translation. Methylation in the 5’UTR is also relevant due to the critical role of the 5’UTR in controlling eukaryotic translation initiation. Under normal growth conditions, eukaryotic cells employ a cap-dependent mechanism to initiate translation for most mRNAs. The 7-methylguanosine (m7G) cap present at the 5’ end of most transcripts is known to recruit 43S ribosomal preinitiation complex via the eIF4F complex. Although our knowledge about the molecular events of cap-dependent translation is steadily increasing, very little is known about the molecular basis of translation initiation independent of the m7G cap. The best characterized cap-independent initiation mechanism involves the recognition of an internal ribosome entry site (IRES) in the mRNA.Citation5 However, beyond a few examples, many cellular mRNAs capable of cap-independent translation do not seem to contain any IRES elements.
It is commonly believed that the 5'UTR of Hsp70 mRNA acts as an IRES, but conflicting results have been described. We previously reported that the translation of heat shock-induced Hsp70 mRNA follows a cap- and IRES-independent manner.Citation6 Given the fact that m6A resembles m7G, we speculated that the stress-induced 5’UTR methylation enables selective translation by acting as a functional cap substitute. This is indeed the case. Using a reporter assay based on the firefly luciferase (Fluc), we demonstrated that the presence of the m6A in the 5’UTR of Hsp70 mRNA markedly increased the Fluc activity, indicating an increased level of translation, particularly under stress conditions. Importantly, when the m7G cap was replaced by a non-functional cap, m6A promotes the Fluc activity in response to heat shock stress. We further demonstrated that a single m6A site is able to facilitate cap-independent translation initiation.
How does m6A modification in the 5’UTR mediate translation initiation independent of the normal m7G cap? It is possible that distinct translation initiation factors are recruited to the methylated 5’UTR, thereby facilitating cap-independent translation. In a relevant study, Meyer et al. reported that 5’UTR m6A interacts with eIF3, a crucial mediator for the recruitment of 40S ribosome subunits to the mRNA.Citation7 Interestingly, m6A-mediated cap-independent translation still requires the scanning process of the pre-initiation complex along the 5’UTR. Further exploration of these concepts will likely yield insight into how m6A is recognized by the translation machinery and facilitates cap-independent initiation.
Our work marks the first time a stress-inducible feature of m6A in the 5’UTR was demonstrated in mammalian cells and introduces a surprising role for 5’UTR methylation in mediating cap-independent translation. Cap-independent translation occurs during normal cellular processes (for example, mitosis and apoptosis) or when the cap-dependent translation machinery is compromised by stress or disease. It has long been suggested that some cellular mRNAs exhibit a relaxed cap dependence without typical IRES structures, and we can now attribute the presence of m6A to this anomaly. The mechanistic connection between 5’UTR methylation and cap-independent translation not only expands the breadth of physiological roles of m6A, but also uncovers a new mode of non-canonical translation initiation.
References
- Dominissini D, et al. Nature 2012; 485:201–6; PMID: 22575960; http://dx.doi.org/10.1038/nature11112
- Meyer KD, et al. Cell 2012; 149:1635–46; PMID: 22608085; http://dx.doi.org/10.1016/j.cell.2012.05.003
- Wang X, et al. Nature 2014; 505:117–20; PMID: 24284625; http://dx.doi.org/10.1038/nature12730
- Zhou J, et al. Nature 2015; 526:591–4; PMID: 26458103; http://dx.doi.org/10.1038/nature15377
- Hellen CU, et al. Genes Dev 2001; 15:1593–1612; PMID: 11445534; http://dx.doi.org/10.1101/gad.891101
- Sun J, et al. J Biol Chem 2011; 286:6791–800; PMID: 21177857; http://dx.doi.org/10.1074/jbc.M110.172882
- Meyer KD, et al. Cell 2015; 163:999–1010; PMID:; http://dx.doi.org/10.1016/j.cell.2015.10.012
