ABSTRACT
Numerous regulatory factors in epidermal differentiation and their role in regulating different cell states have been identified in recent years. However, the genetic interactions between these regulators over the dynamic course of differentiation have not been studied. In this Extra-View article, we review recent work by Lopez-Pajares et al. that explores a new regulatory network in epidermal differentiation. They analyze the changing transcriptome throughout epidermal regeneration to identify 3 separate gene sets enriched in the progenitor, early and late differentiation states. Using expression module mapping, MAF along with MAFB, are identified as transcription factors essential for epidermal differentiation. Through double knock-down of MAF:MAFB using siRNA and CRISPR/Cas9-mediated knockout, epidermal differentiation was shown to be impaired both in-vitro and in-vivo, confirming MAF:MAFB's role to activate genes that drive differentiation. Lopez-Pajares and collaborators integrated 42 published regulator gene sets and the MAF:MAFB gene set into the dynamic differentiation gene expression landscape and found that lncRNAs TINCR and ANCR act as upstream regulators of MAF:MAFB. Furthermore, ChIP-seq analysis of MAF:MAFB identified key transcription factor genes linked to epidermal differentiation as downstream effectors. Combined, these findings illustrate a dynamically regulated network with MAF:MAFB as a crucial link for progenitor gene repression and differentiation gene activation.
Introduction
The skin epidermis is vital for the body to regulate transepidermal water loss as well as to defend against environmental stressors. As a stratified squamous epithelial tissue, the epidermis is composed of multiple cell layers: a basal layer, that contains tissue renewing progenitor cells; a spinous layer that contains cells early in differentiation; and, the granular and cornified layers, which provides the outermost lipid barrier.Citation1 While the process of morphological development in the epidermis is well characterized, our understanding of the molecular mechanisms that underlie differentiation continues to grow. To maintain homeostasis of epidermal tissue a careful balance of progenitor cell self-renewal and terminal differentiation is crucial.Citation1,2 Recent work has identified mechanisms that drive progenitor differentiation, such as the Notch signaling pathway, as well as the involvement of structural proteins that are key for terminal differentiation, such as Keratins 1 and 10.Citation3-9 Conversely, the epigenetic regulators DNMT1, EZH2 and ACTL6a have been shown to enforce the progenitor state.Citation10-12 However, the majority of these regulators have been studied on an individual basis. Progenitor cell differentiation requires initiation of a complex, coordinated, and tightly regulated program, yet the specific mechanisms enforcing stage-dependent gene patterns remain unclear. Thus, further investigation, with particular attention to the role of transcription factors (TFs), is warranted.
This extra views article will focus on a recent study by Lopez-Pajares et al. that characterizes genetic signatures in progenitor and differentiation states and identifies a novel network of transcription factors and lncRNAs that allows for the most complete view to date of the genetic interactions that drive human epidermal differentiation.Citation13
MAF and MAFB are key to progenitor differentiation
Recent studies have identified mechanisms that coordinate gene expression to drive differentiation. For example, Moriyama et al. used loss- and gain-of-function studies to understand the downstream effects of the Notch signaling pathway.Citation14 By conditionally down-regulating Notch signaling in embryonic mouse epidermis, they observed a thinner epidermis characterized by a reduction in the spinous and granular layers. Furthermore, Notch was found to up- and down-regulate genes throughout the epidermis using mechanisms both dependent and independent of the Hes-1 transcription factor. Interestingly, Ctip2, a C2H2 zinc finger transcription factor, has been found to induce Notch1 in differentiating keratinocytes. Chromatin immunoprecipitation (ChIP) analysis revealed that the Ctip2:Notch1 interaction was direct and additional data suggested that Ctip2 positively regulates Notch1 as well.Citation15 Together, these studies provides excellent examples of mechanistic regulation of epidermal differentiation, however additional mechanism remain undefined.
To explore novel mechanisms involved in epidermal differentiation, Lopez-Pajares et al. analyzed dynamic gene expression from progenitors to fully stratified epithelium in regenerated epidermal tissue.Citation13 Three genetic signatures, defined as progenitor, early, and late differentiation, were identified based on transcriptomic analysis throughout the 7-day regeneration period. Expression module mapping was then used to identify transcriptional regulators whose expression patterns correlated with progenitor and differentiation gene signatures. Out of 100 regulators predicted to regulate epidermal differentiation, the transcription factor, MAF, was the most frequently associated with differentiation modules. Its family member, MAFB, was also highly correlated with differentiation modules. Intriguingly, expression modules predicted that MAF and MAFB might serve to activate and repress genes during differentiation.
MAF and MAFB (MAF:MAFB) are basic leucine zipper TFs that are part of the AP-1 superfamily.Citation16,17 The MAF subfamily contains both small (150-160 amino acids) and large (240-340) MAFs which have been linked to controlling cell states and terminal differentiation in cell types such as pancreatic β cells, as well as in numerous tissues such as bone, brain and kidney.Citation16,18-20 MAF and MAFB protein expression has been observed in the epidermis and hair during rat embryonic development, however their mechanistic role in epidermis was unknown.Citation21,22 Lopez-Pajares et al. confirmed the expression of MAF and MAFB in the epidermis, where they were found to be exclusively expressed in the suprabasal layers of normal adult human skin.Citation13
To build on the initial findings that MAF and MAFB may be important TFs for differentiation, loss- and gain-of function experiments in organotypic tissue were employed to establish their role in differentiation. First, single and double siRNA-mediated knockdown of MAF and MAFB demonstrated impaired differentiation characterized by loss of expression of known differentiation marker genes, such as KRT1, FLG and LOR.Citation13 To confirm these results and observe long-term effects of MAF:MAFB depletion the authors turned to CRISPR/Cas9 genome-editing to completely ablate both MAF and MAFB in primary epidermal keratinocytes. These cells were then used to generate tissue xenografts and were followed for 21 d.Citation13 This was the first example of a CRISPR/Cas9 gene-edited human primary epithelial tissue. Similar to short-term studies, the MAF:MAFB ablated cells failed to activate numerous differentiation genes. To determine if MAF:MAFB can drive differentiation gene expression, gain-of-function experiments were carried out that enforced expression of MAF:MAFB in progenitor keratinocytes. Strikingly, expression of keratin-1 was observed in the basal layer of organotypic epidermal tissue overexpressing MAF:MAFB, as well as aberrant expression of loricrin, a marker of late-terminal differentiation, in the spinous layer. Finally, gain-of-function experiments using clonogenic growth assays and MARK-IT, a stem cell competition assay in tissue, demonstrated that progenitors overexpressing MAF:MAFB failed to self-renew and indicated a role for MAF:MAFB in promoting cell cycle exit. Together, these results show that MAF:MAFB are key regulators of epidermal differentiation.
MAF:MAFB in the known epidermal genetic Landscape
To fully understand how the MAF:MAFB TFs function within the epidermis, the effects of MAF:MAFB loss during differentiation was assessed at the transcriptome level. In total, 393 genes were found to be differentially expressed with MAF:MAFB loss, of which 315 were downregulated and 78 were upregulated.Citation13 GO term analysis showed that genes downregulated by MAF:MAFB loss were associated with epidermal differentiation, whereas the upregulated genes were associated with progenitor function.Citation13 Interestingly, the loss of either MAF or MAFB resulted in very little gene expression change, implying that they may functionally compensate for each other.
Further bioinformatic analysis was performed using gene set enrichment analysis (GSEA) to incorporate the MAF:MAFB gene set into the known landscape of epidermal regulators. The authors used 42 published gene sets to create a catalog of which epidermal regulators control the distinct differentiation gene signatures.Citation13 Multi-dimensional GSEA was performed to statistically determine specific target gene signatures for these regulators throughout differentiation. MAF:MAFB were shown to control genes involved in late differentiation, with comparable numbers to p63, KLF4 and ZNF750, known critical regulators of terminal differentiation. Expectedly, the reverse GSEA, assessing repressed genes, found that MAF:MAFB was repressing gene in the progenitor signature.
Beyond MAF:MAFB's repression of the progenitor state and activation of pro-differentiation genes, Lopez-Pajares et al. found that the MAFs regulate other downstream TFs. Using ChIP-seq coupled with transcriptome analysis 80 genes were identified as directly bound and regulated by both MAF and MAFB.Citation13 These genes were found to be enriched for transcriptional regulation based on GO term analysis. Among these genes were GRHL3, KLF4, ZNF750 and PRDM1, TFs known to promote differentiation. MAF:MAFB was shown to be required for expression of these TFs, and ChIP-qPCR verified that MAF:MAFB were bound near the genomic loci encoding these TFs.Citation13 Using FOCIS analysis, a bioinformatic method to interrogate genomic intervals for TF binding, MAF:MAFB bound genomic intervals were enriched for the p63 motif. p63 is a TF known as a master regulator of epidermal homeostasis and these data suggested that MAF:MAFB work cooperatively with p63.Citation23,24 Lopez-Pajares et al. confirmed these findings using sequential ChIP-qPCR to show that MAF:MAFB and p63 were localized to the same genomic regions during differentiation.Citation13 Interestingly, they also showed that MAF:MAFB are themselves regulated by p63, thus generating a complex regulatory network of TFs orchestrating epidermal differentiation gene expression.
Upstream regulators of MAF:MAFB
Several regulators of epidermal progenitor maintenance and terminal differentiation have been identified to date. In particular, epigenetic factors appear to be important for epidermal progenitor maintenance. In addition to DNMT1 and EZH2 mentioned above, Bmi-1, a member of the PRC1 Polycomb group (PcG) complex that modifies chromatin and represses genes through methylation, and other PcG's have been identified as central regulators of keratinocyte function.Citation25,26 Bmi-1 specifically was found to have expression in the basal and suprabasal layers and has been linked to progenitor maintenance. Furthermore, increasing levels of Bmi-1 correlated with a decrease in pro-differentiation factors such as AP-1.Citation25,26
In a similar manner, long non-coding RNAs (lncRNA) have emerged as important regulators in the maintenance of and differentiation of stem cells within various cell types.Citation27 In pluripotent stem cells, lncRNAs have been found to activate or repress many pluripotency-related transcription factors.Citation28 In the epidermis, the lncRNA ANCR has been implicated as a suppressor of progenitor differentiation while another lncRNA, TINCR, is important for terminal epidermal differentiation.Citation29–31
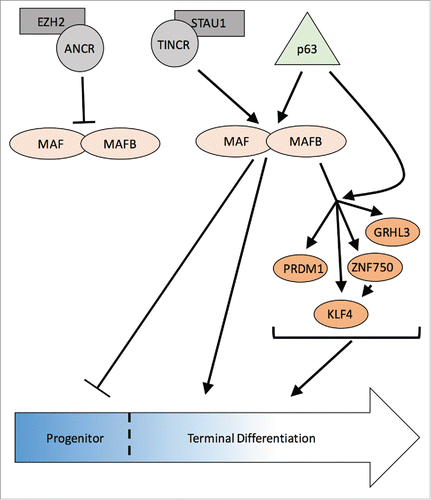
Using the catalog of epidermal gene sets, Lopez-Pajares et al. analyzed MAF:MAFB expression changes within these sets to predict upstream regulators of MAF:MAFB. Unexpectedly, an entire network of factors were predicted to predominantly repress MAF:MAFB expression, and interestingly 2 lncRNAs with opposing function converged on MAF:MAFB.Citation13 One of these was ANCR (an anti-differentiation lncRNA). Indeed, in the absence of ANCR, MAF and MAFB expression was induced in progenitor keratinocytes. Phenotypic rescue experiments of ANCR loss with concomitant MAF:MAFB loss suggested that MAF:MAFB were acting downstream of ANCR, as predicted. Of note, EZH2-mediated gene silencing has been described in epidermis, and recently ANCR has been implicated in osteoblasts to recruit EZH2 to deposit H3K27meCitation3 repressive marks on target genes, analogous to the mechanism described for Bmi-1.Citation12,32 Lopez-Pajares et al. showed this is also the mechanism by which ANCR suppresses MAF:MAFB expression in epidermis. These data suggest that repression of MAF:MAFB in the progenitor state is crucial for epidermal homeostasis.
Another predicted upstream regulators of MAF:MAFB was TINCR, a pro-differentiation lncRNA. TINCR has been found to cooperatively work with STAU1, an RNA-binding protein, to stabilize pro-differentiating mRNAs.Citation29 Together, they bind to mRNAs to ensure expression and drive differentiation through mediation of transcript stability. To confirm the prediction that TINCR is an upstream regulator of MAF:MAFB, TINCR loss-of-function studies showed suppression of MAF:MAFB expression. Furthermore, mRNA stability assays indicated that TINCR was required to stabilize MAF:MAFB mRNA.Citation13 Rescue experiments where TINCR depletion was combined with MAF:MAFB overexpression was able to drive differentiation gene expression, thus placing MAF:MAFB downstream of TINCR. Altogether, the findings by Lopez-Pajares et al. generated an intricate lncRNA-TF network critical for epidermal differentiation ().
Figure 1. MAF:MAFB are central players in an epidermal differentiation regulatory network. In epidermal progenitors, lncRNA ANCR recruits EZH2 to suppress MAF/MAFB expression and enforce the progenitor state. During differentiation, p63 induces MAF/MAFB gene expression and lncRNA TINCR functions with STAU1 to stabilize MAF/MAFB transcripts. MAF/MAFB suppress the progenitor state and induce the expression of downstream transcription factors, ZNF750, KLF4, PRDM1 and GRHL3, in part by cooperating with p63, to promote epidermal differentiation.
Concluding remarks
Throughout epidermal differentiation the network of transcription factors guiding this process is dynamically changing, illustrating the complexity of the mechanisms that drive differentiation.
Lopez-Pajares et al. have presented the most complete view of the epidermal differentiation transcriptional landscape and our understanding of epidermal differentiation has never been more defined. There is still much more to learn about the mechanisms underlying the transformation from progenitor stem cell to fully differentiated progeny. Among regulators of epidermal differentiation, the transcription factors MAF and MAFB have emerged as critical regulators of progenitor exit as well as terminal differentiation. As such, it is likely that additional regulators that may act as activating or repressive partners with MAF:MAFB may provide insight into additional regulatory mechanisms involved in differentiation. Furthermore, the entire scope of MAF:MAFB gene targets remains unclear. Thus, chromosome conformation capture coupled with the current ChIP-seq data will provide a better understanding of the MAF:MAFB gene regulatory network, as well as provide insight into MAF:MAFB mediated epigenomic effects. While these further studies will enhance our insight into gene regulatory networks operating during differentiation, the work presented, along with others, has allowed for the best framework to currently understand mechanisms regulating epidermal differentiation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 2009; 10:207-17; PMID:19209183; http://dx.doi.org/10.1038/nrm2636
- Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol 2006; 22:339-73; PMID:16824012; http://dx.doi.org/10.1146/annurev.cellbio.22.010305.104357
- Massi D, Panelos J. Notch signaling and the developing skin epidermis. Adv Exp Med Biol 2012; 727:131-41; PMID:22399344; http://dx.doi.org/10.1007/978-1-4614-0899-4_10
- Williams S, Beronja S, Pasolli H, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 2011; 470:353-8; PMID:21331036; http://dx.doi.org/10.1038/nature09793
- Watt FM, Estrach S, Ambler CA. Epidermal Notch signalling: differentiation, cancer and adhesion. Curr Opin Cell Biol 2008; 20:171-9; PMID:18342499; http://dx.doi.org/10.1016/j.ceb.2008.01.010
- Blanpain C, Lowry WE, Pasolli HA, Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev 2006; 20:3022-35; PMID:17079689; http://dx.doi.org/10.1101/gad.1477606
- Gu L, Coulombe P. Keratin function in skin epithelia: a broadening palette with surprising shades. Curr Opin Cell Biol 2007; 19:13-23; PMID:17178453; http://dx.doi.org/10.1016/j.ceb.2006.12.007
- Pan X, Hobbs RP, Coulombe PA. The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr Opin Cell Biol 2013; 25:47-56; PMID:23270662; http://dx.doi.org/10.1016/j.ceb.2012.10.018
- Lopez-Pajares V, Yan K, Zarnegar BJ, Jameson KL, Khavari PA. Genetic pathways in disorders of epidermal differentiation. Trends Genet TIG 2013; 29:31-40; PMID:23141808; http://dx.doi.org/10.1016/j.tig.2012.10.005
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 2010; 463:563-7; PMID:20081831; http://dx.doi.org/10.1038/nature08683
- Bao X, Tang J, Lopez-Pajares V, Tao S, Qu K, Crabtree GR, Khavari PA. ACTL6a Enforces the Epidermal Progenitor State by Suppressing SWI/SNF-Dependent Induction of KLF4. Cell Stem Cell 2013; 12:193-203; PMID:23395444; http://dx.doi.org/10.1016/j.stem.2012.12.014
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su I -hsi, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 2009; 136:1122-35; PMID:19303854; http://dx.doi.org/10.1016/j.cell.2008.12.043
- Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, Zarnegar BJ, Boxer LD, Rios EJ, Tao S, et al. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell 2015; 32:693-706; PMID:25805135; http://dx.doi.org/10.1016/j.devcel.2015.01.028
- Moriyama M, Durham A-D, Moriyama H, Hasegawa K, Nishikawa S, Radtke F, Osawa M. Multiple roles of Notch signaling in the regulation of epidermal development. Dev Cell 2008; 14:594-604; PMID:18410734; http://dx.doi.org/10.1016/j.devcel.2008.01.017
- Zhang L, Bhattacharya S, Leid M, Ganguli-Indra G, Indra AK. Ctip2 is a dynamic regulator of epidermal proliferation and differentiation by integrating EGFR and Notch signaling. J Cell Sci 2012; 125:5733-44; PMID:23015591; http://dx.doi.org/10.1242/jcs.108969
- Tsuchiya M, Misaka R, Nitta K, Tsuchiya K. Transcriptional factors, Mafs and their biological roles. World J Diabetes 2015; 6:175-83; PMID:25685288; http://dx.doi.org/10.4239/wjd.v6.i1.175
- Kataoka K. Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J Biochem (Tokyo) 2007; 141:775-81; PMID:17569705; http://dx.doi.org/10.1093/jb/mvm105
- Hang Y, Stein R. MafA and MafB activity in pancreatic β cells. Trends Endocrinol Metab TEM 2011; 22:364-73; PMID:21719305; http://dx.doi.org/10.1016/j.tem.2011.05.003
- Nishikawa K, Nakashima T, Takeda S, Isogai M, Hamada M, Kimura A, Kodama T, Yamaguchi A, Owen MJ, Takahashi S, et al. Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. J Clin Invest 2010; 120:3455-65; PMID:20877012; http://dx.doi.org/10.1172/JCI42528
- Aziz A, Soucie E, Sarrazin S, Sieweke M. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Sci N Y NY 2009; 326:867-71; PMID:19892988; http://dx.doi.org/10.1126/science.1176056
- Miyai M, Tanaka Y, Kamitani A, Hamada M, Takahashi S, Kataoka K. c-Maf and MafB transcription factors are differentially expressed in Huxley's and Henle's layers of the inner root sheath of the hair follicle and regulate cuticle formation. J Dermatol Sci 2010; 57:178-82; PMID:20060689; http://dx.doi.org/10.1016/j.jdermsci.2009.12.011
- Ogata A, Shimizu T, Abe R, Shimizu H, Sakai M. Expression of c-maf and mafB genes in the skin during rat embryonic development. Acta Histochem 2004; 106:65-7; PMID:15032330; http://dx.doi.org/10.1016/j.acthis.2003.10.001
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 2006; 20:3185-97; PMID:17114587; http://dx.doi.org/10.1101/gad.1463206
- Mills AA, Zheng B, Wang X-J, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999; 398:708-13; PMID:10227293; http://dx.doi.org/10.1038/19531
- Lee K, Adhikary G, Balasubramanian S, Gopalakrishnan R, McCormick T, Dimri GP, Eckert RL, Rorke EA. Expression of Bmi-1 in epidermis enhances cell survival by altering cell cycle regulatory protein expression and inhibiting apoptosis. J Invest Dermatol 2008; 128:9-17; PMID:17625597; http://dx.doi.org/10.1038/sj.jid.5700949
- Eckert RL, Adhikary G, Rorke EA, Chew YC, Balasubramanian S. Polycomb group proteins are key regulators of keratinocyte function. J Invest Dermatol 2011; 131:295-301; PMID:21085188; http://dx.doi.org/10.1038/jid.2010.318
- Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 2014; 14:752-61; PMID:24905165; http://dx.doi.org/10.1016/j.stem.2014.05.014
- Jia W, Chen W, Kang J. The functions of microRNAs and long non-coding RNAs in embryonic and induced pluripotent stem cells. Genomics Proteomics Bioinformatics 2013; 11:275-83; PMID:24096129; http://dx.doi.org/10.1016/j.gpb.2013.09.004
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013; 493:231-5; PMID:23201690; http://dx.doi.org/10.1038/nature11661
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GXY, Chow J, Kim GE, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 2012; 26:338-43; PMID:22302877; http://dx.doi.org/10.1101/gad.182121.111
- Hombach S, Kretz M. The non-coding skin: exploring the roles of long non-coding RNAs in epidermal homeostasis and disease. BioEssays News Rev Mol Cell Dev Biol 2013; 35:1093-100; http://dx.doi.org/10.1002/bies.201300068
- Zhu L, Xu P-C. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun 2013; 432:612-7; PMID:23438432; http://dx.doi.org/10.1016/j.bbrc.2013.02.036
