ABSTRACT
The ribosomal GTPase associated center constitutes the ribosomal area, which is the landing platform for translational GTPases and stimulates their hydrolytic activity. The ribosomal stalk represents a landmark structure in this center, and in eukaryotes is composed of uL11, uL10 and P1/P2 proteins. The modus operandi of the uL11 protein has not been exhaustively studied in vivo neither in prokaryotic nor in eukaryotic cells. Using a yeast model, we have brought functional insight into the translational apparatus deprived of uL11, filling the gap between structural and biochemical studies. We show that the uL11 is an important element in various aspects of ‘ribosomal life’. uL11 is involved in ‘birth’ (biogenesis and initiation), by taking part in Tif6 release and contributing to ribosomal subunit-joining at the initiation step of translation. uL11 is particularly engaged in the ‘active life’ of the ribosome, in elongation, being responsible for the interplay with eEF1A and fidelity of translation and contributing to a lesser extent to eEF2-dependent translocation. Our results define the uL11 protein as a critical GAC element universally involved in trGTPase ‘productive state’ stabilization, being primarily a part of the ribosomal element allosterically contributing to the fidelity of the decoding event.
Introduction
One of the important elements on the ribosome is the GTPase Associated Center (GAC) situated on the large ribosomal subunit, which represents a platform for binding and stimulation of translational GTPases (trGTPases).Citation1 The protein part of the GAC forms a protruding structure on the 60S subunit, which is composed of P-proteins forming a pentameric complex uL10-(P1-P2)2,Citation2 and protein uL11, which together with uL10 constitute a base of the structure. This lateral protuberance is called the P-stalk, and, contrary to the L1-stalk, which is situated in the vicinity of the ribosomal E-site, is located close to the A-site, also known as the A/T-site, and is responsible for mediating interactions of the ribosome with trGTPases.Citation3-5 The pentameric stalk-complex has attracted a lot of attention, but other GAC elements, such as uL11, have been characterized in less detail. The uL11 protein is anchored to the rRNA thiostreptone loop via a globular rRNA binding domain located at the C-terminus, whereas the N-terminal domain faces trGTPase. Both N- and C-terminal domains of the uL11 protein are connected with a flexible spacer, which enables their movement in respect to each other and conformational rearrangement.Citation6 Such flexibility is necessary for the full biological properties, because different orientation of the N-terminal domain in respect to the ribosome-anchored C-terminus was observed in different states of the translating ribosome.Citation7 Structural analyses of the bacterial ribosome showed that during the translation elongation cycle uL11 works through the so-called double-switching mechanism and can be considered as a molecular sensor to monitor and control the translational state of the ribosome, allowing cyclic interactions of trGTPases with the ribosome.Citation8 Although it is considered a conserved protein across all domains of life in respect to its primary and tertiary structure, uL11 is not functionally interchangeable between themCitation9, suggesting functional divergence. High-resolution structural models of the uL11 protein were obtained for both prokaryotic and eukaryotic ribosomes; especially, the structural organization of bacterial uL11 on the ribosomal particles stalled at various steps of the translational cycle has been described.Citation10-12 The bacterial uL11 protein was shown to interact with the EF-G (homolog of eukaryotic eEF2) via its N-terminal domain and together with the C-terminal domain of bacterial ribosomal stalk protein bL12 (analog of eukaryotic P1/P2 proteins) interact with the G’-domain of EF-G, regarded as the so-called Arc-Like Connection (ALC).Citation13 This structure, which seems to be crucial for trGTPase action, is formed directly after GTP hydrolysis as a result of allosteric structural rearrangement within the GAC and EF-G. The structural dynamics of uL11 was also shown upon EF-Tu (eukaryotic eEF1A) binding, where the N-terminal domain of uL11 is flipped away in respect to the ribosomal core, taking part in anchoring EF-Tu to the ribosome.Citation11,14 Thus, a majority of structural data connect uL11 with the elongation cycle of protein synthesis. However, several structural analyses implicate this protein in interaction with IF2 and RF3, indicating involvement of uL11 in the initiation and termination steps of translation,Citation15,16 which is also supported by several functional analyses.Citation17,18 The large ribosomal subunit depleted of this protein is able to bind initiation factor IF2 in vitro but it cannot stimulate GTP hydrolysis required for the subunit joining.Citation19 Similarly, the RF3-dependent release of RF1 and RF2 is drastically decreased when the uL11 protein is absent on the ribosome during termination.Citation20 In the case of eukaryotic uL11, its structural and functional mode of action is still less understood; however, its structure was determined on eukaryotic ribosomes, and interaction between yeast uL11 protein and eEF2 was postulated.Citation21 So far, there are scattered biochemical data showing that the absence of the uL11 protein impairs the translation elongation step, in both efficiency and accuracy meanings.Citation22 The in vitro translation elongation rate measured by the poly-phenylalanine synthesis is decreased, whereas the decoding error rate, shown as a higher frequency of misincorporation events measured in vivo by a specific dual-luciferase system, is increased upon uL11 deletion.Citation23 Although the involvement of eukaryotic uL11 in ribosomal action was documented, the exact impact on the translational apparatus and metabolic fitness is still beyond full understanding. Moreover, apart from the direct role in the translational cycle, many eukaryotic ribosomal proteins were found to play an important role in ribosome biogenesis, a multistep process involving myriads of auxiliary trans-acting factors helping the ribosome to gain its final shape.Citation24 One of the pivotal points during this process is the formation of mature GAC, which is considered as a sine qua non for structural/functional verification of pre-60S subunit correctness.Citation25 The early nucleolar GAC is composed of uL11 and uL10-like protein (trans-acting factors called Mrt4), which occupies the uL10 binding site.Citation26,27 It is postulated that uL11 and Mrt4 are important for proper spatial organization of early GAC and subsequent Yvh1 driven P-stalk loading, which gives rise to mature GAC formation.Citation28 Correctly assembled GAC becomes a functional checkpoint for verification of 60S translational capacity. Two consecutive steps of ribosome biogenesis, namely Tif6 release by the Efl1 GTPase and the “translation-like cycle” catalyzed by the eIF5B, might be GAC dependent.Citation28,29 Based on sequence homology, high-resolution structures, and previous biochemical data, it is tempting to assume that the mode of functioning of Efl1 and eIF5B is similar to other trGTPases (e.g. eEF2 or eEF1A), and consequently uL11 dependent.
In spite of the richness of structural work on uL11, especially using a bacterial model, the role of the uL11 in the GAC functioning has never been fully explored. In this work, the function of the eukaryotic ribosomal uL11 protein in the modus operandi of the translational apparatus was analyzed in vivo. A null yeast strain depleted of both RPuL11A and RPuL11B gene copies was subjected to functional analyses. We have shown that uL11, being an important element in the translation elongation step, has more extended function far beyond the currently anticipated one, discovering its new activity connected with the pre-elongation steps of protein synthesis (e.g., initiation and ribosomal biogenesis). Surprisingly, although uL11 is supposed to be loaded onto maturating ribosomes at the very early nucleolar step of biogenesis, it seems not to be critical for proper pre-rRNA processing and at early stages of 60S biogenesis. Importantly, as a core GAC component, uL11 is involved in very late steps of 60S assembling, coupling biogenesis with the translational cycle.
Results
Translational fitness
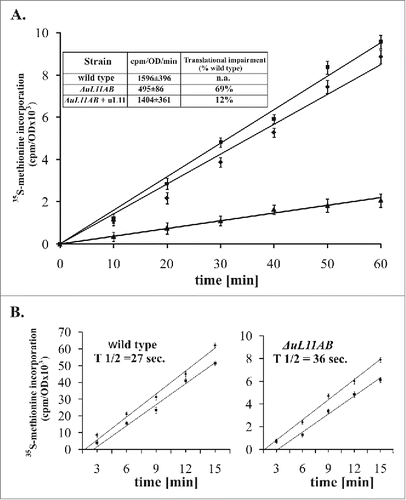
The ribosomal uL11 protein, being part of the ribosomal interaction platform with trGTPases, is not essential for cell survival; however, yeast strain lacking this r-protein exhibits a slow growth phenotype.Citation9 It has been suggested that the growth perturbations can be mainly attributed to a defect in the translation elongation step, as was measured by poly(U)-dependent polyphenylalanine synthesis in vitro Citation22, but the exact impact of uL11 on the translational apparatus in vivo has never been investigated. To get insight into the uL11 modus operandi during protein synthesis in living yeast cells, we prepared a double disrupted yeast strain ΔuL11AB (lacking RPuL11A and RPuL11B genes, former name RPL12A and RPL12B). First, the strain was subjected to translational fitness measurement in vivo, quantified by the 35S-Methionine (35S-Met) incorporation into newly synthesized peptides. As shown in , the ΔuL11AB strain exhibits translational impairment in comparison to the parental wild-type strain, showing 69% inhibition (cpm/OD/min. 1596 ± 396 and 495 ± 86 for the wild type and ΔuL11AB cells, respectively ). This is also reflected in the slow-growth phenotype of yeast mutant cells with their doubling time exceeding 200 min. (data not shown). The plasmid-borne expression of the uL11 protein almost completely restored translational capacity (), along with growth restoration (). The impairment of the overall translational fitness raises a question about the step in which the translational cycle is mostly affected. Since the uL11 protein, being a GAC element, was implicated in the elongation step, we have determined the so-called ribosomal half-transit time for cells deprived of uL11 protein. This parameter refers to the time required for the ribosome to traverse an average-sized mRNA and release a complete polypeptide, therefore the elongation and termination defects might be demonstrated.Citation30 The analysis was performed by measurement of the kinetics of 35S-Met incorporation into total proteins (nascent and released peptides) and peptides released from the ribosomes. The results shown in are expressed as 2 corresponding lines of data related to total synthesized and released polypeptides, plotted as a function of time. The determined half-transit time was 27 and 36 seconds for the wild type and mutant strain, respectively. This shows 25% reduction in the elongation speed of ribosomes deprived of the uL11 protein.
Figure 1. Ribosomal uL11 protein is indispensable for full translational capacity. (A) translational fitness determination by the radiolabeled 35S-methionine incorporation measurement. The results are an average of 3 independent experiments; ▪ - wild type stain, ▴ - ΔuL11AB mutant yeast strain, ♦ - complementation with the uL11 protein. Inset, the slope value determined for individual yeast strains, with the translation inhibition, which was quantified as a percent of translational impairment in respect to wild type cells. (B) ribosomal half-transit time determination. The incorporation of 35S-methionine into total proteins (nascent and completed - diamonds) and completed (squares) are shown for each strain. The average half-transit time was determined by linear regression analysis as displacement between 2 lines and presented on the graph as inset - T 1/2. The radioactivity at each time point is presented as a mean of 3 independent measurements.
Polysome profile analysis
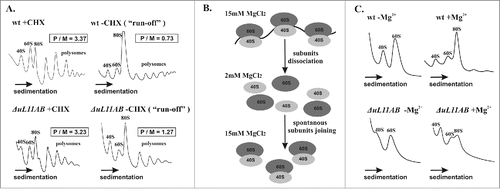
The translational fitness analysis showed significant impairment of the translational machinery deprived of the uL11 protein. This result was further confirmed through polysome profile inspection where modest reduction of 60S and polysomal fractions with concurrent appearance of half-mers was seen for the ΔuL11AB cells (). The polysome to monosome ratio (P/M) determined as 3.37 and 3.23 for the wild type and mutant cells, respectively, does not directly suggest any defects in elongation; however, such a defect is clearly seen when the cycloheximide (CHX) is omitted during cell extract preparation (). In such conditions, translating ribosomes are not stalled by the CHX and are able to continue the elongation cycle (so-called polysome ‘run-off’ conditions). The P/M ratio determined for the ‘run-off’ conditions was 0.73 and 1.27 for the wild type and mutant cells, which indicates a defect of the translation elongation speed. Special attention should be placed on half-mers, which are present in the ΔuL11AB yeast cell polysome profile. They are a fraction containing mature ribosomes with an additional initiatory 43S particle bound to the mRNA. Appearance of half-mers usually occurs when there is physiological imbalance of initiatory 40S subunits over 60S subunits.Citation31 This might be attributed to pre-60S maturation impairment lowering the amount of 60S accessible for initiation. The half-mers might also appear when the total amount of 60S is not reduced but they are not able to bind 43S and efficiently initiate translation. The polysome profile indicates that there is only modest disproportion between 40S and 60S fractions; however, this imbalance (excess of 40S over 60S) becomes more prominent in ‘run-off’ conditions (). This suggests that 60S abundance is indeed moderately reduced but additionally subunits are not able to efficiently join each other to enter the elongation cycle. Since the process of 40S binding to uL11 deprived of 60S needs to take more time, the subunit-joining kinetics is probably delayed. There is enough time during ‘run-off’ experiments to “saturate” all initiating 43S with uL11-deficient 60S, depicted as disappearance of half-mers. To further prove the initiation defect, a subunit-joining analysis was performed (the experiment is schematically presented in ). As shown in , lowering the magnesium ion concentration (2 mM MgCl2) during cell extract preparation led to complete disassembling of translating ribosomes into free 40S and 60S subunits. In the case of the wild-type strain, the typical 40S and 60S distribution was observed, but the mutant strain exhibited subunit imbalance, with 60S reduction in respect to 40S. An intriguing finding was noted at restoration of 80S ribosomes by supplementation of the disassembled polysomal fraction with 15 mM MgCl2, which induces spontaneous ribosomal subunit joining. Ribosomal subunits from the wild-type strain efficiently restored the 80S ribosome, contrary to the mutant strain, where building of the 80S ribosomal fraction was severely diminished. These results suggest that there is superimposition of a pre-60S maturation defect combined with a translation initiation delay caused by the uL11 depletion.
Figure 2. Polysome profile and subunit-joining analysis. (A) the polysome profiles from wild-type and ΔuL11AB yeast cells with (+CHX) and without (−CHX) cycloheximide treatment are shown on the left and right panels, respectively. The polysome to monosome (P/M) ratio was calculated for each profile by dividing the area of the first 3 polysomal peaks by the area of the peak for the 80S monosome. The sedimentation vector of the ribosomal fractions is indicated by an arrow. (B) schematic representation of the ribosomal subunit-joining experiment. (C) ribosomal subunit-joining experiment, upper panel - wild type ribosomes, lower panel - ΔuL11AB mutant strain, -MgCitation2+, +MgCitation2+, in the presence of 2 mM and 15 mM MgCl2, respectively.
Translation accuracy
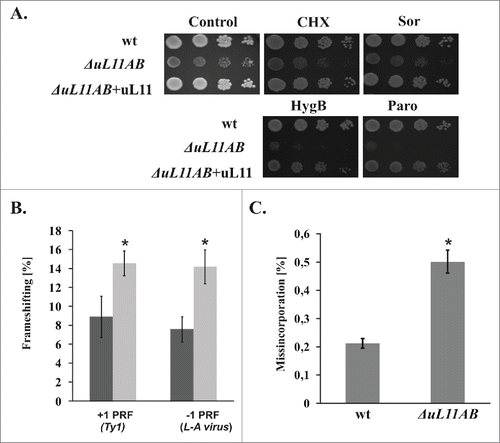
Since the elongation step of the translational cycle is quantitatively affected in the mutant strain, we also focused our attention on the qualitative issue of the elongation cycle. We measured translation fidelity especially reading frame maintenance (programmed ribosomal frameshifting - PRF) as well as the accuracy of decoding during translation elongation (near-cognate misincorporation). Programmed −1 and +1 ribosomal frameshifting has been linked to regulation of the expression of specific genes, including those in viruses, yeast, and humans, thus involvement of uL11 in regulation of this process may represent an interesting aspect of the GAC function. PRF was described as events when the reading frame of translated mRNA is slipped one nucleotide toward 5′ (−1 PRF) or 3′ (+1 PRF).Citation32 To analyze the involvement of uL11 in the PRF, we used a reporter system designed for quantification of ribosomal frameshifting directly in vivo.Citation33 The system applies dual-luciferase reporters harboring a specific yeast L-A viral -1 PRF signal and a Ty1 +1 PRF signal, respectively. The results presented in show that ribosomes deprived of uL11 proteins exhibit a significant ORF maintenance defect. Both -1 and +1 PRFs were affected to the same extent, and the percentage of PRF was increased roughly 2-fold by the ribosomes lacking uL11. Also, the frequency of the erroneous decoding event, namely misincorporation was elevated in ΔuL11AB (). The ribosomal translational accuracy defect may be probed in vivo by growth sensitivity tests using specific antibiotics, interfering with particular steps of protein synthesis. As shown in , uL11-deficient cells are hypersensitive to paromomycin (Paro), an aminoglycoside antibiotic specifically binding to the 40S decoding center at the A-site and inducing an increase in the ribosomal misincorporation rate for near-cognate amino acids. Paro at a concentration 500 μg/ml does not exert a significant effect on the wild type strain growth, whereas such a concentration of Paro was lethal for ΔuL11AB cells, which probably reflects synergism between the increased rate of decoding errors induced by Paro and error prone ribosomes with uL11 deficit. A similar effect was observed for hygromycin B, which also affects the elongation cycle in a similar manner. The uL11-depleted yeast strain is not significantly affected by the sub-lethal level of cycloheximide, a blocker of the translocation step, which acts through the E-site interference. Interestingly, the analyzed yeast deletion strain is only modestly sensitive toward sordarin, an antibiotic that specifically blocks the translocation step by stalling eEF2 in the GDP bound state on the 80S ribosome, .Citation34
Figure 3. Ribosomal uL11 protein involvement in translational accuracy maintenance. (A) antibiotic sensitivity test with specific inhibitors of various steps of the elongation cycle. Yeast cells were spotted onto agar plates with YPD medium, as a serial tenfold dilution of original cell culture with OD600 = 0.1, growth was continued for 3 d (wt - wild type strain, ΔuL11AB mutant strain, and ΔuL11AB+uL11 mutant strain complemented with uL11 full-length protein. The medium was supplemented with 20 ng/ml cycloheximide (CHX), 4 μg/ml sordarin (Sor), 20 μg/ml hygromycin B (HygB), and 500 μg/ml paromomycin (Paro). (B) ribosomal recoding quantification using a dual luciferase reporter assay. The frequency of +1 PRF (Ty1 derived frameshifting signal) and -1 PRF (L-A virus derived signal) efficiency were measured and expressed as a percentage of frameshifting events. (C) Translational accuracy disorder quantification using a dual luciferase reporter assay. The frequency of misincorporated near-cognate amino acids is shown as a percentage of all decoding events. The results are an average of 3 independent experiments. Asterisks indicate a statistically significant result with p > 0.05 calculated with Student's t-test.
Ribosomal biogenesis
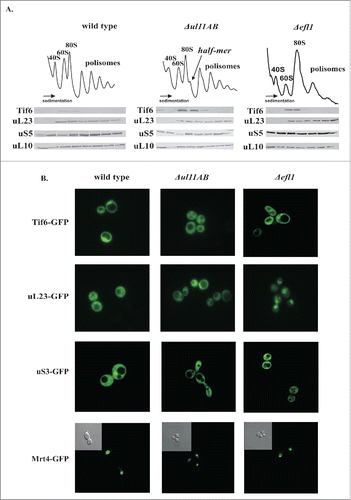
The polysome profile analysis suggested that maturation of the 60S subunit might be affected in ribosomes lacking uL11. As depicted in , the polysome profile from the uL11-depleted yeast cells exhibits reduction in the polysomal fractions and appearance of prominent half-mer structures with concomitant modest reduction of 80S and 60S fractions. As proposed previously.Citation25 the GAC may serve an important function during a very late step of pre-60S biogenesis, taking part in the release of the Tif6 trans-acting factor. Thus, we have analyzed the distribution of Tif6 on the polysomal profile. The pattern of marker protein distribution from both large uL23 and small uS5 proteins traced by western blotting is similar in wild type and mutant strains; however, the accumulation of the Tif6 anti-association factor on the uL11-deficient 60S subunits can be seen (). It has been proposed that the release of Tif6 by Efl1 occurs in a GAC-dependent manner and is considered as one of the last events during pre-60S biogenesis.Citation35-37 Therefore, the Tif6 retention suggests a ribosome biogenesis defect and implicates perturbation in the Efl1-ribosome interplay. Noteworthy, the analysis of the polysome profiles derived from the Efl1-depleted yeast cells used as a control element showed also Tif6 accumulation, as is the case in ΔuL11AB yeast strain, confirming the late biogenesis defect (). To further proof the defect in Tif6 release, we traced the sub-cellular localization of GFP-tagged Tif6. Microscopic observations using a control system (Efl1 depleted cells) showed that the nuclear localization of Tif6-GFP in steady state conditions becomes strictly cytoplasmatic when Efl1-driven Tif6 release is blocked in Efl1 depleted cells (). Interestingly, Tif6 cytoplasmic retention is also observed in ΔuL11AB cells. It should be noted that the pre-60S maturation delay is usually accompanied by nuclear retention of premature subunits, which is seen as nuclear entrapment of ribosomal proteins. Unexpectedly, we saw no nuclear entrapment of the uL23-GFP fusion protein in any tested conditions (optimal and restrictive temperatures - data not shown) for the uL11-depleted cells; however, we saw clear uL23-GFP nuclear entrapment in the Efl1-depleted cells. Additionally, we have analyzed the cellular behavior of Mrt4-GFP, a trans-acting factor involved in ribosomal stalk maturation. It was postulated that the uL11 is responsible for the Yvh1 binding, a trigger factor involved in Mrt4 release from pre-60S.Citation28 However, our microscopic observations do not support this hypothesis. The predominant phenotype associated with Yvh1 malfunction or other conditions affecting Mrt4-P-stalk exchange is re-localization of the Mrt4 protein from the nucleus/nucleolus into the cytoplasm.Citation28,38,39 We saw no signs of such abnormal Mrt4 behavior, which was found in the nuclear compartment in the wild type and in both mutants analyzed, namely the ΔuL11AB and ΔEfl1 strains ().
Figure 4. Involvement of ribosomal uL11 protein in the pre-elongation steps of the translational apparatus. (A) analysis of polysome profiles, left panel - wild-type strain, middle panel - ΔuL11AB analyzed mutant strain, and right panel - Δefl1 mutant strain lacking the Efl1 biogenesis factor. Half-mers are indicated by an arrow. Below, protein gel blotting of polysomal fractions analyzed with antibodies against Tif6, uL23, uS5, uL10. (B) sub-cellular localization of indicated GFP-tagged proteins expressed in wild type and mutant cells. In the case of Mrt4-GFP, an additional phase contrast picture is provided as an inset.
Pre-rRNA processing
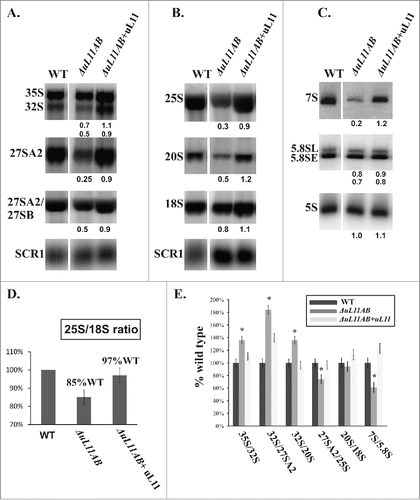
The issue of uL11 involvement in 60S biogenesis was also probed by analysis of pre-rRNA processing in ΔuL11AB yeast mutant cells. The common precursor for 25S, 18S, and 5.8S rRNA is synthetized by the RNA PolI as a primary transcript 35S rRNA, which is then subjected to multistep processing leading to production of mature ribosomes. The 5S rRNA component is independently synthesized as a separate transcript by the RNA PolIII.Citation40 In general, in mutant strains defective in particular steps of ribosomal biogenesis, accumulation of the corresponding pre-rRNA population with concomitant reduction of its downstream products is observed. Interestingly, in the case of the uL11-depleted strain, no accumulation of any pre-rRNA intermediate was observed. Instead, the amount of all intermediates as well as mature rRNAs was reduced (). The efficiency of each pre-rRNA processing step was determined by analyzing the ratio of the pre-rRNA intermediate to its direct maturation product. Most of the processing steps were delayed by the same factor according to wild type efficiency (). The 35S/32S, 32S/27SA2, and 32S/20S processing steps seem to be more affected, but this is caused rather by the decrease in downstream product than the upstream substrate accumulation. Such a phenomenon has never been observed for a yeast mutant strain defective in ribosome biogenesis where strong accumulation of particular pre-rRNAs was usually noticed. In ΔuL11AB cells, the 27S pre-rRNA and 7S pre-rRNA intermediates were significantly underrepresented (4-fold and 5-fold decrease, respectively). Additionally, the analysis of 25S/18S ratio shows a decrease in 60S subunit abundance confirming the 40S-60S imbalance already observed in the polysome profiles. Thus, the absence of uL11 does not cause significant perturbation in pre-rRNA processing in the particular steps of 60S maturation, but rather contributes to specific degradation of pre-60S intermediates.
Figure 5. Pre-rRNA processing analysis using yeast cell mutant deprived of uL11 protein. (A, B) RNA gel blotting analysis of high-molecular weight pre-rRNA intermediates. (C) Northern blot analysis of low-molecular weight pre-rRNA intermediates. For analysis, total RNA was prepared and equal amounts of RNA were subjected to analysis. The signals of specific rRNA populations were counted and calculated in respect to the wild-type cells; values provided below each blot represent the fraction of the wild-type value arbitrarily set at 1.0. All data were normalized against SCR1 loading control. (D) the relative amount of 25S vs. 18S rRNA, provided as a 25S/18S ratio. For calculation, the signal intensity from 25S rRNA-specific and 18S rRNA-specific probes were quantified for each strain. The data for mutant cells were quantified in respect to wild type cells and presented as a percentage of the wild type ratio set as 100%. (E) efficiency of the pre-rRNA processing steps, shown as a ratio of the amount of the particular pre-rRNA intermediate to its direct product. The value calculated for the wild-type strain was set as 100%. All results are presented in respect to wild type cells. The asterisks indicate statistically significant differences in respect to control cells, with p < 0.05.
Discussion
uL11 as a universal element of the GTPase associated center
The ribosome operates as a Brownian machine that harnesses thermal fluctuations into directed motion, where the GTP hydrolysis catalyzed by trGTPases confers unidirectional trajectory for the translational apparatus.Citation41 The pivotal role in trGTPase binding and stimulation has been attributed to the GAC region composed of an rRNA fragment called a sarcin-ricin loop (SRL) and a protein element, i.e. an oligomeric P-stalk structure. Despite numerous trials, including both structural and biochemical approaches, the mode of the GAC action is still far from being exhaustively understood. The multi-component construction of the GAC implies that, on the one hand, all elements cooperate to promote trGTPase activity, but on the other hand every constituent has a specialized function, allowing the GAC to specifically interplay with trGTPases and promote several steps of the translational cycle. In this work, we present functional in vivo characterization of the uL11 ribosomal protein.
uL11 involvement in ribosomal speed and accuracy
We used double disrupted yeast strain ΔuL11AB where 2 gene copies uL11A and uL11B had been deleted from genomic DNA. The mutant strain was viable but exhibited a severely slow growth phenotype, as reported before.Citation22 The growth rate reduction was accompanied by a ca. 70% decrease in overall translation efficiency, shown by the pulse-labeling experiment. The analysis was performed in time-scale exceeding temporal resolution of a single round of protein synthesis. The observed phenotype presumably reflects the superimposition of the impairments of several events associated with the translational apparatus activity. Initial analyses indicate that ribosomes deprived of uL11 elongate 30% less efficiently than the wild type, as shown by the increased half-transit time or in the polysome ‘run-off ’ experiment. This slow-down might be attributed to the detrimental interplay with the elongation factors eEF1A or eEF2. The translation elongation cycle is divided into 2 major steps: eEF1A-dependent decoding and eEF2 dependent translocation. The activity of both trGTPases is stimulated by the GAC, but to a different extent. Previous results have shown that eEF1A impairment is accompanied by a decrease in translation fidelity also manifested in the sensitivity toward specific antibiotics interfering with the decoding process, e.g. paromomycin.Citation42,43 In contrast, the eEF2 defect might affect the sensitivity toward another translation inhibitor, namely sordarin.Citation44 We showed that the uL11 protein is involved in the reading frame maintenance and translation fidelity. The lack of the uL11 protein on the ribosome induces both +1 and -1 PRF. The integrated model of PRF predicts that +1 PRF occurs when the ribosomal A-site is empty with the P-site occupied, whereas -1 PRF might take place or would be promoted when the ribosome adopts a ‘rotated’ state with both A- and P-sites occupied.Citation45 As a rule of thumb, all perturbations affecting stabilization of such ribosomal states would affect PRF efficiency. Assuming that the A-site is empty only when the ribosome is in its post-translocation state awaiting for ternary complex (TC) binding, only a defect in TC binding (originating from both eEF1A or GAC malfunctions) would extend the time of A-site emptiness. Thus, the increased probability of a +1 PRF event indicates that uL11 plays an important role in TC binding and/or its stabilization on the ribosome, probably by promoting A/T hybrid state formation. The role of uL11 in the -1 PRF process seems to be more complex, because the repertoire of ribosomal states when both A- and P-sites are occupied is much broader, encompassing decoding and translocation steps. Thus, the time when both A- and P-sites are occupied on the ribosome might be extended by impairment of (i) stimulation of GTP hydrolysis, inorganic Pi release, and subsequent eEF1A dissociation, (ii) peptidyl-transferase reaction, and (iii) eEF2 driven translocation. We have found that ΔuL11AB cells are not hypersensitive toward anisomycin, a specific inhibitor of peptide bond formation.Citation46,47 This suggests that the peptidyl-transferase reaction is not affected by the lack of the uL11 protein. Complementary, none of the perturbations within the GAC was shown to exert an effect on the peptidyl transferase reaction in bacterial ribosomes.Citation48 Moreover, as reported already, neither sordarin treatment nor mutations within the eEF2 protein conferring sordarin resistance increased -1 PRF efficiency Citation49, indicating that none of the perturbations in eEF2-catalyzed translocation is connected with -1 PRF stimulation. Therefore, the abnormal interplay between eEF1A and the uL11-deficient ribosome is supposed to be responsible for increased -1 PRF, indicating direct involvement of uL11 not only in TC binding (initial selection and codon recognition), but also in later steps of decoding, probably including proper spatial positioning of its catalytic center, a prerequisite for eEF1A-driven GTP hydrolysis. Noteworthy, the -1 PRF is strategically applied by the L-A virus for propagation.Citation50 As shown in our previous report, the lack of P1 and P2 proteins on the ribosome, which form the lateral stalk component of the GAC, was associated with increased L-A virus replication. Interestingly, this was accompanied with increased eEF1A association with the GAC region,Citation51 indicating a functional interplay between P1/P2, uL11, and eEF1A during the ribosomal decoding step. In contrast, the uL10 protein, another stalk base element, seems to be functionally coupled with eEF2 to a greater extent, underscoring the functional divergence of the GAC elements in the interplay with 2 elongation trGTPases.Citation49,52 The dynamics of stalk proteins is well described by the existence of the so-called Arc-Like-Connection (ALC), composed of the N-terminal domain (NTD) of uL11 and the C-terminal domain of bL12, seen on the G-domains of EF-Tu or EF-G. The spatial orientation of uL11 NTD in respect to the large ribosomal subunit was shown to be correlated with different functional states of the ribosome. Two basic conformations can be ascribed to uL11 NTD - “closed” conformation which is characteristic for factor binding, predominantly in the GTP stateCitation10,11 and “open” conformation after GTP hydrolysis when factors transform into the GDP form.Citation7 Since several reports indicate that the "closed" conformation of the uL11 is correlated with bL12 movement,Citation53 it is reasonable that GTPase activation is a result of simultaneous structural rearrangement of uL11 and bL12 enabling proper orientation of the G domain in respect to SRL. Our results show that both eEF2 and eEF1A activity are affected by the lack of uL11, but the abnormal GAC-TC interplay represents the most prominent ribosomal malfunction contributing to cellular fitness.
Based on our in vivo analyses and published results, we postulate a mechanism of uL11 action during the elongation step, which is presented in Figure S1. The “open” conformation of uL11 NTD facilitates TC binding and the NTD closing occurs upon correct codon-anticodon recognition. This collars cognate TC in the A-site, preventing its dissociation, where together with the lateral stalk structure it contributes to formation of the GTPase productive state. Accordingly, the non-cognate TC is not able to induce the uL11 NTD closing at all, whereas the near-cognate TC is able, but at a severely reduced rate. Thus, uL11 might be regarded as one of the ribosomal elements perceiving codon-anticodon recognition and locking the cognate TC in a GTPase activated state. In the case of EF-G/eEF2 translocase, the closing of uL11 NTD was also observed upon factor binding, but functionally it seems to be more coupled with release of inorganic phosphate, a prerequisite for eEF2 structural rearrangement and dissociation.Citation54 According to the proposed model, uL11-deficient ribosomes mimic permanently "open" conformation, allowing equal binding for all TCs, as the initial binding step is codon independent.Citation55 Such ribosomes are not able to properly perceive the codon-anticodon interaction through the uL11 “closed” conformation, thus cognate and near-cognate TCs are not efficiently distinguished by ribosomes. Thus, according to the kinetic model of decoding, the rate of dissociation of cognate and near-cognate TCs becomes equalized on the uL11-deficient ribosomes. Consequently, it takes much more time for the ribosome to establish productive GTPase-activated state for any TC and this time is equalized for both cognate and near-cognate TCs, disfavoring the cognate one and reducing translation fidelity at the same time. We are aware that the translational fitness defect observed in the uL11-deficient strain might also be a result of the defect in the translation termination step. It was reported earlier using a dual-luciferase reporter system that in uL11-depleted cells the frequency of reading-through events is elevated.Citation23 This phenomenon can also be explained by the loss of cognate vs. near-cognate discrimination (as discussed above). In eukaryotic cells, recognition of the STOP codon is subjected to the decoding process analogously to TC recognition, with the eRF1:eRF3:GTP complex (termination ternary complex tTC) as a main player, which structurally mimics TC, where eRF3 works as trGTPase analogously to EF-Tu.Citation56 Similarly to the decoding event during elongation, eRF1 binding to the STOP codon is disfavored when uL11 is absent, and near-cognate TC binding may take place, making termination less accurate. Thus, uL11-depleted cells lose this discriminative capacity, equalizing the probability for cognate, near-cognate, and termination TC binding. This mechanism resembles an analogous action of the uL3 protein described as a “gatekeeper” to the ribosomal A-site. This protein is responsible for allosteric repositioning of rRNA structural elements and, at the same time, coordinating the rearrangements of the factor-binding state in the neighborhood of the GAC, opening and closing the accommodation corridor for aa-tRNA.Citation57 However, the mechanism of action is totally different from that proposed for uL11. uL3 works as a ‘molecular clamp’ contributing to the ‘induced fit’ function of the peptidyltransferase center.Citation58 uL11 might be considered as a “keymaster” to the ribosomal GAC site, responsible for accommodation and formation of GTPase productive states of the aa-tRNA-eEF1A-GTP ternary complex during the decoding steps of elongation. Thus, the uL11 protein allosterically contributes to the decoding event, showing that the ribosome action is subjected to a multiple level of functional redundancy, being a warrant of high fidelity.
uL11 and ribosomal biogenesis
As shown previously, many ribosomal proteins (r-proteins) act in concert with dozens of auxiliary trans-acting factors to drive the maturation of ribosomal particles. They work as co-chaperones involved in proper rRNA folding and constitute a docking platform for external factors.Citation59 Also, an interplay of the GAC with trans-acting factors such as Efl1 was postulated.Citation60 Efl1 has a high degree of homology with eEF2, and it was shown to be involved in GTP-dependent Tif6 release from the pre-60S subunit.Citation61 Recently, structural analyses suggested that uL11 might take part in Efl1-driven Tif6 release, indicating involvement of the GAC in final tuning of pre-60S.Citation37 Indeed, we observed a defect in Tif6 release in ΔuL11AB yeast cells, exhibited as Tif6 accumulation on the 60S fraction and retention of the Tif6-GFP hybrid protein in the cytoplasmic compartment. Therefore, we have provided physiological evidence of the GAC involvement in the late 60S biogenesis step, linking the structural data with physiological ones. Additionally, we have noticed the half-mers in the polysome profile, which is often associated with a pre-60S maturation defect.Citation62-64 Although in such cases nuclear entrapment of proteins from large ribosomal subunits is usually observed, we did not see any mislocalization of the marker uL23 protein in microscopic observations. Additionally, we saw only small reduction of 60S, which departed from the prominent behavior observed in cells with a 60S biogenesis defect.Citation65 What is more, we saw no specific defect in any particular step of pre-rRNA processing from both large (25S, 5.8S and 5S) and small (18S) subunits, in the form of accumulation of intermediate rRNA. Instead, we found that the abundance of 3 rRNA populations, namely 27S, 7S, and mature 25S, was reduced. The only explanation of such an observation may involve selective partial degradation of uL11-deficient pre-60S by the pre-ribosome quality control system, a phenomenon never observed in the studies on 60S biogenesis. Thus, the modest defect in biogenesis is not the only source of the presence of the half-mers in ΔuL11AB yeast cells, but, as we have shown in the subunit-joining experiment, perturbations in the translation initiation which is dependent on another trGTPase, eIF5B.Citation66 Interestingly, involvement of the GAC in eIF5B actions was also shown for the uL3 protein. The uL3 was implicated into eIF5B dependent 3′ end processing of 18S rRNA in the context of 80S ribosomes that have not yet engaged in translation, underscoring the fact that the GAC and its neighborhood actively participate in the interplay with trGTPases at various steps of ribosomal life.Citation67
In summary, in this paper, we have clarified the role of the ribosomal protein uL11 showing its modus operandi in the translational apparatus. Using an in vivo approach, we have brought a missing link between structural and biochemical studies, providing the functional picture of the uL11 interplay with trGTPases during translation. We propose that the ribosomal protein uL11, within the GAC, is involved in all aspects of ‘ribosomal life’, starting from very late steps of 60S biogenesis and Tif6 release, eIF5B-dependent subunit joining during initiation of translation. Especially, uL11 plays an essential role in accommodation and formation of GTPase productive states of the TC, allosterically contributes to the decoding event during elongation, and lastly is probably involved in the termination step. uL11 represents a very conserved GAC element, which together with trGTPases confer the unidirectional trajectory for the translation, but especially contributing to the fidelity of the translational machinery.
Materials and methods
Genetic manipulations and plasmid construction
The mutant strain depleted of 2 genes encoding the uL11 protein was constructed on the basis on the BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) parental strain by 2 rounds of PCR-based gene targeting. RPL12A and RPL12B genes (new name RPuL11A and RPuL11B according to the nomenclature proposed recently by Ban et al.Citation68) were replaced by the auxotrophic marker HIS3 and URA3 ORFs using homologous recombination. The resulting URA+ ΔuL11AB strain (MATa; RLP12A::HIS3; RLP12B::URA3) was converted into the URA-phenotype by homologous recombination using a PCR-amplified URA3 gene fragment where 3 TAA stop codons were introduced after the GCG codon for Ala107. ΔuL11AB cells, transformed with the URA3107STOP PCR fragment, were then plated on the 5-FOA medium to select clones with an inactive URA3 gene. The Efl1-depleted strain (W303; EFL1::HIS3) was a gift from prof. F. Fasiolo. The plasmids used for complementation of ΔuL11AB cells with uL11 as well as the Tif6 protein were constructed on the basis of a tetracycline-repressible pCM190 vector, using PCT cloning. Plasmids for expression of GFP tagged Tif6 (pRS315-Tif6eGFP) under a native TIF6 promoter were kindly provided by prof. F. Fasiolo. Vectors encoding markers of subcellular localization of ribosomal subunits pRS315-uL23eGFP and pRS315-uS3eGFP were provided by prof. E. Hurt. Plasmid pAJ2457 expressing the GFP-tagged Mrt4 protein was kindly provided by prof. A. Johnson.
Yeast polysome profile analysis and immunoblotting
Polysome profiles were obtained by 7–47 % sucrose gradient centrifugation of total cell extracts. Cells were grown to OD600 0.4–0.6 in YPD or appropriate minimal medium (SD) and treated with cycloheximide (CHX) to the final concentration 100 μg/ml for 20 minutes for preservation of the polysomes. Cells were harvested by centrifugation and re-suspended in lysis buffer [10 mM Tris-HCl pH 7.5, 100 mM NaCl, 15 mM MgCl2, 100 μg/ml CHX, 1 mM PMSF, 6 mM β-Me, 1 nM pepstatin A, 10 nM leupeptin, 10 ng/ml Aprotinin, 200 μg/ml heparin, and RNase Inhibitor (Sigma-Aldrich)] and disrupted by vigorous shaking with glass beads at 4°C. The cell lysate was pre-cleared by centrifugation at 13000×g (Rotor 12154-H; SIGMA). Twelve OD260 units were loaded on each sucrose gradient, centrifuged for 4,5 h at 26500 rpm and 4°C in a SW32Ti rotor (Beckman-Coulter), and analyzed using an ISCO Brendel Density Gradient Fractionator. For the polysome run-off experiment, the polysomes were not preserved by CHX treatment and accordingly CHX was omitted in the lysis buffer. Cell extracts were incubated for 20 minutes at 30°C to complete the elongation round. For immunodetection, the proteins from each fraction were precipitated with 10% TCA and analyzed by the SDS-PAGE. Specific antibodies directed against uL23 (1:1000x dilution, gift from prof. E. Hurt), uS2 (1:3000, gift from prof. Seedorf), Tif6 (1:4000, gift from prof. F. Fasiolo), uL10, and P1/P2 1:2000Citation51 were used. Monoclonal antibodies against the uL11 protein were kindly provided by prof. J.P. Ballesta (dilution used 1:500).
Ribosomal subunit-joining assay
The assay was performed as described previously.Citation69 Briefly, for subunit dissociation, cell extracts from the wild type and ΔuL11AB were prepared in low-concentration magnesium buffer where 2 mM (instead of 15 mM) of MgCl2 was used and CHX was omitted, as described above for the polysome run-off experiment. The equivalents of 6 OD260 units were loaded on 7–47% sucrose gradients containing 2 mM of MgCl2. To induce spontaneous ribosomal subunit joining, the MgCl2 was added to the cell extract to the final concentration 15 mM and incubated for 30 min. at 25°C. Extracts were subsequently loaded on 7–47% sucrose gradients supplemented with 15 mM of MgCl2
Fluorescence microscopy
Wild type and mutant strains were transformed with an appropriate reporter vector and grown on liquid selection medium to OD600 0.6–0.8 and transferred into YPD medium (OD600 0.1) for 3 hours at permissive (30°C) or non-permissive (23°C or 37°C) temperatures. Cells were then collected by centrifugation, washed in ice-cold PBS buffer, and re-suspended in mounting medium (Sigma-Aldrich). Acquisition was done under a Zeiss LSM780 confocal microscope, using oil immerse objective Plan Apo 100X magnification; 1.4 NA DIC controlled with Zen 2010 Software (Zeiss).
Translational fitness determination - 35S-radiolabelled methionine and 3H-leucine incorporation
Cells were grown to OD600 0.5–0.7, washed and re-suspended with methionine- or leucine-depleted SD minimal medium (SD -Met and SD -Leu). The cells were cultivated at 30°C for 15 minutes and unlabeled-methionine or -leucine was added to the final concentration 50 μM. 37 kBq of 35S-Methionine (37 TBq/mmol, Hartmann Analytics) or 7.4 kBq of 3H-Leucine (2.22–4.44 TBq/mmol, Hartmann Analytics) was added at time 0 (T0). At 10-min intervals, the OD600 (A600) of the cultures was measured and 1 ml aliquots of the cultures were collected, proteins were precipitated with ice-cold 50% TCA, collected on Whatman GF/C filters, and counted in a scintillation counter (Beckman LS6000SE). The translation impairment was determined by comparison of the incorporation rate (cpm/OD600/min.) of mutant cells with wild type cells, plotted as a function of time. The results were expressed as a percent of wild-type activity.
Ribosome half-transit time determination is based on comparison of the rates of 35S-Methionine incorporation into total (nascent and completed) and completed proteins. Briefly, 10 ml of yeast culture (grown in conditions as described above) was harvested at each time point (0 min, 3min, 6 min, 9 min, 12 min, 15 min) and re-suspended in 500 μl of lysis buffer (10 mM Tris-HCl ph 7.4, 50 mM NaCl, 15 mM MgCl2, 5 mM NH4Cl, 200 μg/ml heparin, 100 μg/ml CHX, 1 mM PMSF, 5 mM β-Me). The cells were then disrupted by vigorous shaking with an equal volume of glass beads. Cell debris was removed by centrifugation (12000 rpm, 4°C, 20 min). The supernatant was then divided into 2 parts and 300 μg of proteins were used for further analyses. One part was used for total protein analysis for translational fitness determination. Another part was ultracentrifuged at 100000 xg for 45 minutes to obtain a post-ribosomal fraction. The radioactivity (cpm) of the total protein fraction and the post-ribosomal fraction was counted and expressed as a function of time. Ribosome half transit time (T½) was determined from the displacement in time between 2 trendlines delineated for each data series and obtained by linear regression analysis as described previously.Citation70
Antibiotic sensitivity tests
Wild type and mutant cells were grown at 30°C with vigorous shaking to the logarithmic phase of growth (OD600 1–2) in YPD or appropriate selective medium and then diluted to OD600 = 0.1. Serial tenfold dilutions of the culture were prepared and spotted onto agar plates supplemented with the indicated concentrations of antibiotics. The cells were grown at 30°C up to 5 d.
RNA electrophoresis and northern hybridization
Total RNA was isolated using the hot phenol method (Schmitt et al., 1990) from the logarithmic phase of cell growth. High-molecular-mass RNAs were analyzed on 1.2% agarose gels with addition of 1.7% Formaldehyde in 1x NBC buffer (5 mM NaOH, 50 mM Boric acid, 10 mM Tris-sodium citrate pH 7.5). Six µg of total RNA in 5 µl of RNase-free water was mixed with 10 µl of Formamide, 3 µl of Formaldehyde, 2 µl of 10x NBC buffer, 2 µl of 10x loading (15% Ficoll, 100 mM EDTA pH 8, 0.25% Bromophenol blue) and 2 µl of Ethidium bromide (2 mg/ml). Low-molecular-mass RNAs were separated on 8% polyacrylamide gels (PAGE; Acrylamide:Bis-acrylamide 19:1) containing 7 M Urea in 0.5x TBE buffer (89 mM Tris, 89 mM Boric acid, 2 mM EDTA). Eight µl of total RNA was mixed with loading buffer (98% Formamide, 0.025% Xylene cyanol; 0.025% Bromophenol blue). Oligonucleotide probes used for hybridization are listed below. Probes (10 pmol) were labeled in the presence of 5 µCi [γ-32P]-ATP (Hartmann Analytics) using T4 Phage Polynucleotide Kinase (New England Biolabs). Membranes were exposed for 12–14 h using the FujiFilm Imaging Plate (BAS-IP MS 2040). Quantification of northern blots was performed using a Typhoon PhosphorImager 9000 (Fuji) and analyzed using ImageQuant software (Molecular Dynamics).
Quantification of programmed ribosomal frameshifting (PRF) and near-cognate amino-acid incorporation using a dual-luciferase assay
The PRF quantification was done using a dual-luciferase assay according to a previously established procedure.Citation33 Briefly, each yeast strain was transformed with a control plasmid pYDL and with an appropriate reporter plasmid for -1 PRF (pYDL-LA) and +1 PRF (pYDL-Ty1), respectively. Transformants were grown on SD minimal medium without Uracil to OD600 0.5–0.7. Cells were then collected by centrifugation and disrupted by vortexing with glass beads in ice-cold PBS buffer supplemented with 1 mM PMSF (Sigma). The cell extracts were pre-cleared by centrifugation at 10000 xg. Five μl of each extract was used for luciferase activity quantification using the Dual-luciferase Assay System (Promega) according to manufacturer's instructions. The Firefly/Renilla (F/R) ratio was calculated for cells bearing control as well as reporter plasmids. Frameshifting efficiency was calculated by dividing the F/R of the control plasmid by the F/R ratio from each reporter plasmid and multiplying by 100%. The results are an average of 3 independent experiments. For the misincorporation assay, the reporter plasmid pDB868 kindly provided by prof. David Bedwell was used. In such a reporter, codon encoding functionally relevant Firefly luciferase His245 was replaced by its near-cognate derivative.Citation71 The ratio of Firefly/Renilla luciferase activity was determined in both mutated and wild type forms of the reporter plasmids. The percentage of misincorporation was expressed for each tested strain as the ratio of Fmut/R and Fwt/R.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Authors' contributions
LW designed and performed experiments for , analyzed and interpreted data, wrote and graphically prepared the manuscript; EM contributed to ; MS contributed to ; BMW contributed to ; AB contributed to ; LB contributed to ; VL contributed to ; JK contributed to ; MT conceived the study, analyzed and interpreted data, wrote the manuscript.
2015CC6950R-s02.docx
Download MS Word (20.6 MB)Acknowledgments
We would like to thank prof. F. Fasiolo for anti-Tif6, anti-uL23 antibodies, Tif6-GFP expressing plasmid, and fruitful discussion; prof. E. Hurt for uS3-GFP and uL23-GFP expressing vectors; prof. A.W. Johnson for yMrt4-GFP expressing plasmid; prof. M. Seedorf for anti-uS5 antibodies; prof. J.P.G. Ballesta for anti-uL11 antibodies; prof. D. Bedwell, prof. J. Dinman for PRF dual-luciferase reporter vectors. We would also like to thank Dr D. Krokowski for fruitful discussions.
Funding
This work was supported by the National Science Center in Poland [UMO-2014/13/B/NZ1/00953 to MT and UMO-2012/05/N/NZ1/00044 to LW,], the European Regional Development Fund under the Operational Program Innovative Economy, project: “National Multidisciplinary Laboratory of Functional Nanomaterials” - ‘NanoFun’, the Project No. POIG.02.02.00-00-025/09 and partially by Polish-Swiss Research Program [PSPB-183/2010 to J.K. and V.L.]
References
- Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 2005; 121:991-1004; PMID:15989950; http://dx.doi.org/10.1016/j.cell.2005.04.015
- Grela P, Krokowski D, Gordiyenko Y, Krowarsch D, Robinson CV, Otlewski J, Grankowski N, Tchorzewski M. Biophysical properties of the eukaryotic ribosomal stalk. Biochemistry 2010; 49:924-33; PMID:20058904; http://dx.doi.org/10.1021/bi901811s
- Gonzalo P, Reboud JP. The puzzling lateral flexible stalk of the ribosome. Biol Cell 2003; 95:179-93; PMID:12867082; http://dx.doi.org/10.1016/S0248-4900(03)00034-0
- Ballesta JP, Remacha M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog Nucleic Acid Res Mol Biol 1996; 55:157-93; PMID:8787610; http://dx.doi.org/10.1016/S0079-6603(08)60193-2
- Tchorzewski M. The acidic ribosomal P proteins. Int J Biochem Cell Biol 2002; 34:911-5; PMID:12007628; http://dx.doi.org/10.1016/S1357-2725(02)00012-2
- Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell 1999; 97:491-502; PMID:10338213; http://dx.doi.org/10.1016/S0092-8674(00)80759-X
- Kavran JM, Steitz TA. Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: analysis of L11 movements. J Mol Biol 2007; 371:1047-59; PMID:17599351; http://dx.doi.org/10.1016/j.jmb.2007.05.091
- Wang L, Yang F, Zhang D, Chen Z, Xu RM, Nierhaus KH, Gong W, Qin Y. A conserved proline switch on the ribosome facilitates the recruitment and binding of trGTPases. Nat Struct Mol Biol 2012; 19:403-10; PMID:22407015; http://dx.doi.org/10.1038/nsmb.2254
- Garcia-Marcos A, Morreale A, Guarinos E, Briones E, Remacha M, Ortiz AR, Ballesta JP. In vivo assembling of bacterial ribosomal protein L11 into yeast ribosomes makes the particles sensitive to the prokaryotic specific antibiotic thiostrepton. Nucleic Acids Res 2007; 35:7109-17; PMID:17940088; http://dx.doi.org/10.1093/nar/gkm773
- Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science (New York, NY 2013; 340:1235490; PMID:23812720; http://dx.doi.org/10.1126/science.1235490
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science (New York, NY 2009; 326:688-94; PMID:19833920; http://dx.doi.org/10.1126/science.1179700
- Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J 2009; 28:755-65; PMID:19229291; http://dx.doi.org/10.1038/emboj.2009.26
- Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G' domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol Cell 2005; 20:723-31; PMID:16337596; http://dx.doi.org/10.1016/j.molcel.2005.10.028
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science (New York, NY 2010; 330:835-8; PMID:21051640; http://dx.doi.org/10.1126/science.1194460.
- Pallesen J, Hashem Y, Korkmaz G, Koripella RK, Huang C, Ehrenberg M, Sanyal S, Frank J. Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1. Elife 2013; 2:e00411; PMID:23755360; http://dx.doi.org/10.7554/eLife.00411
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 2005; 121:703-12; PMID:15935757; http://dx.doi.org/10.1016/j.cell.2005.03.023
- Sato H, Ito K, Nakamura Y. Ribosomal protein L11 mutations in two functional domains equally affect release factors 1 and 2 activity. Mol Microbiol 2006; 60:108-20; PMID:16556224; http://dx.doi.org/10.1111/j.1365-2958.2006.05094.x
- Naaktgeboren N, Schrier P, Moller W, Voorma HO. The involvement of protein L11 in the joining of the 30-S initiation complex to the 50-S subunit. Eur J Biochem 1976; 62:117-23; PMID:765132; http://dx.doi.org/10.1111/j.1432-1033.1976.tb10104.x
- Huang C, Mandava CS, Sanyal S. The ribosomal stalk plays a key role in IF2-mediated association of the ribosomal subunits. J Mol Biol 2010; 399:145-53; PMID:20385143; http://dx.doi.org/10.1016/j.jmb.2010.04.009
- Van Dyke N, Xu W, Murgola EJ. Limitation of ribosomal protein L11 availability in vivo affects translation termination. J Mol Biol 2002; 319:329-39; PMID:12051910; http://dx.doi.org/10.1016/S0022-2836(02)00304-2
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 2004; 23:1008-19; PMID:14976550; http://dx.doi.org/10.1038/sj.emboj.7600102
- Briones E, Briones C, Remacha M, Ballesta JP. The GTPase center protein L12 is required for correct ribosomal stalk assembly but not for Saccharomyces cerevisiae viability. J Biol Chem 1998; 273:31956-61; PMID:9822666; http://dx.doi.org/10.1074/jbc.273.48.31956
- Salas-Marco J, Bedwell DM. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol 2005; 348:801-15; PMID:15843014; http://dx.doi.org/10.1016/j.jmb.2005.03.025
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochimica et biophysica acta 2010; 1803:673-83; PMID:19879902; http://dx.doi.org/10.1016/j.bbamcr.2009.10.009
- Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci 2010; 35:260-6; PMID:20137954; http://dx.doi.org/10.1016/j.tibs.2010.01.001
- Michalec B, Krokowski D, Grela P, Wawiorka L, Sawa-Makarska J, Grankowski N, Tchorzewski M. Subcellular localization of ribosomal P0-like protein MRT4 is determined by its N-terminal domain. Int J Biochem Cell Biol 42:736-48; PMID:20083226; http://dx.doi.org/10.1016/j.biocel.2010.01.011
- Rodriguez-Mateos M, Abia D, Garcia-Gomez JJ, Morreale A, de la Cruz J, Santos C, Remacha M, Ballesta JP. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res 2009; 37:3514-21; PMID:19346338; http://dx.doi.org/10.1093/nar/gkp209
- Lo KY, Li Z, Wang F, Marcotte EM, Johnson AW. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J Cell Biol 2009; 186:849-62; PMID:19797078; http://dx.doi.org/10.1083/jcb.200904110
- Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Bottcher B, Granneman S, Watkins NJ, Tollervey D. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mole Biol 19:744-53; PMID:22751017; http://dx.doi.org/10.1038/nsmb.2308
- Sivan G, Kedersha N, Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol 2007; 27:6639-46; PMID:17664278; http://dx.doi.org/10.1128/MCB.00798-07
- Helser TL, Baan RA, Dahlberg AE. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide. Mol Cell Biol 1981; 1:51-7; PMID:6765595; http://dx.doi.org/10.1128/MCB.1.1.51
- Dinman JD. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev 3:661-73; PMID:22715123; http://dx.doi.org/10.1002/wrna.1126
- Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA (New York, NY 2003; 9:1019-24; PMID:12869712; http://dx.doi.org/10.1261/rna.5930803
- Gomez-Lorenzo MG, Spahn CM, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JP, Lavandera JL, Garcia-Bustos JF, Frank J. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 A resolution. EMBO J 2000; 19:2710-8; PMID:10835368; http://dx.doi.org/10.1093/emboj/19.11.2710
- Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell 2001; 8:1363-73; PMID:11779510; http://dx.doi.org/10.1016/S1097-2765(01)00403-8
- Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, Gonzalez Fernandez A, Simpson P, D'Santos CS, Arends MJ, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev 25:917-29; PMID:21536732; http://dx.doi.org/10.1101/gad.623011
- Weis F, Giudice E, Churcher M, Jin L, Hilcenko C, Wong CC, Traynor D, Kay RR, Warren AJ. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol 2015; 22:914-9; PMID:26479198
- Rodriguez-Mateos M, Garcia-Gomez JJ, Francisco-Velilla R, Remacha M, de la Cruz J, Ballesta JP. Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res 2009; 37:7519-32; PMID:19789271; http://dx.doi.org/10.1093/nar/gkp806
- Kemmler S, Occhipinti L, Veisu M, Panse VG. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J Cell Biol 2009; 186:863-80; PMID:19797079; http://dx.doi.org/10.1083/jcb.200904111
- Woolford JL, Jr., Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195:643-81; PMID:24190922; http://dx.doi.org/10.1534/genetics.113.153197
- Dashti A, Schwander P, Langlois R, Fung R, Li W, Hosseinizadeh A, Liao HY, Pallesen J, Sharma G, Stupina VA, et al. Trajectories of the ribosome as a Brownian nanomachine. Proc Natl Acad Sci U S A 2014; 111:17492-7; PMID:25422471; http://dx.doi.org/10.1073/pnas.1419276111
- Plant EP, Nguyen P, Russ JR, Pittman YR, Nguyen T, Quesinberry JT, Kinzy TG, Dinman JD. Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae. PloS one 2007; 2:e517; PMID:17565370; http://dx.doi.org/10.1371/journal.pone.0000517
- Carr-Schmid A, Durko N, Cavallius J, Merrick WC, Kinzy TG. Mutations in a GTP-binding motif of eukaryotic elongation factor 1A reduce both translational fidelity and the requirement for nucleotide exchange. J Biol Chem 1999; 274:30297-302; PMID:10514524; http://dx.doi.org/10.1074/jbc.274.42.30297
- Botet J, Rodriguez-Mateos M, Ballesta JP, Revuelta JL, Remacha M. A chemical genomic screen in Saccharomyces cerevisiae reveals a role for diphthamidation of translation elongation factor 2 in inhibition of protein synthesis by sordarin. Antimicrob Agents Chemother 2008; 52:1623-9; PMID:18285480; http://dx.doi.org/10.1128/AAC.01603-07
- Harger JW, Meskauskas A, Dinman JD. An "integrated model" of programmed ribosomal frameshifting. Trends Biochem Sci 2002; 27:448-54; PMID:12217519; http://dx.doi.org/10.1016/S0968-0004(02)02149-7
- Ioannou M, Coutsogeorgopoulos C, Synetos D. Kinetics of inhibition of rabbit reticulocyte peptidyltransferase by anisomycin and sparsomycin. Mol Pharmacol 1998; 53:1089-96; PMID:9614213
- Garreau de Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, Yusupov M. Structural basis for the inhibition of the eukaryotic ribosome. Nature 2014; 513:517-22; PMID:25209664; http://dx.doi.org/10.1038/nature13737
- Clementi N, Chirkova A, Puffer B, Micura R, Polacek N. Atomic mutagenesis reveals A2660 of 23S ribosomal RNA as key to EF-G GTPase activation. Nat Chem Biol 2010; 6:344-51; PMID:20348921; http://dx.doi.org/10.1038/nchembio.341
- Harger JW, Meskauskas A, Nielsen J, Justice MC, Dinman JD. Ty1 retrotransposition and programmed +1 ribosomal frameshifting require the integrity of the protein synthetic translocation step. Virology 2001; 286:216-24; PMID:11448174; http://dx.doi.org/10.1006/viro.2001.0997
- Dinman JD, Icho T, Wickner RB. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci U S A 1991; 88:174-8; PMID:1986362; http://dx.doi.org/10.1073/pnas.88.1.174
- Krokowski D, Tchorzewski M, Boguszewska A, McKay AR, Maslen SL, Robinson CV, Grankowski N. Elevated copy number of L-A virus in yeast mutant strains defective in ribosomal stalk. Biochem Biophys Res Commun 2007; 355:575-80; PMID:17307145; http://dx.doi.org/10.1016/j.bbrc.2007.02.024
- Gomez-Lorenzo MG, Garcia-Bustos JF. Ribosomal P-protein stalk function is targeted by sordarin antifungals. J Biol Chem 1998; 273:25041-4; PMID:9737960; http://dx.doi.org/10.1074/jbc.273.39.25041
- Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell 2008; 30:26-38; PMID:18406324; http://dx.doi.org/10.1016/j.molcel.2008.01.009
- Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J 2005; 24:4316-23; PMID:16292341; http://dx.doi.org/10.1038/sj.emboj.7600884
- Rodnina MV, Fricke R, Wintermeyer W. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry 1994; 33:12267-75; PMID:7918447; http://dx.doi.org/10.1021/bi00206a033
- Taylor D, Unbehaun A, Li W, Das S, Lei J, Liao HY, Grassucci RA, Pestova TV, Frank J. Cryo-EM structure of the mammalian eukaryotic release factor eRF1-eRF3-associated termination complex. Proc Natl Acad Sci U S A 2012; 109:18413-8; PMID:23091004; http://dx.doi.org/10.1073/pnas.1216730109
- Meskauskas A, Dinman JD. Ribosomal protein L3: gatekeeper to the A site. Mol Cell 2007; 25:877-88; PMID:17386264; http://dx.doi.org/10.1016/j.molcel.2007.02.015
- Meskauskas A, Dinman JD. A molecular clamp ensures allosteric coordination of peptidyltransfer and ligand binding to the ribosomal A-site. Nucleic Acids Res 2010; 38:7800-13; PMID:20660012; http://dx.doi.org/10.1093/nar/gkq641
- Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol 2004; 340:141-77; PMID:15184028; http://dx.doi.org/10.1016/j.jmb.2004.03.076
- Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 2010; 39:196-208; PMID:20670889; http://dx.doi.org/10.1016/j.molcel.2010.06.018
- Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, Gonzalez Fernandez A, Simpson P, D'Santos CS, Arends MJ, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev 2011; 25:917-29; PMID:21536732; http://dx.doi.org/10.1101/gad.623011
- Adams CC, Jakovljevic J, Roman J, Harnpicharnchai P, Woolford JL, Jr. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA (New York, NY 2002; 8:150-65; PMID:11911362; http://dx.doi.org/10.1017/S1355838202010026
- Bassler J, Kallas M, Hurt E. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60 S pre-ribosomal particles. J Biol Chem 2006; 281:24737-44; PMID:16803892; http://dx.doi.org/10.1074/jbc.M604261200
- Babiano R, de la Cruz J. Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 38:5177-92; PMID:20392820; http://dx.doi.org/10.1093/nar/gkq260
- Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol 2009; 7:e1000213; PMID:19806183
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 2000; 403:332-5; PMID:10659855; http://dx.doi.org/10.1038/35002118
- Garcia-Gomez JJ, Fernandez-Pevida A, Lebaron S, Rosado IV, Tollervey D, Kressler D, de la Cruz J. Final pre-40S maturation depends on the functional integrity of the 60S subunit ribosomal protein L3. PLoS Genet 2014; 10:e1004205; PMID:24603549; http://dx.doi.org/10.1371/journal.pgen.1004205
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol 2014; 24:165-9; PMID:24524803; http://dx.doi.org/10.1016/j.sbi.2014.01.002
- Burwick N, Coats SA, Nakamura T, Shimamura A. Impaired ribosomal subunit association in Shwachman-Diamond syndrome. Blood 2012; 120:5143-52; PMID:23115272; http://dx.doi.org/10.1182/blood-2012-04-420166
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 2005; 19:2199-211; PMID:16166381; http://dx.doi.org/10.1101/gad.351605
- Salas-Marco J, Fan-Minogue H, Kallmeyer AK, Klobutcher LA, Farabaugh PJ, Bedwell DM. Distinct paths to stop codon reassignment by the variant-code organisms Tetrahymena and Euplotes. Mol Cell Biol 2006; 26:438-47; PMID:16382136; http://dx.doi.org/10.1128/MCB.26.2.438-447.2006
