abstract
E-cadherin-p120 catenin complexes are essential for adherens junction (AJ) formation and for the maintenance of the normal epithelial phenotype. PLEKHA7 was originally identified as a member of this complex that tethers microtubules to the AJs and supports their overall integrity. Recently, we revealed that PLEKHA7 regulates cellular behavior via miRNAs by associating with the microprocessor complex at the apical zonula adherens (ZA). We have also identified a new set of PLEKHA7 interacting partners at the apical ZA, via proteomics. Our analysis shows that the main groups of proteins associating with PLEKHA7 are cytoskeletal-related and RNA-binding proteins. Here, we provide extended evidence for association of PLEKHA7 with several of these proteins. We also show that PLEKHA7 loss activates the actin regulator cofilin in a p120-dependent manner, providing an explanation for the effects of PLEKHA7 on the cortical actin ring. Interestingly, PLEKHA7 regulates the levels and associates with PP1α, a phosphatase responsible for cofilin activation. Finally, we clarify the mode of regulation of the oncogenic miR-19a by PLEKHA7. Overall, our findings support a multi-layered role of PLEKHA7 in converging cytoskeletal dynamics and miRNA-mediated growth regulation at the ZA, with potentially critical implications in cancer that warrant further investigation.
Introduction
The adherens junctions (AJs) are critical for the development and maintenance of the epithelial phenotype. The core element of the AJs is the protein family of cadherins,Citation1,2 with E-cadherin (Ecad) being the main cadherin member in epithelial tissues. p120 catenin (p120) is an essential constituent of the AJs that interacts and stabilizes Ecad at areas of cell-cell contact.Citation2,3 Mature AJs form at apical regions of polarized epithelia, at the zonula adherens (ZA).Citation1 The ZA associates with a sub-membrane acto-myosin circumferential ring, which stabilizes the cellular architecture.Citation4 It has been well demonstrated that p120 regulates the activity of RhoGTPases, and thus the organization of the actomyosin cytoskeleton.Citation5-10 p120 also interacts with the microtubule network.Citation11-13
Recently, a new member of the ZA was identified, called PLEKHA7.Citation13 PLEKHA7 was originally described as ZA stabilizer and as an adaptor protein that links the AJs to the minus ends of the microtubules.Citation13 Although PLEKHA7 was identified as a p120-binding partner, it localizes specifically at the apical ZA, whereas p120 and Ecad localize also at the baso-lateral areas of cell-cell contacts.Citation14,15 We performed proteomics analysis of the PLEKHA7 interactions and reported a novel function of PLEKHA7 in regulating cell growth.Citation15 Our proteomics confirmed that PLEKHA7 forms an Ecad-p120-based complex specifically at the apical ZA, which is distinct from a basolateral-specific Ecad-p120 junctional complex.Citation15 Notably, this analysis revealed an extensive association of PLEKHA7 with cytoskeletal components.Citation15 In accordance, PLEKHA7 loss resulted in compromised cortical actin ring and decreased junctional impedance.Citation15 Importantly, we identified a novel interaction of PLEKHA7 with the microprocessor complex and its core components DROSHA and DGCR8 at the ZA.Citation15 Through this association, PLEKHA7 regulates the levels of certain miRNAs at the processing level, such as miR-30b, to suppress expression of pro-tumorigenic markers such as SNAI1, MYC, CCND1, p-Src and anchorage-independent growth (AIG). The findings revealed a novel function of the ZA in general and PLEKHA7 in particular in regulating cellular behavior, and connected two previously unrelated fields, cell adhesion and RNAi biology.
In this report, we extend our prior findings regarding PLEKHA7 with an emphasis on: a) its apparently broad association with the cytoskeleton; and b) its regulation of cell growth via changes in miRNA levels.
Results and discussion
PLEKHA7 is an apical junctional marker with broad cytoskeletal interactions
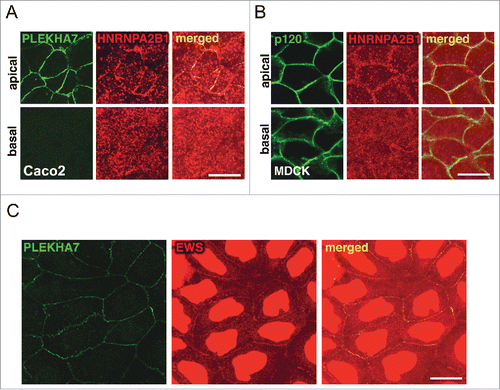
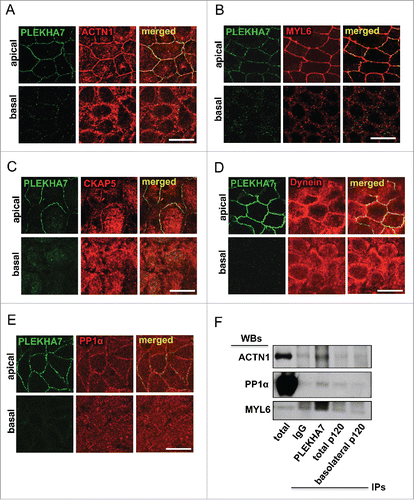
Our cross-linked proteomics revealed enrichment specifically of the apical PLEKHA7 junctional complex with several cytoskeletal proteins,Citation15 including: a) actin binding proteins, such as actin (ACTB), α-actinin (ACTN1), myosin light chain 6 (MYL6), actin-related protein 2/3 complex, subunit 5 (ARPC5), IQ motif containing GTPase activating protein 1 (IQGAP1), Filamin a, α FLNA; b) microtubule-related proteins, such as cytoskeleton associated protein 5 (CKAP5), Dynein, Cytoplasmic 1, Heavy Chain 1 (DYNC1H1); and c) intermediate filament proteins, like Periplakin (PPL) and Filaggrin family member 2 (FLG2). We already demonstrated the association of PLEKHA7 with IQGAP1, both by immunofluorescence (IF) and co-immunoprecipitation (co-IP).Citation15 We have also confirmed that PLEKHA7 co-IPs with ACTN1. We show here that PLEKHA7 also co-localizes with ACTN1 only apically at the ZA in polarized Caco2 cells (). In addition, MYL6 shows strong apical junctional co-localization with PLEKHA7 by IF (). PLEKHA7 was initially described as a protein that associates with the minus ends of microtubules through a direct interaction with Nezha and KIFC3.Citation13 Surprisingly, our proteomics revealed the presence of CKAP5 in the PLEKHA7 complex,Citation15 a centrosome-organizing protein that also binds and stabilizes the plus-ends of the microtubules.Citation16,17 IF of polarized Caco2 cells confirm that CKAP5 localizes at the apical ZA, together with PLEKHA7 (). A similar apical junctional co-localization is also evident for Dynein (), a protein thought to associate directly with β-catenin and to tether microtubules to the AJs.Citation18 Interestingly, protein phosphatase 1α (PP1α; PPP1CA), a major phosphatase that regulates actin dynamics,Citation19-21 was also identified to be specifically associated with the apical complex in our proteomics. PP1α regulates actin dynamics by dephosphorylating cofilin.Citation19-21 IF of polarized Caco2 cells confirmed the apical junctional localization of PP1α and its co-localization with PLEKHA7 (). Western blot of the isolated apical and basolateral immunoprecipitates, obtained as previously described,Citation15 confirmed the specific binding of PLEKHA7 with ACTN1, MYL6 and PP1α (). The data further support an extensive interaction of PLEKHA7 not only with the microtubules, but also with the actin network at the apical ZA.
Figure 1. PLEKHA7 associates with several cytoskeletal regulators at the ZA. Caco2 cells were grown for 21 days to polarize and subjected to IF for PLEKHA7 and co-stained for: (A) ACTN1; (B) MYL6; (C) CKAP5; (D) Dynein; and (E) PP1. In all cases, stained cells were imaged by confocal microscopy and image stacks were acquired, covering the entire polarized monolayer between the basal and the apical level. Representative x-y image stacks and merged composite x-z images are shown. Scale bars: 20 μM. (F) Western blot of the lysates from the immunoprecipitated fractions of PLEKHA7 (apical complex), total p120, and basolateral p120, isolated as previously described,Citation15 for the markers shown. IgG is the negative immunoprecipitation control.
PLEKHA7 maintains the integrity of the apical cortical actin ring
We and others have shown that PLEKHA7 knockdown results in decreased cell-cell junction impedance and compromised cortical actin ring.Citation13,15 Concomitantly, PLEKHA7 overexpression strengthens the actin ring in Caco2 cells (). It also induces robust α-catenin localization to the apical junctions (), which is the main link of cadherin complexes to cortical actin.Citation22-24 While the mechanism of α-catenin recruitment and/or stabilization is currently unclear, PLEKHA7 seems to actively promote the association of the actin ring at the ZA.
Figure 2. PLEKHA7 stabilizes the actin cytoskeleton at the ZA. Caco2 cells were transiently transfected with a PLEKHA7-GFP construct and stained by IF for PLEKHA7 and (A) Actin (phalloidin) or (B) α-catenin. Scale bars: 20 μM. Enlarged image parts are shown to the right. (C) Control, PLEKHA7 knockdown (shPLEKHA7), and PLEKHA7-p120 (shPLEKHA7, shp120) double knockdown Caco2 cells were analyzed by western blot for the markers shown. Actin is the loading control. (D) Control (NT) or PLEKHA7 knockdown (shPLEKHA7#8 and #10) Caco2 cells were analyzed by protein gel blot for PP1α expression. Actin is the loading control.
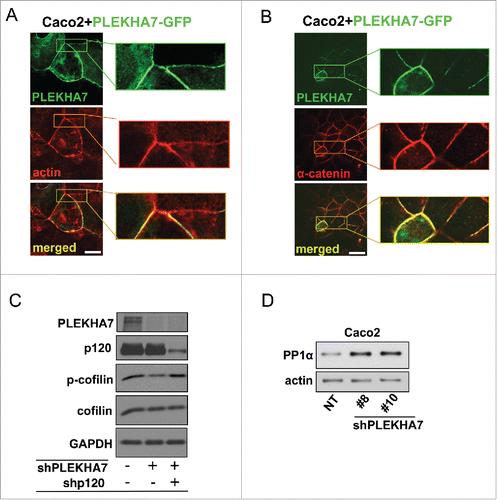
Actin dynamics play a key role in junction formationCitation23,25,26 and regulate cellular signaling.Citation27,28 Through its ability to sever actin filaments and produce monomeric G-actin, cofilin is a major regulator of actin dynamics.Citation29 It has been demonstrated that p120 promotes AIG via cofilin activation.Citation30 We reported that basolateral p120 mediates the AIG induced by PLEKHA7 depletion.Citation15 Here, we show that PLEKHA7 knockdown results in cofilin activation, as indicated by decreased phosphorylation of cofilin at the inhibitory S3 (). Importantly, S3 cofilin phosphorylation is restored to its normal levels in the PLEKHA7-depleted cells, when p120 is simultaneously knocked down. This indicates that cofilin activation upon PLEKHA7 depletion is indeed p120-dependent (). Importantly, PP1α is a major serine phosphatase that dephosphorylates cofilin at S3Citation20,21 and PP1α co-precipitated and co-localized with PLEKHA7 at the ZA. It is likely that the select recruitment of PP1α at the ZA regulates overall cofilin activation and actin filament severing, thus promoting an epithelial phenotype. Additionally, PLEKHA7 knockdown results in increased PP1α levels (), suggesting that the decreased cofilin phosphorylation and increased actin severing upon PLEKHA7 depletion may be mediated, at least in part, by PP1α. Collectively, the data further strengthen the notion that PLEKHA7 is essential for the integrity of the cortical actin ring.
PLEKHA7 associates with several RNA-binding proteins at the ZA
In our recent work, we revealed association of PLEKHA7 with the two core components of the microprocessor complex, DROSHA and DGCR8 at the ZA.Citation15 However, our cross-linked proteomics also revealed association of junctional complexes with several other accessory members of the microprocessor and of other RNA complexes.Citation31-34 Most of them were identified to specifically co-precipitate with the apical complex, such as DDX1, DDX4, DDX17, RALY, HNRNPA2B1, HNRNPA1L2, HNRNPA3, whereas EWS and FUS were found in both the apical and basolateral complexes. We previously confirmed binding of HNRNPA2B1 specifically with the apical complex and of EWS with both complexes.Citation15 Here, we show the co-localization of HNRNPA2B1 with PLEKHA7 at the apical ZA, in polarized Caco2 and MDCK cells (). We also confirm the junctional localization of EWS (). Interestingly, it has been shown that CKAP5, an apical-specific PLEKHA7 partnerCitation15 (), associates with HNRNPA2B1Citation35 and mediates RNA granule trafficking via the microtubules.Citation35 Although we reported that localization of DROSHA and DGCR8 at the junctions is microtubule-independent,Citation15 this observation could provide a mechanism for the transport of particular pri-miRNAs to the junctions. In addition, several of the RNA-binding proteins specifically identified with the apical complex at the ZA are known to play roles in pri-miRNA transport and microprocessor function.Citation32,34,36,37 Overall, the interaction of PLEKHA7 with RNA-binding proteins seems to be extensive and implies for multiple levels of regulation of the junctional microprocessor.
PLEKHA7 loss induces increased expression of miR-19a at the transcriptional level
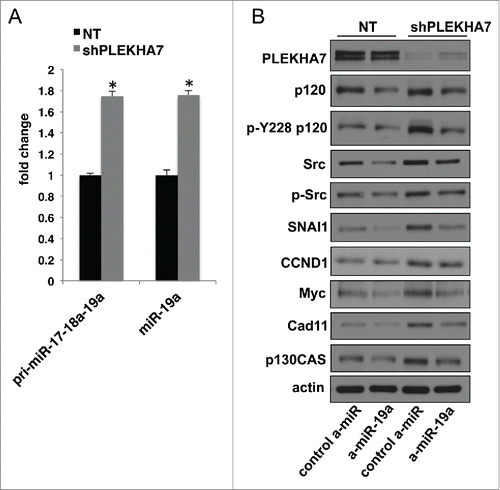
PLEKHA7 regulates the levels of a select set of 29 miRNAs in Caco2 cells; some are downregulated and some upregulated upon PLEKHA7 loss.Citation15 Notably, those decreased were known anti-tumorigenic miRNAs, whereas those upregulated were pro-tumorigenic, corroborating the net signaling and phenotypic effects of PLEKHA7 loss toward a pro-tumorigenic phenotype. We have provided a mechanism that explains how PLEKHA7 loss results in down-regulation of certain anti-tumorigenic miRNAs, such as miR-30b. In particular, we reported that PLEKHA7 promotes miRNA processing at the ZA via its association with the microprocessor complex.Citation15 However, this could not directly explain why some miRNAs are upregulated upon PLEKHA7 knockdown, such as miR-19a.Citation15 miR-19a is expressed as part of the miR-17-92 polycistronic miRNA, which is considered pro-tumorigenic by multiple studies and alternatively referred to as Oncomir-1.Citation38-40 To examine which level at the miR-19a biogenesis was affected by PLEKHA7 loss, we performed a qPCR analysis using probes that detect the pri-miR-17-18a-19a region within the pri-miR-17-92 primary transcript, which contains the miR-19a.Citation32 The analysis showed that PLEKHA7 knockdown results in almost the same upregulation of the pri-miR-17-18a-19a as of the mature miR-19a (), indicating that upregulation of miR-19a occurs at the transcriptional level. It has been shown that Myc promotes expression of miR-19a.Citation41 Myc expression is induced upon PLEKHA7 knockdown, in a miR-30b-dependent manner.Citation15 Therefore, it is likely that upregulation of miR-19a expression is an indirect effect of PLEKHA7 depletion, suggesting that PLEKHA7 affects miRNA expression at multiple levels. Interestingly, miR-19a knockdown using an anti-miR in PLEKHA7-depleted cells reversed the increased levels of a series of pro-tumorigenic markers that are induced upon PLEKHA7 knockdown,Citation15 such as SNAI1, total Src, pY228-p120 or p130CAS (). This argues that miR-19a regulation has functional effects, although its regulation by PLEKHA7 is probably indirect. Interestingly, a recent report has indicated the involvement of miR-19a in colon tumorigenesis,Citation42 which is consistent with our findings in the colonic Caco2 cells. Together, the data further support the anti-tumorigenic role of PLEKHA7, via regulation of miRNA expression at multiple levels.
Figure 4. PLEKHA7 loss results in upregulation of growth-related markers via miR-19a (A) qRT-PCR analysis of Caco2 control (NT) or PLEKHA7 knockdown (shPLEKHA7) cells for pri-miR-17-18a-19a and mature miR-19a (mean ± s.d. from n = 3 independent experiments; *P < 0.0001, Student's two-tailed t-test). (B) Western blot of NT and shPLEKHA7 Caco2 cells transfected with the either control or anti-miR-19a constructs and blotted for the markers shown; Actin is the loading control.
Conclusions
PLEKHA7 is becoming a protein of increasing interest due to its multi-layered effects in cellular architecture and cell growth. The multiple associations of PLEKHA7 with actin-binding proteins and its critical role in the maintenance of the cortical actin ring warrant further investigation of the details of this mechanism. The emerging involvement of PLEKHA7 in glaucomaCitation43,44 and hypertension,Citation45,46 both conditions that stem from compromised epithelial or endothelial monolayer tension and integrity, could be explained on the basis of its effects on the actin ring. It is likely that several of the cytoskeletal PLEKHA7 partners identified at the apical ZA play important roles in these conditions. In addition, association of PLEKHA7 with the microprocessor and with several accessory RNA-binding proteins further supports the function of PLEKHA7 in regulating miRNA levels, which is critical for suppression of pro-tumorigenic signaling ().Citation15 We and others have shown loss of PLEKHA7 in tumor tissues,Citation15,47 which together with the well-established roles of miRNAs in tumorigenesisCitation48,49 implies for a regulatory role of this protein in tumor progression. The accessory RNA-binding proteins that we identified, together with the associated cytoskeletal components in the same apical complex, may be key to the selectivity of PLEKHA7 action toward a particular subset of cellular miRNAs. Thus, investigation of the cross talk between the adhesive, cytoskeletal, and signaling functions of PLEKHA7 may be important in fully delineating its role in cell growth. Taken together, these findings suggest that the ZA is not only a structural component but also a regulatory hub in epithelial cells, orchestrating a convergence of cellular architecture with epithelial behavior, via PLEKHA7.
Materials and methods
Cell culture
In all comparisons, cells were used strictly at the same confluences. All cell lines were obtained from ATCC, used at low passage (<20), and tested negative for mycoplasma contamination. Caco2 colon epithelial cells were cultured in MEM (Cellgro) supplemented with 10% FBS (Invitrogen), 1 mM sodium pyruvate (Invitrogen) and 1x non-essential amino-acid supplement (Mediatech). MDCK canine kidney epithelial cells were cultured in DMEM supplemented with heat-inactivated 10% FBS.
Constructs, shRNAs, and anti-miRs
Cells were transfected using Lipofectamine 2000 (Invitrogen) or Lipofectamine RNAiMAX (Invitrogen) for anti-miR transfection, according to the manufacturer's protocols. The PLEKHA7-GFP construct was a gift of the M. Takeichi Lab (RIKEN, Japan).Citation13 Lentiviral shRNAs were derived from the pLKO.1-based TRC1 (Sigma - RNAi consortium) shRNA library (pLKO.1-puro Non-Target shRNA Control: SHC016; PLEKHA7 #8: TRCN0000146289; PLEKHA7 #10: TRCN0000127584. Lentiviruses were produced in HEK 293FT cells and used to infect cells according to standard protocols. Retroviruses were prepared in Phoenix-Ampho cells, as described previously.Citation50 mirVana (Life technologies, cat# 4464084) anti-miRs used: anti-hsa-miR-19a, ID MH10649; negative control #1 (cat# 4464076).
Antibodies
PLEKHA7 (Sigma, HPA038610); p120 (15D2, F1aSHCitation51); p-p120: Y228 (BD Transduction Labs, P21420); α-catenin (BD Transduction Labs, 610193); Cad11 (Zymed, 32-1700 and 71-7600); Src (Cell Signaling, 2123, clone 32G6), p-Src: Y416 (Cell Signaling, 2101); GFP (Invitrogen, A11120 - clone 3E6); Actin (Sigma, A2066); p130CAS (BD Transduction Labs, p27820); SNAI1 (Cell Signaling, L70G2); CCND1 (Cell Signaling, 2922); MYC (Zymed 13-2500 - clone 9E10); cofilin (Cell Signaling, 3312); p-cofilin (S3, Cell Signaling, 3313 - clone 77G2); EWS (Santa Cruz, sc-48404); HNRNPA2B1 (Sigma, R4653 - clone DP3B3); ACTN1 (Santa Cruz, sc-15335); Dynein (Novus Biologicals, H00064446-M01); PP1α (Invitrogen, 438100 - clone 10C6.3); CKAP5 (Novus Biologicals, NB500-182); MYL6 (LifeSpan, LS-C102726); GAPDH (Cell Signaling, 2118, clone 14C10). Antibodies were used at 1:250-1:2000 dilutions for western blot and at 1:50-1:500 for IF. Secondary antibodies used: HRP-anti-mouse (Jackson ImmunoResearch, 715-035-150); HRP-anti-rabbit (Jackson ImmunoResearch, 711-035-152); HRP-anti-goat (Jackson ImmunoResearch, 705-035-003); Alexa 488 anti-mouse (Invitrogen, A-11029); Alexa 488 anti-rabbit (Invitrogen, A11034); Alexa 594 anti-mouse (Invitrogen, A-11005); Alexa 594 anti-rabbit (Invitrogen, A-11037); Alexa 594 anti-goat (Invitrogen, A-11058). HRP-conjugated secondaries were used for protein gel blot at 1:2000 dilution. Alexa-conjugated secondaries were used for IF at 1:500 dilution. Alexa 594 Phalloidin (Invitrogen, A12381) was used to detect actin filaments in IF experiments at 1:100 dilution.
Immunofluorescence
Caco2 and MDCK cells were grown on transwell inserts (Costar 3413) for 7 or 21 days respectively, until they polarized, or on sterile glass coverslips until they reach full confluence. Cells were washed once with PBS and then fixed with either: i) 100% methanol (Fisher) for 7 min; or ii) 4% formaldehyde (EMS) for 20 min, followed by 0.02% Triton-X 100 permeabilization for 10 min; or iii) 10% TCA (Sigma) for 15 min on ice, particularly for MYL6 and PP1α stainings, and permeabilized as above. Cells were blocked with either 3% non-fat milk (Carnation) in PBS, or Protein-Block reagent (Dako, X090930-2) for 30 min and stained with primary antibodies diluted either in milk or Antibody Diluent (Dako, S302281-2) for 1 h. Cells were then washed 3x with PBS, stained with the fluorescent-labeled secondary antibodies for 1 h, washed 3x with PBS, co-stained with DAPI (Sigma) to visualize the nuclei, mounted (Aqua Poly/Mount, Polysciences), and imaged using a Zeiss LSM 510 META laser confocal microscope, under a 63x objective, with an additional 1.6x zoom. z-stacks were acquired in 0.5 μM intervals. Images were processed using the Zen software (Zeiss).
Immunoblotting
Whole cell extracts were obtained using RIPA buffer (Tris, pH 7.4, 50 mM NaCl, 150 mM, NP-40, 1%, Deoxycholic Acid, 0.5%, SDS, 0.1%) supplemented with protease (cocktail III, RPI) and phosphatase inhibitors (Pierce). Lysates were homogenized through a 29g needle and cleared by full speed centrifugation for 5min. Protein extracts were mixed with LSB and separated by SDS-PAGE, transferred to nitrocellulose membranes (Bio-Rad), blotted according to standard protocols, detected by luminescence using ECL (GE Healthcare) and imaged using X-ray films (Pierce).
Immunoprecipitations (IPs)
Cross-linked IPs were performed as previously described.Citation15 Caco2 cells were grown on 10cm plates with transwell inserts (Costar 3419) for 21 days to polarize. Cells were then washed 2x with PBS and proteins were cross-linked, using 0.75 mM of the reversible cross-linker DSP (Lomant's Reagent; Pierce) for 30 min at RT, which was then neutralized using 20 mM Tris pH 7.5 for 15 min. Cells were finally lysed using RIPA (see recipe above) containing 2x concentration of proteinase (protease inhibitor cocktail III, RPI) and phosphatase inhibitors (Pierce) and 1mM EDTA. Four 10 cm transwells (˜3×106 cells) were used per IP. In parallel, 4 μg of antibody or IgG control (rabbit, Jackson ImmunoResearch, 011-000-003) were incubated with 40 μl Protein G Dynabeads (Invitrogen) O/N and cross-linked to the beads using 5 mM BS3 (Pierce) according to the manufacturer's protocol. Cross-linked lysates were finally incubated with the beads-conjugated antibodies O/N. Following incubation, beads were washed 3x with lysis buffer and proteins were extracted using 50 mM of DTT (Sigma) in lysis buffer. Eluates were mixed with LSB and separated by SDS-PAGE.
Total RNA isolation and qRT-PCR
Cells were lysed using Trizol (Invitrogen) and subjected to the Trizol Plus Total Transcriptome Isolation protocol of the PureLink RNA mini kit (Ambion - Life Technologies) specified to isolate both mRNAs and miRNAs. Final RNA concentrations were determined using a NanoDrop spectrophotometer. RNA was converted to cDNA using the High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems). qPCR reactions were performed using the Taqman FAST Universal PCR master mix (Applied Biosystems), in a 7900 HT or ViiA™ 7 Thermocycler (Applied Biosystems). Data were analyzed using RQ Manager (Applied Biosystems). U6 was used as a control for miRNA expression normalization and GAPDH, β-actin for pri-miRNA normalization. TaqMan assays used for miRNAs (Applied Biosystems, cat# 4427975): hsa-miR-19a, 000395; U6, 001973. TaqMan assays for: pri-miR-17-18a-19a (Applied Biosystems, cat# 4427012): Hs03295901_pri. TaqMan assays for mRNAs (Applied Biosystems, cat# 4331182): GAPDH, Hs99999905_m1; β-actin, Hs99999903_m1. Averages and standard deviations (s.d.) were calculated and presented as error bars, from three independent experiments. The Student's two-tailed t-test was employed for P value calculations since all comparisons were between two groups, control and experimental condition.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Masatoshi Takeichi for constructs.
Funding
This work was supported by NIH R01 CA100467, R01 NS069753, P50 CA116201 (PZA); AK is supported by the Jay and Deanie Stein Career Development Award for Cancer Research at Mayo Clinic.
References
- Nishimura T, Takeichi M. Remodeling of the adherens junctions during morphogenesis. Curr Top Dev Biol 2009; 89:33-54; PMID:19737641; http://dx.doi.org/10.1016/S0070-2153(09)89002-9
- Kourtidis A, Ngok SP, Anastasiadis PZ. p120 catenin: an essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Prog Mol Biol Transl Sci 2013; 116:409-32; PMID:23481205; http://dx.doi.org/10.1016/B978-0-12-394311-8.00018-2
- Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, Anastasiadis PZ, Matrisian L, Bundy LM, Sealy L, et al. A novel role for p120 catenin in E-cadherin function. J Cell Biol 2002; 159:465-76; PMID:12427869; http://dx.doi.org/10.1083/jcb.200205115
- Miyoshi J, Takai Y. Structural and functional associations of apical junctions with cytoskeleton. Biochim Biophys Acta 2008; 1778:670-91; PMID:18201548; http://dx.doi.org/10.1016/j.bbamem.2007.12.014
- Anastasiadis PZ, Moon SY, Thoreson MA, Mariner DJ, Crawford HC, Zheng Y, Reynolds AB. Inhibition of RhoA by p120 catenin. Nat Cell Biol 2000; 2:637-44; PMID:10980705; http://dx.doi.org/10.1038/35023588
- Noren NK, Liu BP, Burridge K, Kreft B. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J Cell Biol 2000; 150:567-80; PMID:10931868; http://dx.doi.org/10.1083/jcb.150.3.567
- Grosheva I, Shtutman M, Elbaum M, Bershadsky AD. p120 catenin affects cell motility via modulation of activity of Rho-family GTPases: a link between cell-cell contact formation and regulation of cell locomotion. J Cell Sci 2001; 114:695-707; PMID:11171375
- Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, Lobdell NA, Settleman J, Reynolds AB. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell 2006; 127:1027-39; PMID:17129786; http://dx.doi.org/10.1016/j.cell.2006.09.046
- Schackmann RC, van Amersfoort M, Haarhuis JH, Vlug EJ, Halim VA, Roodhart JM, Vermaat JS, Voest EE, van der Groep P, van Diest PJ, et al. Cytosolic p120-catenin regulates growth of metastatic lobular carcinoma through Rock1-mediated anoikis resistance. J Clin Invest 2011; 121:3176-88; PMID:21747168; http://dx.doi.org/10.1172/JCI41695
- Smith AL, Dohn MR, Brown MV, Reynolds AB. Association of Rho-associated protein kinase 1 with E-cadherin complexes is mediated by p120-catenin. Mol Biol Cell 2012; 23:99-110; PMID:22031287; http://dx.doi.org/10.1091/mbc.E11-06-0497
- Yanagisawa M, Kaverina IN, Wang A, Fujita Y, Reynolds AB, Anastasiadis PZ. A novel interaction between kinesin and p120 modulates p120 localization and function. J Biol Chem 2004; 279:9512-21; PMID:14676216; http://dx.doi.org/10.1074/jbc.M310895200
- Chen X, Kojima S, Borisy GG, Green KJ. p120 catenin associates with kinesin and facilitates the transport of cadherin-catenin complexes to intercellular junctions. J Cell Biol 2003; 163:547-57; PMID:14610057; http://dx.doi.org/10.1083/jcb.200305137
- Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell 2008; 135:948-59; PMID:19041755; http://dx.doi.org/10.1016/j.cell.2008.09.040
- Pulimeno P, Bauer C, Stutz J, Citi S. PLEKHA7 is an adherens junction protein with a tissue distribution and subcellular localization distinct from ZO-1 and E-cadherin. PloS One 2010; 5:e12207; PMID:20808826; http://dx.doi.org/10.1371/journal.pone.0012207
- Kourtidis A, Ngok SP, Pulimeno P, Feathers RW, Carpio LR, Baker TR, Carr JM, Yan IK, Borges S, Perez EA, et al. Distinct E-cadherin-based complexes regulate cell behaviour through miRNA processing or Src and p120 catenin activity. Nat Cell Biol 2015; 17:1145-57; PMID:26302406; http://dx.doi.org/10.1038/ncb3227
- Nakamura S, Grigoriev I, Nogi T, Hamaji T, Cassimeris L, Mimori-Kiyosue Y. Dissecting the nanoscale distributions and functions of microtubule-end-binding proteins EB1 and ch-TOG in interphase HeLa cells. PloS One 2012; 7:e51442
- Cassimeris L, Morabito J. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol Biol Cell 2004; 15:1580-90; PMID:14718566; http://dx.doi.org/10.1091/mbc.E03-07-0544
- Ligon LA, Karki S, Tokito M, Holzbaur EL. Dynein binds to β-catenin and may tether microtubules at adherens junctions. Nat Cell Biol 2001; 3:913-7; PMID:11584273; http://dx.doi.org/10.1038/ncb1001-913
- Zhang Y, Kim TH, Niswander L. Phactr4 regulates directional migration of enteric neural crest through PP1, integrin signaling, and cofilin activity. Genes Dev 2012; 26:69-81; PMID:22215812; http://dx.doi.org/10.1101/gad.179283.111
- Oleinik NV, Krupenko NI, Krupenko SA. ALDH1L1 inhibits cell motility via dephosphorylation of cofilin by PP1 and PP2A. Oncogene 2010; 29:6233-44; PMID:20729910; http://dx.doi.org/10.1038/onc.2010.356
- Ambach A, Saunus J, Konstandin M, Wesselborg S, Meuer SC, Samstag Y. The serine phosphatases PP1 and PP2A associate with and activate the actin-binding protein cofilin in human T lymphocytes. Eur J Immunol 2000; 30:3422-31; PMID:11093160; http://dx.doi.org/10.1002/1521-4141(2000012)30:12<3422::AID-IMMU3422>3.0.CO;2-J
- Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science 2014; 346:1254211; PMID:25359979; http://dx.doi.org/10.1126/science.1254211
- Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell 2000; 100:209-19; PMID:10660044; http://dx.doi.org/10.1016/S0092-8674(00)81559-7
- Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci U S A 1995; 92:8813-7; PMID:7568023; http://dx.doi.org/10.1073/pnas.92.19.8813
- Chen CS, Hong S, Indra I, Sergeeva AP, Troyanovsky RB, Shapiro L, Honig B, Troyanovsky SM. α-Catenin-mediated cadherin clustering couples cadherin and actin dynamics. J Cell Biol 2015; 210:647-61; PMID:26261181; http://dx.doi.org/10.1083/jcb.201412064
- Quinlan MP, Hyatt JL. Establishment of the circumferential actin filament network is a prerequisite for localization of the cadherin-catenin complex in epithelial cells. Cell Growth Differ 1999; 10:839-54; PMID:10616909
- Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of α-catenin in skin. Cell 2001; 104:605-17; PMID:11239416; http://dx.doi.org/10.1016/S0092-8674(01)00246-X
- Papakonstanti EA, Stournaras C. Cell responses regulated by early reorganization of actin cytoskeleton. FEBS Lett 2008; 582:2120-7; PMID:18325339; http://dx.doi.org/10.1016/j.febslet.2008.02.064
- Bamburg JR, Bernstein BW. Roles of ADF/cofilin in actin polymerization and beyond. F1000 Biol Rep 2010; 2:62; PMID:21173851
- Dohn MR, Brown MV, Reynolds AB. An essential role for p120-catenin in Src- and Rac1-mediated anchorage-independent cell growth. J Cell Biol 2009; 184:437-50; PMID:19188496; http://dx.doi.org/10.1083/jcb.200807096
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004; 432:235-40; PMID:15531877; http://dx.doi.org/10.1038/nature03120
- Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat struct Mol Biol 2007; 14:591-6; PMID:17558416; http://dx.doi.org/10.1038/nsmb1250
- Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci U S A 2006; 103:2647-52; PMID:16477042; http://dx.doi.org/10.1073/pnas.0509333103
- Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 2014; 156:893-906; PMID:24581491; http://dx.doi.org/10.1016/j.cell.2013.12.043
- Kosturko LD, Maggipinto MJ, D'Sa C, Carson JH, Barbarese E. The microtubule-associated protein tumor overexpressed gene binds to the RNA trafficking protein heterogeneous nuclear ribonucleoprotein A2. Mol Biol Cell 2005; 16:1938-47; PMID:15703215; http://dx.doi.org/10.1091/mbc.E04-08-0709
- Guil S, Long JC, Caceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol 2006; 26:5744-58; PMID:16847328; http://dx.doi.org/10.1128/MCB.00224-06
- Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol 2007; 9:604-11; PMID:17435748; http://dx.doi.org/10.1038/ncb1577
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature 2005; 435:828-33; PMID:15944707; http://dx.doi.org/10.1038/nature03552
- Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 2010; 42:1348-54; PMID:20227518; http://dx.doi.org/10.1016/j.biocel.2010.03.004
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev 2009; 23:2839-49; PMID:20008935; http://dx.doi.org/10.1101/gad.1861409
- Loven J, Zinin N, Wahlstrom T, Muller I, Brodin P, Fredlund E, Ribacke U, Pivarcsi A, Pahlman S, Henriksson M. MYCN-regulated microRNAs repress estrogen receptor-α (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci U S A 2010; 107:1553-8; PMID:20080637; http://dx.doi.org/10.1073/pnas.0913517107
- Huang L, Wang X, Wen C, Yang X, Song M, Chen J, Wang C, Zhang B, Wang L, Iwamoto A, et al. Hsa-miR-19a is associated with lymph metastasis and mediates the TNF-α induced epithelial-to-mesenchymal transition in colorectal cancer. Sci Rep 2015; 5:13350; PMID:26302825; http://dx.doi.org/10.1038/srep13350
- Lee MC, Chan AS, Goh SR, Hilmy MH, Nongpiur ME, Hong W, Aung T, Hunziker W, Vithana EN. Expression of the Primary Angle Closure Glaucoma (PACG) Susceptibility Gene PLEKHA7 in Endothelial and Epithelial Cell Junctions in the Eye. Invest Ophthalmol Vis Sci 2014; 55:3833-41; PMID:24801512; http://dx.doi.org/10.1167/iovs.14-14145
- Shuai P, Yu M, Li X, Zhou Y, Liu X, Liu Y, Zhang D, Gong B. Genetic associations in PLEKHA7 and COL11A1 with primary angle closure glaucoma: a meta-analysis. Clin Exp Ophthalmol 2015; 43:523-30; http://dx.doi.org/10.1111/ceo.12516
- Endres BT, Priestley JR, Palygin O, Flister MJ, Hoffman MJ, Weinberg BD, Grzybowski M, Lombard JH, Staruschenko A, Moreno C, et al. Mutation of Plekha7 attenuates salt-sensitive hypertension in the rat. Proc Natl Acad Sci USA 2014; 111:12817-22; PMID:25136115; http://dx.doi.org/10.1073/pnas.1410745111
- Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet 2009; 41:677-87; PMID:19430479; http://dx.doi.org/10.1038/ng.384
- Tille JC, Ho L, Shah J, Seyde O, McKee TA, Citi S. The Expression of the Zonula Adhaerens Protein PLEKHA7 is strongly decreased in high grade ductal and lobular breast carcinomas. PloS One 2015; 10:e0135442; PMID:26270346; http://dx.doi.org/10.1371/journal.pone.0135442
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Ann Rev Pathol 2014; 9:287-314; http://dx.doi.org/10.1146/annurev-pathol-012513-104715
- Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle (Georgetown, Tex 2008; 7:2643-6; PMID:18719391; http://dx.doi.org/10.4161/cc.7.17.6597
- Soto E, Yanagisawa M, Marlow LA, Copland JA, Perez EA, Anastasiadis PZ. p120 catenin induces opposing effects on tumor cell growth depending on E-cadherin expression. J Cell Biol 2008; 183:737-49; PMID:19015320; http://dx.doi.org/10.1083/jcb.200805113
- Wu J, Mariner DJ, Thoreson MA, Reynolds AB. Production and characterization of monoclonal antibodies to the catenin p120ctn. Hybridoma 1998; 17:175-83; PMID:9627058; http://dx.doi.org/10.1089/hyb.1998.17.175