The aggressive nature of many pediatric malignancies appears to be shaped by a relatively simple somatic genome, especially in comparison to adult tumors. However, when one considers the genome of the host from which these tumors arise, the landscape grows more complex. Our laboratory has focused on neuroblastoma, an aggressive childhood cancer of the developing nervous system. We have been guided by the hypothesis that germline variation in critical oncogenes and tumor suppressors leads to deregulated signaling that both initiates and sustains this malignancy. Through an ongoing genome-wide association study (GWAS), we have identified associations between neuroblastoma susceptibility and germline variation in several genes, including Lin-28 Homolog B (LIN28B) ().Citation1
Figure 1. LIN28B influences neuroblastoma susceptibility and disease presentation via a complex oncogenic signaling cascade offering possibilities for future therapeutic intervention. Shown under “Predisposition” is a Manhattan plot depicting the discovery of germline variants within LIN28B associated with neuroblastoma; cells harboring the risk allele express high levels of LIN28B and increased cell proliferation.Citation1 “Disease Presentation” illustrates a complex signaling cascade in neuroblastoma driven by LIN28B that involves RAN, AURKA, and MYCN.Citation3 Targeting of downstream LIN28B signaling with novel combinatorial approaches may improve the treatment of patients with high-risk neuroblastoma, as depicted in “Future Therapies.”
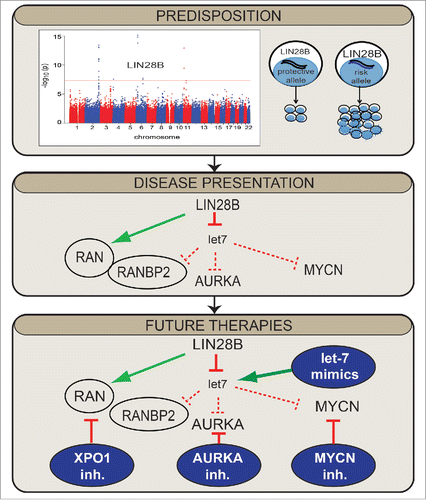
LIN28B is an RNA binding protein that blocks the maturation of let-7, a diverse microRNA family known to inhibit tumorigenesis. LIN28B also binds directly to mRNAs, likely influencing transcription and translation. We previously demonstrated that neuroblastoma cells harboring the germline risk-allele at the LIN28B locus showed high LIN28B expression and increased cell proliferation.Citation1 In addition, we showed that high LIN28B expression is associated with poor patient survival.Citation1 Providing further evidence that LIN28B drives tumorigenesis, mice overexpressing LIN28B in the developing adrenal gland developed neuroblastoma.Citation2
As multiple lines of investigation strongly supported an oncogenic role for LIN28B in neuroblastoma, we recently aimed to define key LIN28B-directed signaling networks.Citation3 We first analyzed gene expression data from neuroblastoma primary tumors and identified a strong positive correlation between LIN28B expression and RAN (RAS-related nuclear protein) signaling. RAN, a member of the Ras family of small GTPases, is well known for its role in nuclear trafficking, is overexpressed in diverse malignancies, and promotes AURKA (Aurora kinase A) phosphorylation. RAN is located at chromosome 12q24, a region of recurrent DNA copy number gain we and others have observed in neuroblastoma. We demonstrated that neuroblastomas with 12q gain were associated with increased RAN expression, suggesting that somatic gain might represent another means of driving RAN expression. We also showed that downregulation of RAN led to decreased cellular proliferation, mimicking LIN28B downregulation, and that RAN expression was correlated with advanced stage neuroblastoma and decreased overall survival.
Next, we demonstrated that LIN28B directly induced the expression of RAN, identifying 2 distinct mechanisms by which it does so (). First, we showed that RAN Binding Protein 2 (RANBP2) is a direct let-7 target. As RANBP2 had been shown to stabilize the expression of various proteins, including RAN, within the murine retina,Citation4 we hypothesized that it played a similar role in neuroblastoma and indeed demonstrated that RANBP2 knockdown led to decreased RAN protein levels. Next, we found that LIN28B binds RAN mRNA, constituting a second and direct mechanism of regulation. Finally, we demonstrated a convergence of LIN28B and RAN signaling on AURKA. LIN28B promoted RAN expression, which in turn induced higher levels of phosphorylated AURKA, leading to kinase activation. Unexpectedly, we also observed higher levels of total AURKA, implying that there might be RAN-independent mechanisms by which LIN28B influenced AURKA. We discovered that AURKA was a direct let-7 target, explaining the effect of LIN28B on total AURKA levels. As AURKA stabilizes MYCN at the level of the protein,Citation5 these findings reveal an intricate and novel signaling cascade connecting LIN28B, RAN, AURKA, and MYCN.
Our findings illustrate the ability of genome-wide association studies to inform our understanding of aberrant oncogenic signaling in neuroblastoma. Thus, LIN28B joins LMO1 and BARD1, as well as the long noncoding RNA CASC15-S, as factors in both initiating and sustaining neuroblastoma tumorigenesis. With the contribution of 12q24 gain to RAN expression in neuroblastoma subsets, the interplay of both germline and somatic variation in shaping the oncogenic phenotype is also illustrated. Of note, although we focused on the interactions between LIN28B and RAN itself, our analyses revealed strong correlations between LIN28B and additional RAN pathway members. These RAN pathway associated genes encode proteins that play key roles in the regulation of the cell cycle and apoptosis, nuclear trafficking, invasion, and metastasis. It will be interesting to dissect the influence of LIN28B on these genes and determine how these genes impact the phenotype of neuroblastoma.
From a translational genomics perspective, the elucidation of this network suggests potential novel combinatorial approaches that could target downstream LIN28B signaling and improve the treatment of patients with high-risk neuroblastoma (). While there are currently no therapies targeting RAN, inhibitors against XPO1/CRM1 (a protein that directly binds RAN and, like RAN, participates in nuclear export) are under investigation for both liquid and solid malignancies. Moreover, AURKA inhibitors, in combination with chemotherapy, are in Phase 2 trials for neuroblastomaCitation6 and Bromodomain and extraterminal (BET) inhibitors that target the MYC family, including MYCN, have demonstrated some efficacy in neuroblastoma preclinical models.Citation7 Novel-novel approaches inhibiting RAN/XPO1, AURKA, and MYCN, and promoting let-7 expression could provide a rational approach to target the LIN28B network, although further work to determine optimal combinations is needed. Given the diverse cancers in which deregulation of LIN28B is seen, it will be intriguing to determine whether this LIN28B-mediated network is seen in the context of other malignancies and whether modulating this pathway might improve the treatment of cancer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Diskin SJ, et al. Nat Genet 2012; 44(10):p. 1126-30; PMID:22941191; http://dx.doi.org/10.1038/ng.2387
- Molenaar JJ, et al. Nat Genet 2012; 44(11):p. 1199-206; PMID:23042116; http://dx.doi.org/10.1038/ng.2436
- Schnepp RW, et al. Cancer Cell 2015; 28(5):p. 599-609; PMID:26481147; http://dx.doi.org/10.1016/j.ccell.2015.09.012
- Patil H, et al. J Biol Chem 2014; 289(43):p. 29767-89; PMID:25187515; http://dx.doi.org/10.1074/jbc.M114.586834
- Otto T, et al. Cancer Cell 2009; 15(1):p. 67-78; PMID:19111882; http://dx.doi.org/10.1016/j.ccr.2008.12.005
- Mosse YP, et al. Clin Cancer Res 2012; 18(21):p. 6058-64; PMID:22988055; http://dx.doi.org/10.1158/1078-0432.CCR-11-3251
- Puissant A, et al. Cancer Discov 2013; 3(3):p. 308-23; PMID:23430699; http://dx.doi.org/10.1158/2159-8290.CD-12-0418
