Eukaryotic cells adapt the endocytic pathway to sort, traffic and deliver neo-synthesized or internalized cargoes to their destinations. Early endosomes comprise vacuolar sorting endosomes and tubular recycling endosomes. By emerging and extending from the vacuole, these tubules detach and are transported along the microtubule network before fusing with their target compartments (plasma membrane or lysosome related organelles [LROs]).
Although recycling endosomes contribute to many cellular functions (cell polarity, division, migration or signaling), how they form is still unclear. Recently, however, the light came from the dark. Pigment producing cells have shed light on the molecular mechanism responsible for the biogenesis of recycling endosomal tubules from sorting endosomesCitation1
Skin melanocytes exploit their endocytic pathway to generate melanosomes, LROs in which melanin pigments are synthesized and stored. While unpigmented pre-melanosomes originate from sorting endosomes, their maturation into pigmented melanosomes requires the transport of specific cargoes (i.e. melanogenic enzymes) via tubular recycling endosomes.Citation2 Impairing the endosome-to-melanosome pathway leads to tissue hypopigmentation illustrated in the Hermansky-Pudlak Syndrome (HPS), a genetic disorder characterized by oculocutaneous albinism. HPS mutated genes encode subunits of intracellular trafficking complexes, like the octameric Biogenesis of LRO Complex-1 (BLOC-1), which results in the most severe hypopigmentation found in human HPS (subtypes 7 to 9). Whereas BLOC-1−/− melanocytes accumulate melanogenic cargoes in endosomes and fail to generate pigmented melanosomes, the molecular function of BLOC-1 remains unsolved.
The endosome-to-melanosome route requires the microtubule-associated motor KIF13A (Kinesin Family member 13A) whose depleted melanocytes harbored BLOC-1-like hypopigmentation phenotype.Citation2 KIF13A generates long recycling endosomal tubules transported toward melanosomes for pigmentation or plasma membrane for conventional recycling.Citation3 Knowing that BLOC-1 and KIF13A are ubiquitously expressed, BLOC-1 might contribute to the modeling of recycling tubules in melanocytes and non-pigment cells.
Live imaging of BLOC-1-depleted HeLa cells showed that KIF13A accumulated in static and aberrant sorting endosomes devoid of tubules. To get insights into this BLOC-1-dependent morphogenetic step, high-pressure freezing and Correlative Light and Electron Microscopy (CLEM) were combined to preserve endosomal membranes and assign an ultrastructure to a fluorescent event of interest. Upon BLOC-1 depletion, quantitative CLEM analysis showed that KIF13A-positive sorting endosomes displayed an increased number of spherical budding profiles. Given that BLOC-1 localize to endosomal tubulesCitation4 and might adopt a structure with affinity for highly curved membranes,Citation5 BLOC-1 promote the KIF13A-dependent elongation of spherical buds into nascent tubules.
Once elongated, the nascent tubule probably detaches via local actin polymerization. Inhibiting the actin-branching activity of the ARP2/3 complex prevented the formation of KIF13A-positive tubules. Among ARP2/3 activators, the WASH (Wiskott-Aldrich Syndrome Homolog) complex and Annexin A2 localize and function on endosomes. WASH interact with BLOC-1,Citation6 but do not contribute to the formation of KIF13A-positive tubules. Yet, live imaging on Annexin A2-depleted cells recapitulated the phenotype of BLOC-1-depleted or ARP2/3-inhibited cells. Surprisingly, CLEM on those cells showed that sorting endosomes extended tubular profiles in continuity of their limiting membranes. Thus, the Annexin A2-dependent actin polymerization controls the fission of recycling tubules from sorting endosomes, with BLOC-1 acting upstream of Annexin A2. This suggests also that different actin-polymerizing machineries (WASH or Annexin A2) contribute to indistinguishable remodeling events that likely operate on distinct endosomal subdomains.
BLOC-1 coordinate the action of microtubule and actin cytoskeletons to elongate and release recycling tubules from sorting endosomes (). In non-pigment cells, the BLOC-1/KIF13A/Annexin A2 triad controls the recycling of endocytic cargoes (transferrin). Is this mechanism transposable to melanogenic cargoes in pigment cells? Using melanin synthesis and melanosome biogenesis as functional readouts of the formation and transport of recycling endosomes, this triad functioned within the same endosome-to-melanosome pathway, explaining the cellular and molecular pigmentary defects observed in patients suffering from HPS7-9. Interestingly, Kif13A KO mice do not display coat color dilution,Citation7 indicating that another kinesin might compensate in vivo for the need of KIF13A in melanocytes. Yet, BLOC-1 and Kif13A KO mice harbor neurological defects and altered surface expression of specific receptors in neurons, reflecting likely the need of BLOC-1 and KIF13A to recycle particular cargoes.
Figure 1. The biogenesis of recycling endosomes starts from sorting endosome with the generation of a bud (1) to which BLOC-1 likely bind (2). By interacting with KIF13A, BLOC-1 promote the extension of nascent tubule along microtubules (3). Then, Annexin A2 and branched actin filaments polymerization (4) release and/ or stabilize recycling tubules directed toward plasma membrane or pigmented melanosomes.
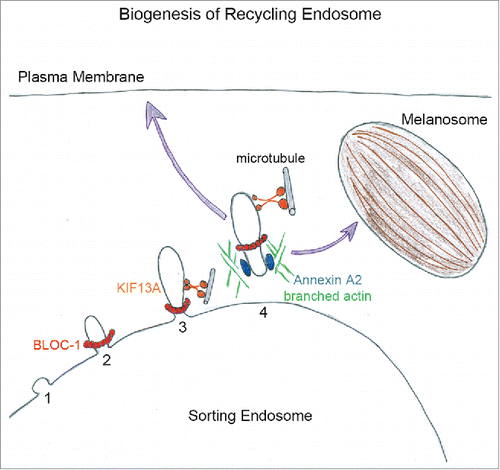
BLOC-1, KIF13A and Annexin A2 function in ubiquitous and specific recycling pathways (). Whether they correspond to unique or distinct populations of tubular endosomes remains unsolved. Beyond their final destinations — plasma membrane or melanosomes — the dynamic of recycling tubules in melanocytes appears singular. By live imaging on melanocytes, recycling tubules “sense” their environment by extending and retracting before stably contacting melanosomes.Citation2 By electron tomography and 3D-reconstruction, these tubules form ‘pipes’ connected to both sorting endosome and melanosome.Citation2 This reveals that detachment of recycling tubules from vacuolar endosomes is not required for their function, and suggests that actin polymerization should be tightly tuned to either stabilize or fission the tubule. 3D reconstruction identified also multiple endosomal tubules emerging from the ‘pipe’. How do they form; where do they go; do they share cargoes, trafficking machineries or functions might be the purpose of intense research.
Deciphering the biogenesis of recycling endosomes reveals the spatiotemporal complexity required to coordinate the emergence of a thin tubule from a large ‘balloon’ concomitantly to the sorting, transport and delivery of molecules. Undoubtedly, we are just starting to crack the recycling mystery.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Delevoye C, et al. Curr Biol 2016; 26, 1-13; PMID:26725201; http://dx.doi.org/10.1016/j.cub.2015.11.020
- Delevoye C, et al. J Cell Biol 2009; 187, 247-64; PMID:19841138; http://dx.doi.org/10.1083/jcb.200907122
- Delevoye C, et al. Cell Rep 2014; 6, 445-54; PMID:24462287; http://dx.doi.org/10.1016/j.celrep.2014.01.002
- Di Pietro SM, et al. Mol Biol Cell 2006; 17, 4027-38; PMID:16837549; http://dx.doi.org/10.1091/mbc.E06-05-0379
- Lee HH, et al. J Biol Chem 2012; 287, 5882-90; PMID:22203680; http://dx.doi.org/10.1074/jbc.M111.325746
- Ryder PV, et al. Mol Biol Cell 2013; 24(14):2269-84; PMID:23676666; http://dx.doi.org/10.1091/mbc.E13-02-0088
- Zhou R, et al. Cell Rep 2013; 3, 509-19; PMID:23438369; http://dx.doi.org/10.1016/j.celrep.2013.01.014
