ABSTRACT
Barrett's esophagus (BE) is essentially a metaplasia in which the normal stratified squamous epithelium is replaced by columnar epithelium. This study focuses on the involvement of OCT4 and SOX2, 2 key cell-reprogramming factors, in the deoxycholic acid (DCA)-induced expression of the intestinal hallmarks Cdx2 and MUC2 using both in vivo and in vitro models. Up-regulated expression of OCT4 and down-regulated expression of SOX2 were observed in BE compared with normal esophagus and esophagitis. Consistent with the data in vivo, DCA induced time-dependent expression of OCT4 at both the mRNA and protein levels and decreased nuclear expression of SOX2 in Het-1A cells. Down-regulation of OCT4 expression by siRNA abrogated DCA-induced expression of Cdx2 and MUC2, whereas siRNA against SOX2 significantly upregulated the expression of both Cdx2 and MUC2. Our data indicate that both OCT4 and SOX2 play important roles in the development of BE triggered by bile acid reflux.
Introduction
Barrett's esophagus (BE) is defined as an acquired metaplastic abnormality in which the normal stratified squamous epithelium is replaced by a specialized or intestinal-like columnar epithelium in the distal part of the esophagus. It represents the main precursor lesion for the development of esophageal adenocarcinoma, the incidence of which has dramatically increased in Western populations in recent years.Citation1-3 Although known to arise because of chronic gastroesophageal reflux, the cellular origin and molecular mechanisms underlying the development of BE remain unclear.
There are several theories to explain the cellular origins of columnar metaplasia in BE: columnar cells that characterize BE develop through the abnormal differentiation of stem cells from (a)the basal layer of the normal squamous epithelium,Citation4,5 (b)the cardia,Citation6 (c)the lining of the esophageal gland duct,Citation7 or (d)the bone marrowCitation8 or through the transdifferentiation of mature squamous cells or the stromal compartment.Citation9 In general, the stem cell theory is favored by most researchers, but up to now there is no solid experimental evidence to exclude the possibility of transdifferentiation. During the past decade, with the breakthrough and progress of somatic cell reprogramming, the transdifferentiation theory has captured researchers' attention. Rather than a stem-cell abnormality, the acidic environment created by chronic reflux might induce transdifferentiation through an epigenetic effect on postmitotic cells. Consistent with this idea, the in vitro treatment of esophageal squamous cells and gastric epithelial cells with bile acid can lead to the expression of intestinal cell markers such as Krt8, Cdx2 and MUC2.Citation10,11
OCT4 and SOX2 are 2 of the key factors involved in somatic cell reprogramming.Citation12 It is reported that SOX2 and p63 cooperate to promote squamous-cell differentiation in the esophagus,Citation13 loss of either results in failed squamous epithelial formation and persistent mucus-producing ciliated columnar epithelial cells in embryonic and adult mice.Citation14 However, the role of SOX2 in the development of BE remains unexplained. OCT4 is an important factor that maintains pluripotency in embryonic stem cells (ESCs). A set of direct and indirect interactions of OCT4 with the bone morphogenetic proteins (BMP) dorsoventral patterning network in the process of embryonic development has been reported.Citation15 Our previous study confirmed that acid and bile salt increased the expression of BMP4, thus inducing villin expression in human esophagus epithelium cells. Therefore, BMP4 may play an important role in the development of BE.Citation16 Recently, Wang et al have found that compared with a normal esophagus, BE displays elevated expression of OCT4.Citation17 These evidence make us speculate that OCT4 may also play a role in the development of BE.
In this study, to explore whether OCT4 and SOX2 are involved in the process of intestinal metaplasia, we examined the expression of OCT4 and SOX2 in BE, normal esophagus and esophagitis tissues, both in human specimens and in a surgical bile acid reflux rat model. We also observed the effects of DCA exposure on OCT4 and SOX2 expression in the human esophagus epithelium cells (Het-1A) and the effects of modulation on the expression levels of OCT4 and SOX2 on intestinal metaplasia. We showed that both OCT4 and SOX2 were involved in BE development.
Results
Evaluation of OCT4 and SOX2 expression in human BE versus normal esophageal squamous epithelium and esophagitis tissues with IHC
As a first step, we examined the expression of OCT4 and SOX2 in patients with BE (10 patients), patients with esophagitis (15 patients) and healthy donors (15 individuals) who underwent a GI cancerization tract endoscopy in the Southwest Hospital with immunohistochemistry. In addition, biopsies of normal colons from 15 patients were stained as comparisons. All of the BE specimens lack either dysplasia or cancerization.
Increased expression of OCT4 was seen in BE compared with normal esophageal tissues and the esophagitis (). As shown in Table S4, substantial nuclear expression of OCT4 staining was observed in 70% of cases (7/10) of BE, both in the intestinal Barrett gland and the squamous epithelium of the BE patient. In contrast, OCT4 expression could not be observed in normal esophageal squamous epithelium (0/15; 0%). Weak expression (+1) was observed in the basal layers of squamous epithelium in the esophagitis (2/15; 13%). Expression of OCT4 in the colon showed a decrease compared with BE (2/15; 13%).
Figure 1. Representative expression of transcription factors (OCT4, SOX2), intestinal hallmarks (Cdx2, MUC2) and squamous hallmark (P63) in human normal esophagus, esophagitis and BE. All of the tissue sections were stained immunohistochemically with a specific antibody (×100 magnification). Colon epithelium was referenced as the control tissue.NC: negative controls.
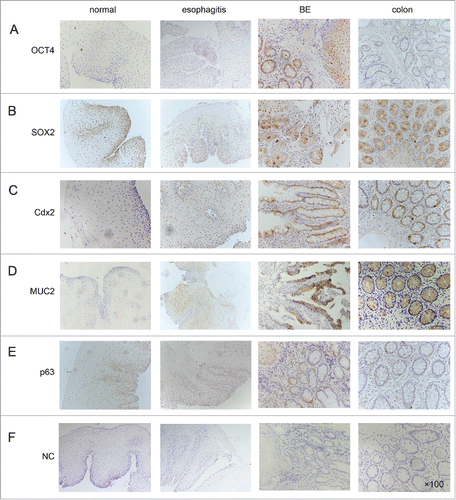
All of the normal esophageal tissues revealed a strong nuclear expression (+3) of SOX2 especially in basal layer cells. Moderate (+2) expression was observed in the esophagitis tissue. In Barrett glands and the colon tissue, nuclear expression of SOX2 was not observed ().
To assess the lineage conversion, we also evaluated the expression of intestinal cell markers Cdx2, MUC2 and a squamous differentiation marker p63. Increased expression of Cdx2 was seen in BE compared with normal esophageal tissues and the esophagitis. In all BE and colon mucosa, Cdx2 was moderately (+2) expressed in glandular areas, whereas in normal esophageal squamous mucosa, Cdx2 immunostaining was not seen, and a small number of cases (2/15; 13%) showed weak staining (+1) in esophagitis mucosa (). Similar results were obtained for MUC2 expression. MUC2 expressed predominantly in the cytoplasm of goblet cells dispersed within the colon and BE glands (). Weak cytoplasm staining (+1) was observed in esophagitis (2/15; 13%), and no expression was observed in the normal squamous epithelium (0/15; 0%). In contrast, p63 showed a decreased nuclear expression in BE compared with normal esophageal tissues and the esophagitis ().
Evaluation of Oct4, Sox2 expression in BE vs. normal esophageal squamous epithelium and esophagitis tissues with IHC in a surgical bile acid reflux rat model
Bile acid is known to play an important part in the development of esophagitis and BE. Next, we explore the expression of Oct4 and Sox2 in a bile regurgitation animal model, as described in the methods section.
Sixty animals underwent esophagoduodenostomy plus gastrectomy, with an overall mortality of 13.3% (8/60). Five of the 52 animals developed Barrett metaplasia with goblet cells (9.6%; 1 at 15 weeks after surgery, 4 at 30 weeks after surgery), and 35 cases of esophagitis were observed by 30 weeks after surgery (details showed in Table S3). Immunohistochemistry studies were performed to analyze the expression of Oct4, Sox2, Cdx2, MUC2 and p63.
As shown in , there was a marked increase in nuclear staining of Oct4 in BE compared with normal esophagus and esophagitis mucosa. Similar to human BE tissue, our bile regurgitation rat model resulted in decreased expression of Sox2 in BE compared with normal esophageal tissues and the esophagitis.
Figure 2. Representative expression of transcription factors (OCT4, SOX2), intestinal hallmarks (Cdx2, MUC2) and squamous hallmark (P63) in normal esophagus, esophagitis and BE of surgical bile acid reflux rats. All of the tissue sections were stained immunohistochemically with a specific antibody (×100 magnification). NC: negative controls.
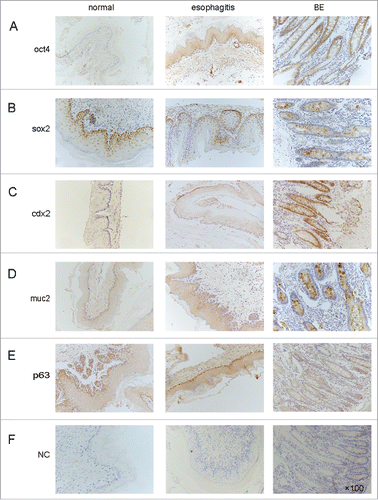
As in human tissue, the immunohistochemistry analysis of the rats' esophageal tissues revealed increased expression of Cdx2 and MUC2 and decreased expression of p63 in BE compared with normal esophageal tissues and the esophagitis.
Taken together, the data suggest that the transcription factors OCT4 and SOX2 may be involved in the process of intestinal metaplasia triggered by bile acid. To confirm this hypothesis, we investigated the effects of bile acids on OCT4 and SOX2 expression using a squamous epithelial cell line Het-1A.
Effect of DCA exposure on OCT4 and SOX2 expression in Het-1A
Het-1A cells were exposed to 200 µM DCA for 2, 4, 8 and 12 hours, and the effects on the expression of transcription factors OCT4 and SOX2 were examined at the mRNA and protein levels.
The expression of OCT4 mRNA showed a 2.11-fold increase after 4 hours of exposure to DCA, the increase intensified after 8 and 12 hours of exposure (2.88- and 3.75-fold, respectively). SOX2 mRNA expression was downregulated to 0.65-fold after 8 hours of DCA treatment and 0.41-fold after 12 hours of DCA treatment. Similar changes in the expression of OCT4 and SOX2 at the protein level were also observed ().
Figure 3. Effect of DCA exposure on OCT4 and SOX2 expression in Het-1AHet-1A cells were treated with 200 μM DCA for 2–12 hours in a pH-neutral medium. Cells were collected for (A) Western Blot utilizing specific antibodies and (B) real-time PCR to analyze the expression of OCT4, Cdx2, MUC2 and P63. (C) Cell viability of Het-1A cells treated with DCA at different time points(2 h, 4 h, 8 h, 12 h, 18 h, 24 h) was analyzed by MTT assay. The results are expressed as the mean ± SEM of 3 experiments. *P < 0.05, **P < 0.01, ***P < 0.001 was the statistically significant difference compared with the control (n = 3 for each group).
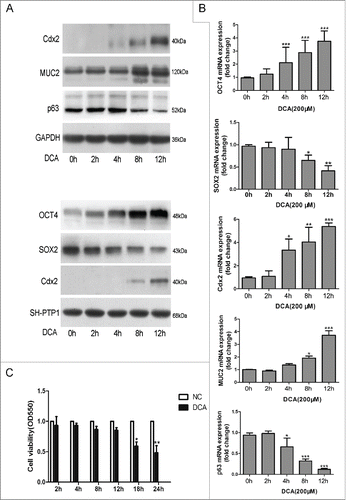
Cdx2, MUC2 and p63 were also tested after exposure to 200 µM DCA for 2–12 hours. Cdx2 expression in mRNA levels significantly increased after treatment for 4 hours and showed a maximum increase of 5.38-fold after a 12-hour of DCA exposure. MUC2 expression was up-regulated by DCA at later time points. After 8 hours, MUC2 was upregulated to 1.92-fold and showed a maximum increase of approximately 3.71-fold after 12 hours of DCA exposure. On the other hand, p63 expression was suppressed in time-dependent manners after 4 to 12 hours of treatment. Similar changes in the expression at the protein level were also observed, except that Het-1A cells expressed trace levels of Cdx2 proteins and p63 was significantly down-regulated after 12-hours of DCA treatment (0.36-fold), which was later compared with mRNA expression ().
To clarify whether the above results are caused by decreased cell activity after DCA exposure, viability of Het-1A cells treated with DCA at different time points(2 h, 4 h, 8 h, 12 h, 18 h, 24 h) was measured by MTT assay. As showed in , the cell viability was not significantly affected after treating with 200µM DCA at time points 2 h, 4 h, 8 h, 12 h and was decreased by 38% and 51% at 18 h and 24 h.
Effects of modulations in the expression levels of OCT4 on esophageal epithelial cells
Our data confirmed that the expression of OCT4 can be augmented by exposure in 200 µM DCA. Simultaneously, intestinal markers Cdx2 and MUC2 were up-regulated. Next, to investigate whether the induction of Cdx2 and MUC2 occurs via activation of OCT4, the siRNA approach against OCT4 was used to inhibit endogenous OCT4 in Het-1A cells. First, we screened 2 effective siRNAs from 4 siRNAs against OCT4 (Fig. S1A), and the optimal concentration of siRNA (50 nM) for transfection and time for OCT4 inactivation (48 hours) were assessed. Next, Het-1A cells were incubated with DCA (at 200 µM) for 12 hours after OCT4-siRNA transfection. The expression of Cdx2, MUC2 and p63 was evaluated through Western Blot analyses and RT-PCR.
Transfection of the Het-1A cell line with OCT4-siRNAs significantly inhibited OCT4 expression (reduced to 22 ± 3% and 8 ± 2%) and led to a reduction in expression for MUC2. The expression of p63 and Cdx2 show no significant change, whereas the knockdown of OCT4 significantly suppressed DCA-induced Cdx2 expression. Similarly, after OCT4-siRNA transfection, the up-regulation effect of MUC2 and the downregulation effect of p63 by DCA were significantly blocked (). Similar results were observed in the mRNA level ().
Figure 4. Effects of modulations in the expression levels of OCT4 on esophageal epithelial cellsHet-1A cells were transfected with either siRNA directed against OCT4 or negative control siRNA. 48 hours after transfection, cells were treated with or without DCA (200 μM) for 12 hours. Then, cells were collected for (A) Western Blot utilizing specific antibodies and (B) real-time PCR to analyze the expression of OCT4, Cdx2, MUC2 and P63. (C): Quantitative analysis of WB showing relative densities of bands normalized to GAPDH levels from 3 separate experiments. N=3, ANOVA, **P < 0.01, ***P < 0.001 compared with the NC-siRNA group, +++P<0.001 compared with the NC-siRNA+DCA group. NC: Negative control without siRNA transfection. Het-1A cells were transfected with either an empty lentivirus vector or OCT4 expression lentivirus vector. Cells were collected 48 hours after transfection, and then subjected to (D) Western Blot analysis and (E) real-time PCR analysis for OCT4, Cdx2, MUC2 and P63. N = 3, ANOVA, ***P < 0.001 compared with the LV-NC group. Blots shown are representative of 3 separate experiments.
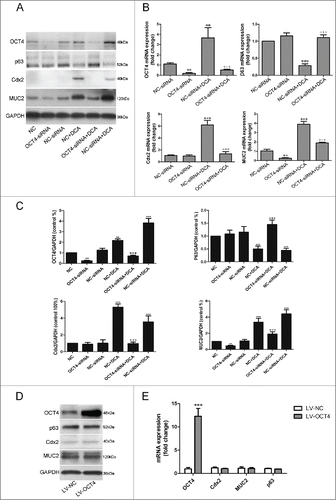
Having demonstrated that OCT4 knockdown blocked the regulation of Cdx2, MUC2 and p63 by DCA, we next questioned whether OCT4 over-expression was sufficient to initiate the intestinal phenotype in squamous epithelium cells Het-1A. To address this we designed a lentiviral construct over-expressing OCT4 (LV-OCT4). LV-NC containing a short noncoding sequence was used as a control. Overexpression of OCT4 was confirmed by Western Blot and RT-PCR, and a 9-12-fold increase in OCT4 protein level was observed. Over-expression of OCT4 had no effect on Cdx2, MUC2 and p63 expression in Het-1A cells, suggesting that OCT4 was not itself sufficient to initiate intestinal phenotype ().
Effects of modulations in the expression levels of SOX2 on esophageal epithelial cells
To better estimate the role of SOX2 in Barrett's metaplasia, a series of experiments with the application of siRNA against SOX2 was performed. Specific knockdown of endogenous SOX2 was observed (Fig. S1B). Knockdown of SOX2 significantly upregulated the expression of both Cdx2 and MUC2, whereas p63 expression was downregulated in cells transfected with SOX2-siRNA. Furthermore, DCA treatment (200 µM, 12 h) of Het-1A cells pretreated with SOX2-siRNA significantly enhanced DCA-induced Cdx2 protein expression versus nonspecific RNA-treated controls (). Similar results were observed in mRNA levels, except that an additive effective on DCA efficiency to stimulate MUC2 expression was also observed ().
Figure 5. Effects of modulations in the expression levels of SOX2 on esophageal epithelial cellsHet-1A cells were transfected with either siRNA directed against SOX2 or negative control siRNA. 48 hours after transfection, cells were treated with or without DCA (200 μM) for 12 hours. Then, cells were collected for (A) Western Blot utilizing specific antibodies and (B) real-time PCR to analyze the expression of SOX2, Cdx2, MUC2 and P63. (C): Quantitative analysis of WB showing relative densities of bands normalized to GAPDH levels from 3 separate experiments. N = 3,ANOVA, **P < 0.01, ***P < 0.001 compared with the NC-siRNA group, +P<0.05, ++P<0.01, +++P < 0.001, compared with the NC-siRNA+DCA group. NC: Negative control without siRNA transfection. Het-1A cells were transfected with either an empty lentivirus vector or SOX2 expression lentivirus vector. Cells were collected 48 hours after transfection and then subjected to (D) Western Blot analysis and (E) real-time PCR analysis for SOX2, Cdx2, MUC2 and P63. N = 3, ANOVA, **P < 0.01, ***P < 0.001 compared with the LV-NC group. Blots shown are representative of 3 separate experiments. Quantitative analysis of whole cellular and nuclear SOX2 levels showing relative densities of bands normalized to GAPDH and SH-PTP1 levels from 3 separate experiments.
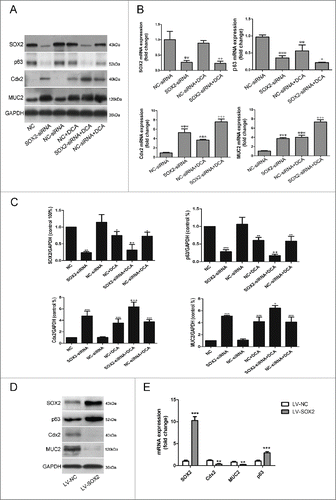
To investigate whether SOX2 upregulation could conversely decrease Cdx2 and MUC2 expression, a SOX2 overexpression lentivirus vector was transfected into the Het-1A cells, and the expression of Cdx2 and MUC2 was determined by RT-PCR and a Western Blot. Over-expression of SOX2 significantly reduced the expression of Cdx2 and MUC2 protein or mRNA compared with control groups (p < 0.01), and it increased the expression of the p63 protein or mRNA by 3- to 5-fold ().
Evaluation of the interaction between OCT4 and SOX2
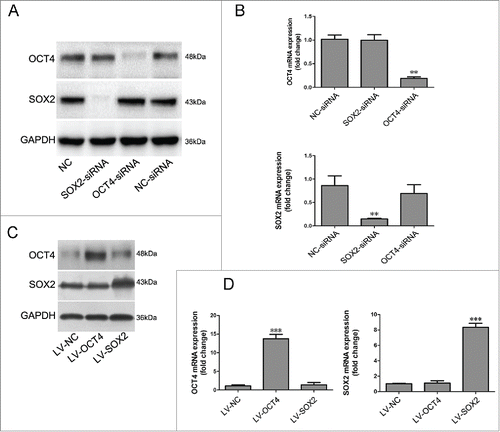
The simultaneous up-regulation of OCT4 and down-regulation of SOX2 were observed in Het-1A cells after DCA exposure. We asked whether OCT4 and SOX2 interact with each other in Het-1A cells. Knockdown or overexpression of the 2 transcription factors was performed as mentioned above. As shown in , neither knockdown nor over-expression of OCT4 affected expression of SOX2. Similarly, neither knockdown nor overexpression of SOX2 affected the expression of OCT4 in Het-1A cells.
Figure 6. Evaluation of interaction between OCT4 and SOX2.Het-1A cells were transfected with SOX2-siRNA, OCT4-siRNA or negative control siRNA. Forty-eight hours after transfection, cells were collected for (A) Western Blot utilizing specific antibodies and (B) real-time PCR to analyze the expression of SOX2 and OCT4. N = 3, ANOVA,**P < 0.01, compared with the NC-siRNA group.Het-1A cells were transfected with the OCT4 expression lentivirus vector and SOX2 expression lentivirus vector or an empty lentivirus vector. Cells were collected 48 hours after transfection and then subjected to (C) Western Blot analysis and (D) real-time PCR analysis for OCT4 and SOX2. N = 3, ANOVA, ***P < 0.001 compared with the LV-NC group. Blots shown are representative of 3 separate experiments.
Discussion
In this study, we successfully established a surgical rat model of BE, and the efficiency is similar to previous studies using a mouse or rat model.Citation18,19 We found that OCT4 expression was up-regulated in BE compared with normal esophageal squamous epithelium and esophagitis, both in human tissue and surgical bile acid reflux rat model. DCA exposure in Het-1A cells also upregulated OCT4 expression and simultaneously induced expression of intestinal differentiation factor Cdx2 and the goblet cell–specific gene MUC2. These effects were blocked when Het-A cells were transfected with a siRNA against OCT4. These results indicate that OCT4 is involved in the transition from squamous epithelium to the intestinal phenotype triggered by gastro-esophageal reflux.
OCT4 has been shown to function as a core transcription factor maintaining pluripotency.Citation20,21 It is essential for the generation of induced pluripotent stem (iPS) cells.Citation22 Because BE is essentially an intestinal metaplasia, it is plausible that transcription factors that play an important role in cell reprogramming may also play a role in the development of BE. However, little data are available regarding the relationship between OCT4 and BE. Several studies have shown that OCT4 related to esophageal squamous cell carcinoma (ESCC) pathobiology and response to treatment.Citation23-25 Recently, Wang et al have reported that the expression of OCT4 in BE was elevated compared with the normal esophagus, but decreased compared with EAC cells. Their study focused on OCT4's role in the transformation of BE to EAC, and no experiment is performed to assess the role in the intestinal metaplasia.Citation17 Coupling our inhibition and over-expression experiment with OCT4, we deduce that the transcription factor OCT4 may be involved in the development of intestinal metaplasia by forming a compound with other factors, because overexpression of OCT4 alone was insufficient to up-regulate the intestinal hallmark Cdx2 and MUC2.
One question that remains concerns the cell origin of OCT4 up-regulated cells in BE. In our immunohistochemistry experiment, OCT4 staining was observed both in Barrett's glands and in the nearby squamous epithelium in BE. In two esophagitis specimens, only traces of OCT4 expression were observed in the basal layers of squamous epithelium. In addition, we confirmed the upregulated effect of DCA exposure on OCT4 expression in Het-1A cells, which is an SV-40-immortalized, non-neoplastic, normal squamous epithelial cell line that has been previously described.Citation26 These results suggest that OCT4 up-regulated cells may originate from squamous stratified epithelium. Indeed, multipotent stem cells have been isolated from basal layers of squamous epithelium,Citation27 and metaplastic Barrett glands are also clonal units maintained by multiple stem cells of themselves.Citation28 The cell of origin for BE has been debated for decades, this question has been difficult to address in part due to the absence of useful in vivo and in vitro lineage tracing models.Citation29 Further lineage tracing is required to determine the cell origin of upregulated OCT4 expression.
It is known that SOX2 promotes the development and maintenance of squamous epithelium. It is expressed in the pharynx, esophagus, and stomach of the gut, but not in the lower tract. SOX2 mutations are associated with esophageal atresia in anophthalmia-esophageal-genital syndrome.Citation30-32 In previous works, SOX2 has been reported to be down-regulated by exposure to acid, DCA or combined media.Citation33 Bhardwaj et al have treated primary squamous and Barrett's epithelial cells, along with HaCaT (a human keratinocyte cell line), with pulses of acid for 15 minutes at pH 4.5 four times daily with bile salts for up to 6 d. As a result, acid+bile decreased SOX2 expression. Those researchers have also found that SOX2 knockdown by siRNA in HaCaT cells resulted in decreased squamous and increased columnar differentiation markers.Citation34 In this study, we exposed Het-1A cells to 200 µM DCA for 2–12 hours, SOX2 showed an decreased expression in a time-dependent manner. This result is consistent with previous studies. We also showed that inhibition of SOX2 expression with siRNA induced Cdx2 and MUC2 expression and had an additive effect on DCA's efficiency at stimulating Cdx2 and MUC2 expression in Het-1A cells.
It is believed that Cdx2 is the nuclear transcription factor that plays a crucial role in the development, proliferation, and differentiation of intestinal epithelium.Citation35-37 In this study, up-regulated expression of Cdx2 and MUC2 was observed after DCA exposure. It is reported that SOX2 and Cdx2 inhibited each other, and a fine balance between SOX2 and Cdx2 expression in the gastrointestinal tract is essential for proper development.Citation38 In our study, DCA exposure resulted in decreased SOX2 expression and increased Cdx2 expression in the nucleus. SOX2 inhibition upregulated Cdx2 expression, inferring that DCA upset the balance between SOX2 and Cdx2 and consequently changed the fate of squamous epithelium to intestinal metaplasia.
Furthermore, we investigated the relationship between OCT4 and SOX2 through knockdown or over-expression of the 2 transcription factors. It turned out that modulations in the expression levels of OCT4 did not affect the expression of SOX2, and vice versa. This result is quite different from that found in ES cells. It is reported that OCT4 and SOX2 combine by forming a complex (OCT4/SOX2), and specific knockdown of either OCT4 or SOX2 by RNA interference leads to the reduction of both genes' enhancer activities and endogenous expression levels in ESCs.Citation39,40 Synergistic action of OCT4/SOX2 was not seen in our study, which implies novel heterodimeric partners of SOX2 and OCT4 in esophageal squamous epithelial cells. Evidence supporting this conclusion may be found in the collaborative interacting pair SOX2-BRN2 discovered in the neural lineage.Citation41 Recently, it is reported that in squamous cell carcinomas (SCCs), SOX2 preferentially interacts with the transcription factor p63, as opposed to the transcription factor OCT4.Citation13
Based on our data, we propose the following mechanism of BE: when esophageal squamous epithelial cells are stimulated by bile reflux, the squamous differentiation pathway may be inactivated through inactivation of squamous transcription factor SOX2 and p63. Meanwhile, the columnar differentiation pathway may be activated through activation of OCT4. These molecular events may lead to conversion of esophageal squamous epithelium into intestinalized columnar epithelium. Our data showed that OCT4 is essential but not itself sufficient to initiate the intestinal phenotype. Further work is needed to explore the factors that collaborate with OCT4 in the development of BE. Because multiple molecular pathways (TGF-β/BMP pathway; WNT pathway; NF-κB pathway; Notch pathway; Hedgehog pathway etc.) and factors are known to contribute to BE, it is quite challenging to assemble all pieces of this puzzle. We believe these pathways may act through the core reprogramming factors mentioned above.
In summary, we showed that stemness-associated reprogramming factors OCT4 and SOX2 is involved in the transition of squamous epithelium to the intestinal phenotype triggered by gastro-esophageal reflux. The identification of OCT4 expressed in BE may have important clinical implications as a novel molecular biomarker for early diagnosis.
Materials and methods
Biopsy specimens
Human tissue samples were obtained from 10 patients with documented BE, 15 patients with normal squamous epithelium and 15 patients with reflux esophagitis undergoing an endoscopic examination at Southwest Hospital (Chongqing, China). In addition, 15 colon biopsies were selected as colonic controls. The ethical regulations of the institute on research conducted on human tissues were followed. Details of the biopsy specimens of BE are given in Table S1.
Sixty Sprague-Dawley (SD) rats (experimental animal center, Third Military University, China) with an average weight of 210 g (± 15 g SD) were used for the study. All animal experiments were approved by the Institutional Animal Care and Use Committee of Third Military Medical University. Esophagoduodenostomy plus gastrectomy was performed as a duodenal reflux model. Animals were sacrificed at 5, 15 and 30 weeks after surgery. The inferior segment of the esophagus was formalin-fixed, paraffin-embedded and subjected to immunohistochemical analysis.
Cell line and deoxycholic acid (DCA) exposure
Het-1A (ATCC) was cultured in a bronchial epithelial cell basal medium (BEBM) culture with supplements (Lonza).
Het-1A cells were grown in a medium containing final concentrations of 200 µM DCA (Sigma) at a neutral pH (pH 7.1) for 2, 4, 8 and 12 hours. Total RNA and cellular protein and nucleoprotein were extracted and subjected to different analyses.
Immunohistochemistry
Immunohistochemistry was performed using paraffin-embedded blocks. Freshly cut sections were deparaffinized in xylene and rehydrated through sequential graded ethanol steps. The tissue sections were incubated with primary antibodies, followed by incubation with secondary biotinylated antibodies (BOSTER). Bound antibodies were detected using an avidin-biotin peroxidase method (ABC kit, Vector Labs). The antibodies and dilutions used are summarized in . Mounted slides were examined through light microscopy. The intensity of staining was graded as +0 (negative), +1 (weak), +2 (moderate) or +3 (strong). Two pathologists examined all of the slides.
Table 1. Primer sequences used for real-time PCR.
RNA extraction and real-time PCR
RNA was extracted using TRIzol (Invitrogen). RNA was reverse-transcribed using a Prime ScriptTM RT reagent Kit with gDNA Eraser (TAKARA). A quantitative real-time PCR was performed using SYBR® Premix Ex Taq™ II (TAKARA) according to the manufacturer's recommendations. Primers are listed in .
Protein extraction and Western Blotting
Proteins from cells with and without treatments were extracted using Total protein Protein Extraction Kit (KeyGENE) and Nuclear Protein Extraction Kit (KeyGENE) according to the instructions. Western Blotting analysis was performed using standard protocols. An equal amount of proteins was loaded and separated using sodium dodecylsulfate polyacrylamide gel electrophoresis and then transferred to PVDF membranes. Image J software was used to measure the band density of each protein. The antibodies and dilutions used are summarized in .
Table 2. Antibody used for WB and IHC.
MTT assay
Het-1A cells were incubated in 96-well plates, after treating with 200 µM DCA (Sigma) at a neutral pH (pH 7.1) for 2, 4, 8, 12, 18 and 24 hours, MTT reagent(20 μL, 5 mg/ml) (Roche) was added to the cells and incubated at 37°C for 4 h. Then, the cells were incubated with 100 μl of solubilisation solution overnight. The next day, the viability of the cells in each well was measured by microplate reader (Bio-rad) at 550 nm(A 550 nm). The control group without DCA treatment was taken as 100% cell survival and all other groups were normalized to this value.
SiRNA inhibition of OCT4 and SOX2
SiRNA against OCT4 and SOX2 was synthesized by the Gene Pharma Co., using the sequence in Table S2. Het-1A cells were equally plated in 6-well tissue culture plates. After 24 hours, the cells were transfected with 100 pmol of the OCT4 or SOX2 siRNA for 12 hours using X-treme GENE HP DNA Transfection Reagent (Roche) according to the manufacturer's instructions. As a negative control, cells were transfected with NC-siRNA, which consists of a sequence that will not lead to specific degradation of any known cellular mRNA (Gene Pharma Co.). Forty-eight hours after transfection, cells were treated with 200 µM DCA or a control medium for 12 hours. To confirm the efficacy of siRNA, OCT4 and SOX2 expression was analyzed using Western Blot.
Lentiviral-mediated overexpression of OCT4 and SOX2
The cDNA coding for OCT4 (GenBank accession number NM-002701.5) or for SOX2 (GenBank accession number NM-003106.3) was cloned into the lentiviral vector pGag/Pol (Gene Pharma Co.) (pLV-OCT4, pLV-SOX2). To produce a negative control virus (LV-NC), a short noncoding sequence was also cloned into the pGag/Pol vector and processed in parallel with the target genes. Lentiviral vectors were generated in 293T cells using a 3-plasmid system according to the manufacturer's instruction.
Het-1A (3 × 105) cells were transduced with the lentivirus containing OCT4, SOX2 and a noncoding sequence using polybrene (5 µg /ml). 48 hours after infection, 2 µg/ml of puromycin was added to the medium for 2 weeks to select the lentivirus-infected cells. Real-time PCR assays and a Western Blot were used to detect the expression of OCT4 and SOX2 in the stable cell lines.
Data analysis
Student's two-tailed t-test was used to compare data between 2 groups. One-way analysis of variance (ANOVA) was used to compare data between 3 or more groups. Values are expressed as the mean ± SEM. P < 0.05 was used to determine statistical significance.
Disclosure of potential conflicts of interest
The authors disclose no potential conflicts of interest.
Supplementary Files
Download Zip (4.9 MB)Funding
The study was supported by the Nature Science Foundation of China (NSFC) (grant numbers 81170356, 81270450).
References
- Bhat S, Coleman HG, Yousef F, Johnston BT, McManus DT, Gavin AT, Murray LJ. Risk of malignant progression in Barrett's esophagus patients: results from a large population-based study. J Natl Cancer Inst 2011; 103:1049-57; PMID:21680910; http://dx.doi.org/10.1093/jnci/djr203
- Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett's esophagus. N Engl J Med 2011; 365:1375-83; PMID:21995385; http://dx.doi.org/10.1056/NEJMoa1103042
- Spechler SJ. Barrett esophagus and risk of esophageal cancer: a clinical review. Jama 2013; 310:627-36; PMID:23942681; http://dx.doi.org/10.1001/jama.2013.226450
- Croagh D, Phillips WA, Redvers R, Thomas RJ, Kaur P. Identification of candidate murine esophageal stem cells using a combination of cell kinetic studies and cell surface markers. Stem Cells (Dayton, Ohio) 2007; 25:313-8; PMID:17038667; http://dx.doi.org/10.1634/stemcells.2006-0421
- Kalabis J, Oyama K, Okawa T, Nakagawa H, Michaylira CZ, Stairs DB, Figueiredo JL, Mahmood U, Diehl JA, Herlyn M, et al. A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification. J Clin Invest 2008; 118:3860-9; PMID:19033657
- McDonald SA, Lavery D, Wright NA, Jansen M. Barrett oesophagus: lessons on its origins from the lesion itself. Nat Rev Gastroenterol Hepatol 2015; 12:50-60; PMID:25365976; http://dx.doi.org/10.1038/nrgastro.2014.181
- Leedham SJ, Preston SL, McDonald SA, Elia G, Bhandari P, Poller D, Harrison R, Novelli MR, Jankowski JA, Wright NA. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett's oesophagus. Gut 2008; 57:1041-8; PMID:18305067; http://dx.doi.org/10.1136/gut.2007.143339
- Sarosi G, Brown G, Jaiswal K, Feagins LA, Lee E, Crook TW, Souza RF, Zou YS, Shay JW, Spechler SJ. Bone marrow progenitor cells contribute to esophageal regeneration and metaplasia in a rat model of Barrett's esophagus. Dis Esophagus 2008; 21:43-50; PMID:NOT_FOUND
- Yu WY, Slack JM, Tosh D. Conversion of columnar to stratified squamous epithelium in the developing mouse oesophagus. Dev Biol 2005; 284:157-70; PMID:15992795; http://dx.doi.org/10.1016/j.ydbio.2005.04.042
- Morrow DJ, Avissar NE, Toia L, Redmond EM, Watson TJ, Jones C, Raymond DP, Litle V, Peters JH. Pathogenesis of Barrett's esophagus: Bile acids inhibit the Notch signaling pathway with induction of CDX2 gene expression in human esophageal cells. Surgery 2009; 146:714-22; PMID:19789031; http://dx.doi.org/10.1016/j.surg.2009.06.050
- Xu YJ, Watanabe T, Okazaki H, Tanigawa T, Watanabe K, Tominaga K, Fujiwara Y, Oshitani N, Arakawa T. Bile Acids Induce Expression of CDx2 and MUC2 in Normal Rat Gastric Epithelial Cells via Activation of Nuclear Receptor FXR - a Possible Mechanism of Intestinal Metaplasia in the Stomach. Gastroenterology 2009; 136:A616-A.
- Masip M, Veiga A, Izpisua Belmonte JC, Simon C. Reprogramming with defined factors: from induced pluripotency to induced transdifferentiation. Mol Hum Reprod 2010; 16:856-68; PMID:20616150; http://dx.doi.org/10.1093/molehr/gaq059
- Watanabe H, Ma Q, Peng S, Adelmant G, Swain D, Song W, Fox C, Francis JM, Pedamallu CS, DeLuca DS, et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clinl Invest 2014; 124:1636-45; PMID:24590290; http://dx.doi.org/10.1172/JCI71545
- Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the developing and adult mouse trachea. Development (Cambridge, England) 2009; 136:1899-907; PMID:19403656; http://dx.doi.org/10.1242/dev.034629
- Belting HG, Wendik B, Lunde K, Leichsenring M, Mossner R, Driever W, Onichtchouk D. Pou5f1 contributes to dorsoventral patterning by positive regulation of vox and modulation of fgf8a expression. Dev Biol 2011; 356:323-36; PMID:21621531; http://dx.doi.org/10.1016/j.ydbio.2011.05.660
- Zhou G, Sun YG, Wang HB, Wang WQ, Wang XW, Fang DC. Acid and bile salt up-regulate BMP4 expression in human esophageal epithelium cells. Scand J Gastroenterol 2009; 44:926-32; PMID:19488929; http://dx.doi.org/10.1080/00365520902998661
- Wang X, Yang S, Zhao X, Guo H, Ling X, Wang L, Fan C, Yu J, Zhou S. OCT3 and SOX2 promote the transformation of Barrett's esophagus to adenocarcinoma by regulating the formation of tumor stem cells. Oncol Rep 2014; 31:1745-53; PMID:24481676
- Clark GW, Smyrk TC, Mirvish SS, Anselmino M, Yamashita Y, Hinder RA, DeMeester TR, Birt DF. Effect of gastroduodenal juice and dietary fat on the development of Barrett's esophagus and esophageal neoplasia: an experimental rat model. Ann Surg Oncol 1994; 1:252-61; PMID:7842295; http://dx.doi.org/10.1007/BF02303531
- Pham TH, Genta RM, Spechler SJ, Souza RF, Wang DH. Development and characterization of a surgical mouse model of reflux esophagitis and Barrett's esophagus. J Gastrointest Surg 2014; 18:234-40; discussion 40–1; PMID:24190247; http://dx.doi.org/10.1007/s11605-013-2386-z
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005; 122:947-56; PMID:16153702; http://dx.doi.org/10.1016/j.cell.2005.08.020
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 2006; 38:431-40; PMID:16518401; http://dx.doi.org/10.1038/ng1760
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663-76; PMID:16904174; http://dx.doi.org/10.1016/j.cell.2006.07.024
- Vaiphei K, Sinha SK, Kochhar R. Comparative Analysis of Oct4 in Different Histological Subtypes of Esophageal Squamous Cell Carcinomas in Different Clinical Conditions. Asian Pac J Cancer Prev 2014; 15:3519-24; PMID:24870750; http://dx.doi.org/10.7314/APJCP.2014.15.8.3519
- Zhou X, Huang G-R, Hu P. Over-expression of Oct4 in human esophageal squamous cell carcinoma. Mol Cells 2011; 32:39-45; PMID:21547540; http://dx.doi.org/10.1007/s10059-011-2315-5
- Wang Q, He W, Lu C, Wang Z, Wang J, Giercksky KE, Nesland JM, Suo Z. Oct3/4 and Sox2 Are Significantly Associated with an Unfavorable Clinical Outcome in Human Esophageal Squamous Cell Carcinoma. Anticancer Res 2009; 29:1233-41; PMID:19414369
- Stoner GD, Kaighn ME, Reddel RR, Resau JH, Bowman D, Naito Z, Matsukura N, You M, Galati AJ, Harris CC. Establishment and characterization of SV40 T-antigen immortalized human esophageal epithelial cells. Cancer Res 1991; 51:365-71; PMID:1703038
- Kushner JA. Development. Esophageal stem cells, where art thou? Science 2012; 337:1051-2; PMID:22936766; http://dx.doi.org/10.1126/science.1227506
- Nicholson AM, Graham TA, Simpson A, Humphries A, Burch N, Rodriguez-Justo M, Novelli M, Harrison R, Wright NA, McDonald SA, et al. Barrett's metaplasia glands are clonal, contain multiple stem cells and share a common squamous progenitor. Gut 2012; 61:1380-9; PMID:22200839; http://dx.doi.org/10.1136/gutjnl-2011-301174
- Krishnadath KK, Wang KK. Molecular Pathogenesis of Barrett Esophagus: Current Evidence. Gastroenterol Clin North Am 2015; 44:233-47; PMID:26021192; http://dx.doi.org/10.1016/j.gtc.2015.02.002
- Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet 2000; 16:182-7; PMID:10729834; http://dx.doi.org/10.1016/S0168-9525(99)01955-1
- Williamson KA, Hever AM, Rainger J, Rogers RC, Magee A, Fiedler Z, Keng WT, Sharkey FH, McGill N, Hill CJ, et al. Mutations in SOX2 cause anophthalmia-esophageal-genital (AEG) syndrome. Hum Mol Genet 2006; 15:1413-22; PMID:16543359; http://dx.doi.org/10.1093/hmg/ddl064
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development (Cambridge, England) 2007; 134:2521-31; PMID:17522155; http://dx.doi.org/10.1242/dev.003855
- Asonuma S, Imatani A, Abe Y, Koike T, Asano N, Ohara S, Shimosegawa T. The down-regulation of a HMG box gene Sox2 by exposure to acid and bile induces the progression of Barrett's esophagus. Gastroenterology 2008; 134:A437-A; PMID:NOT_FOUND; http://dx.doi.org/10.1016/S0016-5085(08)62041-7
- Bhardwaj Y, DeMars CJ, Achra S, Anderson M, Prasad GA, Wang KK, Urrutia RA, Buttar N. SOX2 and CDx2, Transcriptional Regulators of Early Differentation, Play a Key Role in the Development of Acid and Bile-Associated Columnar Metaplasia. Gastroenterology 2009; 136:A596-A; PMID:NOT_FOUND; http://dx.doi.org/10.1053/j.gastro.2008.09.028
- Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology 2000; 119:961-71; PMID:11040183; http://dx.doi.org/10.1053/gast.2000.18142
- Mari L, Milano F, Parikh K, Straub D, Everts V, Hoeben KK, Fockens P, Buttar NS, Krishnadath KK. A pSMAD/CDX2 complex is essential for the intestinalization of epithelial metaplasia. Cell Rep 2014; 7:1197-210; PMID:24794431; http://dx.doi.org/10.1016/j.celrep.2014.03.074
- Clemons NJ, Koh SY, Phillips WA. Advances in understanding the pathogenesis of Barrett's esophagus. Discov Med 2014; 17:7-14; PMID:24411696
- Raghoebir L, Biermann K, Buscop-van Kempen M, Wijnen RM, Tibboel D, Smits R, Rottier RJ. Disturbed balance between SOX2 and CDX2 in human vitelline duct anomalies and intestinal duplications. Virchows Archiv 2013; 462:515-22; PMID:23568430; http://dx.doi.org/10.1007/s00428-013-1405-5
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol 2005; 25:6031-46; PMID:15988017; http://dx.doi.org/10.1128/MCB.25.14.6031-6046.2005
- Lam CS, Mistri TK, Foo YH, Sudhaharan T, Gan HT, Rodda D, Lim LH, Chou C, Robson P, Wohland T, et al. DNA-dependent Oct4-Sox2 interaction and diffusion properties characteristic of the pluripotent cell state revealed by fluorescence spectroscopy. Biochem J 2012; 448:21-33; PMID:22909387; http://dx.doi.org/10.1042/BJ20120725
- Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol 2004; 24:8834-46; PMID:15456859; http://dx.doi.org/10.1128/MCB.24.20.8834-8846.2004
