It is well known that DNA replication is subject to error and that mechanisms are in place to detect and repair such mistakes. Perhaps more surprisingly, recent studies have suggested a similar state of affairs with regard to transcription by RNA Polymerase II (Pol II). Transcription has been associated with the accumulation of DNA damage and tissues with high transcriptional rates such as growing tumors are at risk for increased levels of mutation.Citation1 In addition, some proteins involved in repair of DNA damage appear to play key parts in transcription. These include, factors involved in base excision repair, nucleotide excision repair, mismatch repair and recombination repair.Citation2 Here we will discuss potential mechanisms for transcription-associated DNA damage, subsequent triggering of DNA damage repair (DDR) pathways, and the influence of DDR signaling on transcription.
Genomic DNA is at increased risk for damage after chromatin decondenses close to the transcriptional start site (TSS), where naked DNA, stripped of protective nucleosomal proteins may become accessible to reactive oxygen species in the cell.Citation3 Transcription-associated damage could also be incurred, when the DNA double helix is unwound in general transcription factor (GTF)- RNA polymerase II (Pol II) complexes on gene promoters and copied into RNA sequences.Citation2,3 R loop formation may occur on initiation of transcription; in this process the growing RNA strand hybridizes with the single stranded template DNA sequence. The displaced DNA strand becomes susceptible to chemical damage and formation of secondary structures that are prone to recombination and mutation.Citation2 Melting of the DNA double strand by Pol II also results in torsion, generating negative supercoiling in the upstream DNA, with further potential for oxidative damage and positive supercoiling downstream that may retard Pol II processivity.
In addition to such sporadic damage there appears to be a pathway of “scheduled DNA strand breakage” associated with transcription. Inducible gene transcription has been shown to be accompanied by the generation of DNA double strand breaks (DSB) in cells activated by agents as diverse as heat shock, growth serum, androgens, neuronal activation or estrogens.Citation1,4-7 Increased DSB have been detected directly in transcribing cells, as well as indirectly by the Comet assay and by incorporation into chromatin of the histone γH2Ax, a surrogate marker of DNA damage.Citation5-7 Indeed direct generation of DSB in the promoters of the EGR1 and NPAS4 genes, using a CRISPR-CAS approach, was sufficient to drive transcription even in the absence of external stimuli.Citation7 Repair of DSBs requires DNA recombination repair by mechanisms including homologous recombination (HR) and non-homologous end joining repair (NHEJ).Citation2 It has been shown that some of the components of the NHEJ pathway such as DNA-dependent kinase (DNA-PK), Ku70 and Ku80 become associated with activated genes, including estrogen-induced genes, heat shock genes and serum-inducible immediate early (IE) genes.Citation4,6 The NHEJ intermediates were shown to be associated with ER-induced genes in a complex with Poly (ADP-Ribose) Polymerase 1 (PARP1), Topoisomerase IIβ and transcription appeared to require Topoisomerase IIβ-mediated nicking of sequences within the promoter of the presenilin 2 (pS2) geneCitation4 (). In fact a role for Topoisomerase IIβ in transcriptional activation appeared to be a general finding in a number of the studies.Citation4,6,7 It is not clear which exact stimulus provokes Topoisomerase IIβ to bind and induce DSBs in transcribing genes, although direct recruitment of the enzyme by the androgen receptor has been observed.Citation1
Figure 1. The Role of DNA Double Strand Break and the NHEJ Signaling Pathway in Transcription of Inducible Genes. We depict recruitment to gene promoters, by a transcription factor dimer, of a complex containing NHEJ proteins including DNA-PK, as well as Topoisomerase IIβ and PARP1. Following generation of a DSB in the promoter by Topoisomerase IIβ, DNA repair pathways are triggered, resulting in the activation of DNA-PK that phosphorylates substrates such as H2AX and Kap1/TRIM28 throughout the transcribing gene, permitting RNA elongation by Pol II.
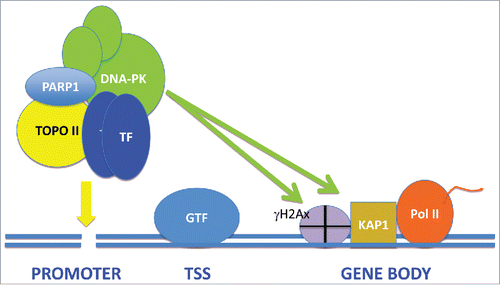
The roles of Topoisomerase IIβ nicking and recruitment of the NHEJ complex in transcription are still to be fully determined. However, PARP1, which forms a part of the complex containing DNA-PK and Topoisomerase IIβ on the pS2 gene, appeared to mediate nucleosome remodeling in the ER bound promoter.Citation4 Topoisomerase IIβ may also relieve the torsion associated with positive supercoiling downstream of Pol II.Citation7 In addition, DNA-PK and Topoisomerase II may play roles in the regulation of transcription by the relief of posttranscriptional Pol II pausing in HSP and IE genes.Citation6 A mechanism involving phosphorylation by DNA-PK of the Pol II pausing regulator Kap1/TRIM28 appeared to be required for transcriptional activation.Citation6 It is notable that in the latter studies, elevated levels of activated phospho-DNA-PK and γH2AX were detected throughout the ORFs of transcribing IE genes.Citation6 This suggested either that DSB are generated by Topoisomerase IIβ throughout the transcribing genes or that a H2Ax kinase, such as activated DNA-PK, may move processively across the gene bodies after Topoisomerase IIβ heerCitation6 (). Other substrates for DNA-PK, in addition to H2AX, including Kap1/TRIM28 may thus play roles in DDR signaling-regulated transcription.Citation6
These studies therefore suggested a primary role for Topoisomerase IIβ–mediated DSB and DDR signaling in the transcription of inducible genes. This novel mechanism is likely to contribute to many areas of biology and medicine.
References
- Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet 2010; 42:668-75; PMID:20601956; http://dx.doi.org/10.1038/ng.613
- Fong YW, Cattoglio C, Tjian R. The intertwined roles of transcription and repair proteins. Mol Cell 2013; 52:291-302; PMID:24207023; http://dx.doi.org/10.1016/j.molcel.2013.10.018
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Ann Rev Genet 2000; 34:77-137; PMID:11092823; http://dx.doi.org/10.1146/annurev.genet.34.1.77
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 2006; 312:1798-802; PMID:16794079; http://dx.doi.org/10.1126/science.1127196
- Williamson LM, Lees-Miller SP. Estrogen receptor alpha-mediated transcription induces cell cycle-dependent DNA double-strand breaks. Carcinogenesis 2011; 32:279-85; PMID:21112959; http://dx.doi.org/10.1093/carcin/bgq255
- Bunch H, Lawney BP, Lin YF, Asaithamby A, Murshid A, Wang YE, Chen BP, Calderwood SK. Transcriptional elongation requires DNA break-induced signalling. Nat Commun 2015; 6:10191; PMID:26671524; http://dx.doi.org/10.1038/ncomms10191
- Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 2015; 161:1592-605; PMID:26052046; http://dx.doi.org/10.1016/j.cell.2015.05.032
