ABSTRACT
Progesterone Receptor Membrane Component 1 (PGRMC1) is expressed in both oocyte and ovarian somatic cells, where it is found in multiple cellular sub-compartments including the mitotic spindle apparatus. PGRMC1 localization in the maturing bovine oocytes mirrors its localization in mitotic cells, suggesting a possible common action in mitosis and meiosis. To test the hypothesis that altering PGRMC1 activity leads to similar defects in mitosis and meiosis, PGRMC1 function was perturbed in cultured bovine granulosa cells (bGC) and maturing oocytes and the effect on mitotic and meiotic progression assessed. RNA interference-mediated PGRMC1 silencing in bGC significantly reduced cell proliferation, with a concomitant increase in the percentage of cells arrested at G2/M phase, which is consistent with an arrested or prolonged M-phase. This observation was confirmed by time-lapse imaging that revealed defects in late karyokinesis. In agreement with a role during late mitotic events, a direct interaction between PGRMC1 and Aurora Kinase B (AURKB) was observed in the central spindle at of dividing cells. Similarly, treatment with the PGRMC1 inhibitor AG205 or PGRMC1 silencing in the oocyte impaired completion of meiosis I. Specifically the ability of the oocyte to extrude the first polar body was significantly impaired while meiotic figures aberration and chromatin scattering within the ooplasm increased. Finally, analysis of PGRMC1 and AURKB localization in AG205-treated oocytes confirmed an altered localization of both proteins when meiotic errors occur. The present findings demonstrate that PGRMC1 participates in late events of both mammalian mitosis and oocyte meiosis, consistent with PGRMC1's localization at the mid-zone and mid-body of the mitotic and meiotic spindle.
Introduction
Progesterone Receptor Membrane Component 1 (PGRMC1) is a multi functional protein that plays important roles in regulating mammalian ovarian function.Citation1,2 Within the ovary, PGRMC1 is expressed and exerts a function in both somatic and germ cells.Citation3-5 Its clinical relevance is indicated by studies showing that altered PGRMC1 expression correlates with defective follicular development and infertility in women.Citation6-8
Primary evidence that PGRMC1 has a fundamental role in ovarian somatic cells comes from in vivo studies in mice, in which conditional knockout of PGRMC1 in granulosa cells impairs antral follicle development.Citation2,9 Accordingly, in vitro studies using different ovarian cell lines have shown that depleting PGRMC1 expression suppresses cell proliferation.Citation10-12 However the mechanism of action by which PGRMC1 controls ovarian cell proliferation is poorly understood.
So far, PGRMC1 is known as a mediator of progesterone's antiapoptotic action in ovarian cell lines.Citation3,13,14 When apoptosis is induced by serum starvation in rat spontaneously immortalized granulosa cells (SIGCs), PGRMC1 mediates progesterone's anti-apoptotic function, at least in part, through the regulation of the expression of apoptosis-related genes.Citation13,15 This genomic action is exerted by high molecular weight forms of PGRMC1 that localize to the nucleus in interphasic cells.Citation13,15 However, PGRMC1 is also found in other sub-cellular compartments where it probably exerts additional functions. In particular PGRMC1 associates to the mitotic spindle,Citation11,16,17 where it directly interacts with β tubulin Citation11 suggesting a role in the regulation of mitosis. Immunofluorescence studies have shown that PGRMC1 changes its localization dynamically during mitosis: it associates with the spindle apparatus in metaphase, while it localizes to the midzone and the midbody in anaphase and telophase/cytokinesis.Citation11 These studies indicate an involvement in mitosis, however the molecular mechanism by which PGRMC1 regulates mitosis has not been fully characterized and further studies are needed to better understand its function.
PGRMC1 is also expressed in oocytes of several mammalian species.Citation3-5 Previous experimental evidence obtained in in vitro matured bovine oocytes supports the hypothesis that PGRMC1 regulates meiotic chromosome segregation during meiosis I.Citation5,18 In the period that spans from meiotic cell cycle reentry to metaphase II, also known as oocyte maturation, PGRMC1's localization dramatically changes. Specifically, PGRMC1 begins to associate with the condensing chromosomes after nuclear envelope break down and localizes to the centromeric region of the metaphasic chromosomes at Metaphase I (MI) and MII stage, while at Anaphase/Telophase I (Ana/Telo I) it concentrates between the separating chromosomes.Citation5 Interestingly, in an oocyte model characterized by increased aneuploidy and embryonic developmental failure, PGRMC1 fails to properly associate with the MII chromosomes.Citation18 Remarkably, PGRMC1 co-localizes with phosphorylated (active) form of AURKB in all the different stages of maturation,Citation5 suggesting an interaction between the two proteins. However, as in the case of somatic cells mitosis, the precise role of PGRMC1 during oocyte meiosis is not known.
Strikingly, PGRMC1 localization in the maturing oocytes mirrors its localization in ovarian mitotic cells, suggesting a possible common function in both mitotic and meiotic cell division. The present study investigates the hypothesis that interfering with PGRMC1 function leads to similar defects in mitosis and meiosis in primary culture of bovine granulosa cells (bGC) and maturing bovine oocytes respectively. bGC were cultured in the presence of serum to stimulate cell growth and PGRMC1 function was altered using small interfering RNA (RNAi) mediated gene silencing approach. Bovine oocytes were in vitro matured and PGRMC1 function was impaired by using either a known PGRMC1 inhibitor (AG 205)Citation19 or RNAi. In addition, a possible relationship between PGRMC1 and AURKB has been investigated in both systems.
Results
PGRMC1 silencing affects bovine granulosa cells (bGCs) proliferation
To determine the effect of PGRMC1 silencing on the proliferation of cultured bGCs, cells were treated with PGRMC1 small interfering RNA (RNAi) or control (CTRL) RNAi and cultured for 24, 48 or 72 h in serum-supplemented medium to stimulate cell growth. Over the course of 72 h, treatment with PGRMC1 RNAi significantly reduced PGRMC1 mRNA levels compared to CTRL RNAi treated group, as assessed by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR, ). Western blot analysis confirmed the presence of multiple PGRMC1 bands, similar to what observed in human and rat granulosa cells.Citation15 All the PGRMC1 bands decreased at 72 h after PGRMC1 RNAi treatment (). Quantification of bands intensities revealed an overall 52.7 ± 10.9% significant protein reduction in PGRMC1 RNAi treated cells, when compared to the CTRL RNAi treated group (one sample t-test, p < 0.05).
Figure 1. Effect of RNAi mediated PGRMC1 silencing on bGC growth. (A) Graph showing PGRMC1 mRNA silencing in PGRMC1 RNAi treated bGC as assessed by qRT-PCR. PGRMC1 expression level was normalized using GAPDH as reference gene. Data were analyzed by one way ANOVA, followed by Tukey's Multiple Comparison Test, Values are means ± SEM (n = 3). *indicates significant differences between groups (P < 0.05). (B) Representative Western blot showing the decreased expression of all PGRMC1 bands at 72 h in PGRMC1 RNAi treated bGC; β tubulin was used as loading control. (C) Graph showing the effect of PGRMC1 down regulation on cell growth. Data were analyzed by two way ANOVA followed by Bonferroni post-hoc test. Values are means ± SEM (n = 3). *indicates significant differences between groups P < 0.05. (D) Graph showing the increase in the percentage of cells arrested at G2/M phase after 72 h from transfection, as assessed by flow cytometry analysis. Data were analyzed by unpaired Student's t-test, Values are means ± SEM (n = 3). *indicates significant differences between groups (P < 0.05).
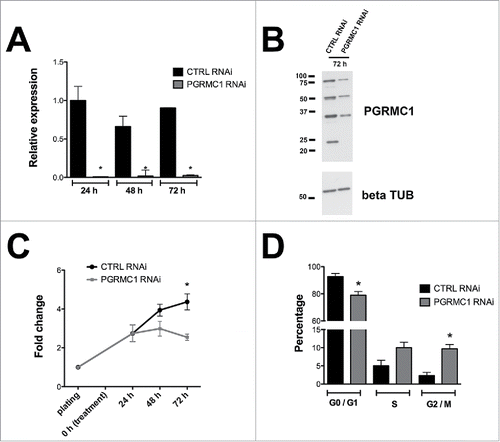
To evaluate the effect of reduced PGRMC1 expression on bGC growth, cells were harvested at each time point and total cell number was counted. PGRMC1 depleted cells were significantly less compared to CTRL RNAi treated group (). Moreover, flow cytometry analysis revealed that the decrease in total cell number after 72h of culture in PGRMC1 RNAi-treated bGCs was accompanied by a decrease in the frequency of G0/G1 stage cells and an increase in the frequency of cells arrested at G2/M phase of the cell cycle ( and supplemental Fig. S1).
The decreased cell number and parallel increase in G2/M rates in PGRMC1 RNAi treated bGC suggests a defective proliferation, which can be due to an arrested or prolonged M-phase of the cell cycle. This hypothesis was confirmed using time-lapse imaging, in which PGRMC1 and CTRL RNAi treated bGCs were stained with the supravital Hoechst 33342 fluorochrome and imaged every 5 minutes, for a total of 6 h. In these experiments, morphological evaluation of dividing nuclei throughout the course of mitosis revealed that cells treated with CTRL RNAi progressed from prophase to telophase giving rise to two daughter nuclei ( and Supplemental Movie 1). In contrast, cells exposed to PGRMC1 RNAi started to divide but their further progression through the cell cycle was impaired and three main phenotypes were observed. In the first phenotype, cells undergo prophase/metaphase but do not progress beyond the Ana/Telo phase, reforming a single nucleus ( and Supplemental Movie 2); in the second phenotype, cells undergo prophase/metaphase and progress through the following stages in an irregular manner that lead to the formation of aberrant nuclei (Supplemental Movie 3), in which small clumps of DNA remain excluded from the reforming nuclei. In the third phenotype, cells undergo prophase/metaphase but DNA remains interconnected through the following stages leading to incomplete karyokinesis (Supplemental Movie 4). Collectively, we defined these phenotypes as ‘abnormal mitosis’ since they differed from the phenotype observed in CTRL RNAi treated bGC. As shown in nearly 70% of the PGRMC1 RNAi treated cells that started dividing showed these morphological alterations of mitotic progression. We can assume that errors leading to the first phenotype occurred earlier during mitotic progression when compared to the second and the third phenotypes. Interestingly, the sum of phenotype 2 and 3 represented the majority of ‘abnormal’ mitosis (78.9%) suggesting that PGRMC1 RNAi treatment mostly affected events occurring during late mitosis.
Figure 2. Effect of PGRMC1 silencing on bGC mitosis by time-lapse analysis of transfected bGC. (A) Example of normal mitosis occurring in bGC transfected with CTRL RNAi. The cell goes through all the mitotic phases from prophase to telophase giving rise to two daughter nuclei (see Supplemental Movie 1). (B) Example of abnormal mitosis in bGC transfected with PGRMC1 RNAi, in which the cell starts the division process undergoing prophase but then fails to proceed beyond the Ana/Telophase (see Supplemental Movie 2). Additional examples of abnormal mitosis occurring in PGRMC1 RNAi treated bGC are shown in Supplemental Movies 3 and 4. (C) Graph showing the frequency of normal and aberrant mitotic events assessed in CTRL and PGRMC1 RNAi treated cells. Data were analyzed by Fisher exact test. *indicates significant differences between groups P < 0.05. (D) Graph showing the time to complete mitosis in CTRL and PGRMC1 RNAi treated cells. Data were analyzed by unpaired Student's t-test, *indicates significant differences between groups (P < 0.05). This experiment was replicated four times; the total number of cells analyzed is shown in brackets.
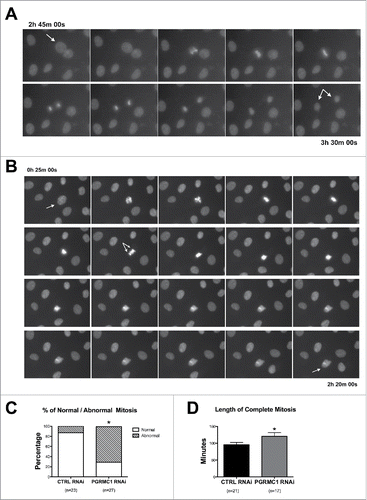
Moreover, the duration of cell cycle progression, from the beginning of prophase to the formation of two daughter nuclei, was significantly increased (). As expected, under these experimental conditions (i.e. culture in serum), cellular death of non-mitotic cells was low in both CTRL and PGRMC1 RNAi treated bGC (2.37 ± 0.42 and 2.41± 0.65 % on a total of 1585 and 1209 observed cells, respectively). Therefore apoptosis does not likely account for the lower cell number observed in PGRMC1 depleted cells.
PGRMC1 co-localizes and interacts with Aurora kinase B during bGC mitosis
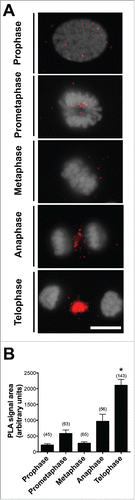
Since cell division is dependent on Aurora kinase B (AURKB),Citation20-22 we performed double immunofluorescence staining to examine the relationship between the localization of AURKB and PGRMC1 during bGC mitosis. As shown in , the two proteins colocalize, particularly during telophase. Furthermore, in situ Proximity Ligation Assay (PLA) was conducted to test whether PGRMC1 directly interacts with AURKB during the different stages of mitosis. This technique uses a pair of oligonucleotide-labeled secondary antibodies (PLA probes), which generate a signal only when the two probes have bound in close proximity. The signal from each detected pair of PLA probes is visualized as an individual fluorescent spot. As shown in , PLA revealed an interaction between AURKB and PGRMC1. Moreover, the degree of interaction, evaluated as the mean of the total fluorescence area in each mitotic cell, was low from prophase to anaphase and then increased significantly at telophase ().
Figure 3. Colocalization PGRMC1-AURK in bGC. Representative images showing PGRMC1 (red) and AURKB (green) colocalization during the different mitotic phases of cultured bGC; DNA was stained with DAPI (blue). PGRMC1 and AURKB start to colocalize to the mitotic spindle region during prophase and the colocalization is more pronounced in the central spindle during ana/telophase. Scale bar is 10 µm.
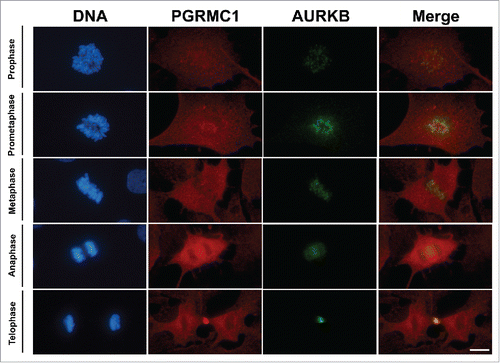
Figure 4. Interaction between PGRMC1 and AURKB during mitosis in bGC. (A) Representative images of bGC showing PGRMC1 - AURKB interaction as assessed by In situ proximity ligation assay (PLA) from prophase until telophase. DNA was stained with DAPI (white). The red spots indicate PGRMC1-AURKB interactions. Scale bar is 10 µm. (B) Graph showing the increased interaction between these two proteins during telophase. Data were analyzed by one way ANOVA followed by Tukey's Multiple Comparison Test. Values are means ± SEM; *indicates significant differences between groups (P < 0.05). This experiment was replicated three times with the total number of cells analyzed shown in brackets.
Pharmacological inhibition of PGRMC1 function during bovine oocyte meiosis impairs karyokinesis and polar body emission
During meiosis I, homolog chromosome segregation and cytokinesis are spatiotemporally coordinated in order to ensure the proper ploidy but asymmetric cytoplasmic division between the MII oocyte and the first polar body (PBI) (reviewed inCitation23). To determine whether PGRMC1 plays a role in PBI cytokinesis, cumulus cell-oocyte complexes (COCs) and denuded oocytes (DOs) were cultured in in vitro maturation (IVM) medium in the presence or absence of increasing concentrations of the PGRMC1 inhibitor AG 205.Citation19 After culture, the effect on PBI emission was evaluated under bright field microscopy, while the effect on chromosome segregation was assessed under epifluorescence microscopy after fixation and DNA staining.
As shown in , PBI extrusion rate decreased when COCs or DOs were cultured with 20 and 40 µM AG205 compared to the control group (0 µM, P<0.05). This effect was more pronounced in DOs, where PBI formation was already lower with 10 µM AG205. As shown in , AG205 treatment also impaired chromosome segregation determining a decrease in the percentage of oocytes that reached the MII stage and an increase in oocytes showing aberrant meiotic figures. In particular, these aberrancies included the presence of DNA clumps scattered within the ooplasm (, aberrant) as well as the coexistence of two metaphase plates or telophases (these examples of aberrant meiotic figures observed after AG205 treatment are shown in Fig. S2). Oocytes in which the DNA was collapsed into a single clump, or in which DNA was not detectable within the ooplasm, were classified as degenerated. Samples observation under bright field combined to UV light illumination confirmed the cytoplasmic localization of DNA clumps (Fig. S3). AG205 effect appeared more pronounced in DOs ().
Figure 5. Effect of AG205 treatment on PBI emission. (A) Representative images showing in vitro matured oocytes with (left, arrow) or without (right) extruded PBI. (B, C) Graphics showing the effect of AG205 treatment on the percentage of oocytes that extruded the PBI in COC and DO, respectively. Data were analyzed by one way ANOVA followed by Tukey's Multiple Comparison Test. Values are means ± SEM (n = 3); a,b different letters indicate significant differences between groups (P < 0.05).
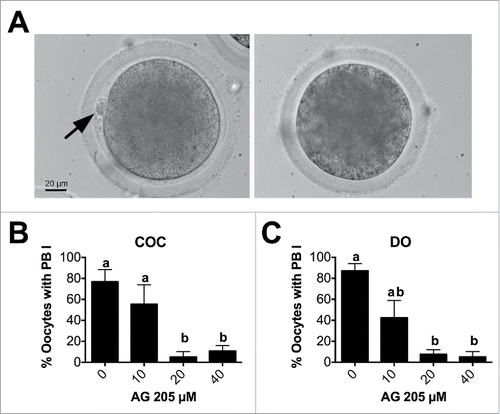
Figure 6. Effect of AG205 treatment on meiotic progression. (A) Representative images showing oocytes classified as MI, Ana/Telophase, MII, aberrant or degenerated after AG205 treatment. Aberrant mitotic figure shown in this figure is characterized by the presence of DNA scattered within the ooplasm (arrow), while additional examples of aberrant meiotic figures observed after AG205 treatment are shown in Figure S1. DNA was stained with DAPI (white). Scale bar is 50 µm. Insets show the DNA at 2X magnification. Graphs in (B) and (C) show the percentages of oocytes at each stage of the first meiotic division (shown in A) in COC and DO respectively. Data were analyzed by one way ANOVA followed by Tukey's Multiple Comparison Test. Values are means ± SEM (N = 3); a,b different letters indicate significant differences between groups (P < 0.05).
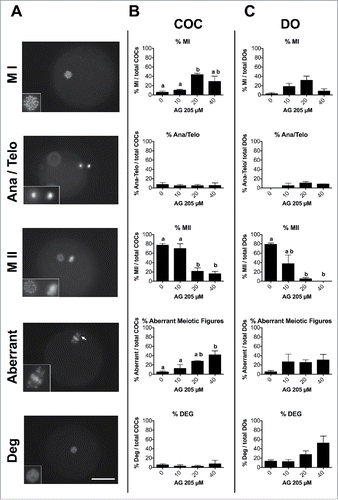
Finally, in order to assess whether AG205 treatment affected AURKB localization, PGRMC1 and AURKB localization was assessed in AG205 treated COCs by double immune fluorescence experiments. PGRMC1 and AURKB localization in the oocytes was classified as regular or irregular according to the previously published criteria.Citation18 Localization was judged as regular when the proteins localized at the centromeric region of each chromosome or irregular when one or more of the following configurations was observed: more than one point on a chromosome, shape different from the punctuated, not in the centromeric region, and/or lack of signal. As shown in aberrant meiotic figures showed irregular AURKB and PGRMC1 localization, while both PGRMC1 and AURKB showed a focused centromeric localization in oocytes with MII plate.
Figure 7. Effect of AG205 on PGRMC1 and AURKB localization. Representative images showing PGRMC1 and AURKB localization in MII plates of matured oocytes or in oocytes showing aberrant meiotic figures. COC were treated with 0 or 20 µM AG205 for 24 h. After AG205 treatment oocytes were fixed, immunostained with anti- PGRMC1 (red) and AURKB (green) antibodies; DNA was stained with DAPI (blue). A total of 97 oocytes from 2 independent experiments were analyzed. Both PGRMC1 and AURKB showed a focused centromeric localization in oocytes with MII plate, while they often showed a more diffused localization in aberrant meiotic figures with scattered chromosomes. When clumps of chromatin were present within the ooplasm, none of them were associated with AURKB and/or PGRMC1. Finally, when double meiotic plates were present, both AURKB and PGRMC1 showed a focused localization on metaphasic chromosomes of both plates.
RNAi mediated PGRMC1 silencing during oocyte maturation mirrors AG205 effects impairing karyokinesis and polar body emission
PGRMC1 function in oocytes was also assessed by microinjecting RNAi into germinal vesicle (GV) stage oocytes surrounded by cumulus cells. After microinjection with either CTRL or PGRMC1 RNAi and culture for 18 h under in vitro meiotic arrest conditions (IVA), allowing sufficient time for the RNAi to effectively deplete the gene of interest, oocytes were cultured in IVM medium for further 24 h. Preliminary optimization experiments in which a fluorescent tracker was used to assess microinjection efficiency indicated that ≈80% of the oocytes were successfully microinjected. Moreover, survival rate, assessed as the percentage of viable COCs after IVA, did not differ in CTRL and PGRMC1-RNAi microinjected COCs (96.06 ± 2.10 and 96.23 ± 2.73% respectively, p > 0.05 t-test, n = 11 independent experiments).
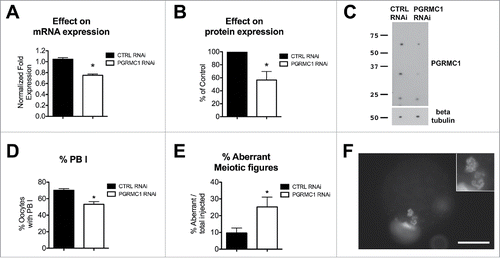
As reported in , PGRMC1 mRNA and protein were reduced approximately by 40% as assessed by qRT-PCR and Western blotting, respectively. Depleting PGRMC1 significantly reduced the percentage of oocytes that extruded the PBI (). Moreover, fluorescence microscopy analysis revealed that even though the formation of MII plate was not blocked in PGRMC1 RNAi treated oocytes, there was an increase in the frequency of oocytes with aberrant meiotic figures (), which were similar to that observed after AG205 treatment. Additional examples of aberrant meiotic figures upon PGRMC1 RNAi treatment are shown in Figure S4.
Figure 8. Effect of PGRMC1 RNAi treatment on PGRMC1 expression level, PBI extrusion and meiotic progression. (A) Graph showing PGRMC1 mRNA expression level in CTRL and PGRMC1 RNAi treated oocytes by RT-qPCR; PGRMC1 expression level was normalized using GAPDH and HIST1H2A as reference genes and differences in gene expression levels were assessed with the Delta-Delta Ct method. Data were analyzed by Student's t-test. Values are means ± SEM (N = 3); * indicates significant differences between groups (P < 0.05). (B and C) graph and representative Western blotting showing PGRMC1 protein expression levels in CTRL and PGRMC1 RNAi treated oocytes; β tubulin was used as loading control. PGRMC1 protein expression in PGRMC1 RNAi treated oocytes is expressed as a percentage of the CTRL RNAi treated group. Data were analyzed by one sample t-test. Values are means ± SEM (n = 6). *indicates significant differences (P < 0.05). (D) Graph showing the percentage of oocytes that extruded the PBI after CTRL or PGRMC1 RNAi treatment and IVM. Data were analyzed by t-test. Values are means ± SEM (n = 14); *indicates significant difference between groups (P < 0.05). (E) Graph showing the percentage of oocytes showing aberrant meiotic figures. Data were analyzed by t-test. Values are means ± SEM (n = 6); *indicate significant differences between groups (P < 0.05). Image in (F) is representative of an aberrant meiotic figure observed after PGRMC1 RNAi treatment, in which chromosomes and DNA clumps are dispersed in the ooplasm and not organized in a MII plate (additional examples of aberrant meiotic figures are shown in Fig. S3). Scale bar is 50 µm. Insets show the DNA at 2X magnification.
Discussion
Proper completion of cell division is an essential process in both somatic and germ cells. In somatic cells, after DNA replication and sister chromatids separation, cytokinesis ensures that the genetic material as well as the cytoplasmic organelles are evenly distributed into two daughter cells.Citation24 In contrast, cell division in oocytes allows for the elimination of half of the genetic material through PB emission while retaining most of the cytoplasm to support the early stages of embryo development. Extrusion of the PB is essential for the formation of the mature egg.Citation23,25 Nonetheless late mitotic and oocyte meiotic division share many similarities.Citation23 In both systems, microtubules of the anaphase spindle play important roles in the formation of the actino-myosin contractile ring and the formation of the cytokinetic furrow at this site, allowing the physical separation of the two daughter cells (reviewed in Citation23). Moreover, the essential role of the chromosomal passenger complex (CPC) in orchestrating late mitotic events Citation20,21 is likely conserved in meiosis.Citation26-29
The present study adds new insights into the role of PGRMC1 as a regulator of both mitotic and meiotic cell division. Our data indicate that when PGRCM1 function is impaired both somatic cells and oocytes fail to divide properly. Specifically, knocking down PGRMC1 by RNAi in cultured bGC results in the inability to successfully complete mitosis and/or to form two normal nuclei in dividing cells. Similarly in the maturing oocytes, perturbing PGRMC1 function by using pharmacological or RNAi approaches results in the impairment of polar body emission together with the formation of aberrant meiotic figures. As a result, a second metaphasic plate and/or scattered chromatin are usually observed.
Previous works demonstrate that PGRMC1 specifically localizes to the mitotic spindle apparatus of different cell lines,Citation11,16,17 where it directly interacts with β tubulin and controls spindle microtubule stability.Citation11 The present study demonstrates that PGRMC1 directly interacts also with AURKB and that the extent of this interaction is highest during the final phase of granulosa cell division. This is important since events occurring at the central spindle during the formation of the midbody are crucial for proper cell division. For example, the central spindle plays an important role in keeping separated chromosomes apart prior to cytokinesis completion, because when microtubules are depolymerized in late anaphase, the nuclei collapse back together.Citation30,31 Thus our findings suggest that an additional mechanism by which PGRMC1 controls mitosis involves a direct interaction with AURKB at the mid-body. This concept is consistent with the finding that interfering with PGRMC1 function affects the localization of AURKB in oocytes that failed to properly complete meiosis. Furthermore, previous studies in bovine oocytes reveal that altering AURKB function by using the AURKB inhibitor ZM447439 also alters PGRMC1 localization, which was associated with meiotic defects.Citation18 Thus PGRMC1 and AURKB functional association seems to be reciprocal, at least in the oocyte. However, the precise mechanism of action by which PGRCM1-AURKB interaction affects AURKB and CPC function remains to be elucidated. PGRMC1 could act as an adaptor protein in many different biological processes as proposed by Aizen et al.Citation32 If this hypothesis were confirmed, PGRMC1 action would depend on the proteins with which it interacts in different cellular and subcellular systems. In this view, altering PGRMC1 function in our studies could have had an effect on different effector proteins, such as AURKB and other CPC components, in the spindle midzone/midbody, thereby determining kariokinesis/cytokinesis disturbance and/or failure. However, this hypothesis remains to be confirmed.
Most probably, the fate of PGRMC1 depleted granulosa cells that do not properly complete cell division would be cellular death, which would contribute to the lower proliferation observed in PGRMC1 RNAi treated cells. This is consistent with effects of depleting PGRMC1 in SIGCs, where cells accumulate in metaphase and then undergo cell death.Citation12 Another possible outcome of cytokinesis failure is the formation of bi- or multi-nucleated cells. This is not the case for bGCs since multinucleated granulosa cells were not observed in PGRMC1 RNAi treated cells (data not shown).
Cell death resulting from aberrant mitosis is generally referred to as ‘mitotic catastrophe’.Citation33 Interestingly, this phenomenon has been functionally re-defined as an ‘atypical mechanism that sense mitotic failure and respond to it by driving the cell to an irreversible fate, be it apoptosis, necrosis or senescence’.Citation34,35 Thus, it has been proposed that mitotic catastrophe can be considered as an ‘onco-suppressive mechanism for the avoidance of genomic instability’.Citation34 Our time-lapse fluorescent microscopy experiments suggest that mitotic catastrophe might occur in PGRMC1 depleted bGC. This finding is in accordance with the experimental evidences that PGRMC1 depletion suppresses cancer cells proliferation in vitro and tumor growth in vivo in all the types of cancers studied so far.Citation10,19,36-38 Moreover, the observation that PGRMC1 is overexpressed in a wide range of tumors when compared to corresponding normal tissuesCitation19,39-42 further support this hypothesis. In this view PGRMC1 overexpression would sustain propagation of abnormal cancer cells, helping them to escape mitotic catastrophe. However, further analyses are required to confirm this hypothesis.
It is possible that, in previous studies, the involvement of PGRMC1 in mitotic catastrophe events has been underestimated. In these studies, indeed, end-point methods have been used to assess the effect of PGRMC1 silencing. Clearly, end-point methods are inappropriate to detect this phenomenon, as they do not take into account the ‘history’ of the cell death.Citation34 On the contrary, time lapse-imaging used in the present study overcomes this limitation giving experimental evidences for the first time of a possible involvement of PGRMC1 in mitotic catastrophe events, which must be deeply investigated in future studies.
Similarly, it is likely that oocytes that fail to extrude PBI and form aberrant meiotic figure would not undergo fertilization and/or second meiotic division, thus degenerate. In a previous study, injecting a PGRMC1 antibody into the ooplasm of immature bovine oocytes revealed a role for PGRMC1 in bovine oocyte maturation Citation5 impairing the transition from pro-MI to MI stages with the majority of the oocytes arresting at the pro-MI stage. Only a very small percentage of oocytes reached MII, thus obscuring a putative PGRMC1 function during the final phase of oocyte maturation. In the present study the use of RNAi gene silencing and a specific PGRMC1 inhibitor, AG205 reveals a role for PGRMC1 during the final phase of maturation (MI to MII transition). In particular, the RNAi study clearly indicates that PGRMC1 is required to allow proper PBI emission. That PGRMC1 RNAi was effective also suggests that PGRMC1 is translated during this period. Moreover, it is known that PGRMC1 can undergo post-translational modifications such as phosphorylation and sumoylation.Citation15,40 Whether translational regulation and/or post-translational modifications are involved in the mechanism that ensures a proper localization of PGRMC1 during oocyte maturation remains to be established.
AG205 experiments have been conducted on both COCs and DOs because cumulus cells also express PGRMC1.Citation4,43,44 The observation that AG205 was more effective on DOs than COCs confirms PGRMC1s specific role within the oocyte. AG205 is a small aromatic compound that acts as a PGRMC1 ligand. It has been used to functionally assess PGRMC1 activity in several biological systems including mammalian cells with effective concentrations in the µM range.Citation19,32,45 It was originally identified as a ligand for AtMAPR2 of the Arabidopsis thaliana,Citation46 which shares homology with the cytochrome b5/heme binding domain of PGRMC1.Citation1,19,46 Thus, although the precise biochemical mechanism through which AG205 inhibits PGRMC1 activity is not known, it likely acts by binding to PGRMC1s heme-binding domain, thereby disrupting PGRMC1s ability to interact with yet to be identified heme proteins. This hypothesis is supported by the experimental evidence that AG205 alters the spectroscopic properties of the PGRMC1-heme complex.Citation19 Thus, it is likely that the PGRMC1 heme binding domain is an important component in the mechanism by which PGRMC1 regulates meiosis.
In conclusion the present findings reveal a new role of PGRMC1 in late stages of the mitotic division and oocytes meiosis. This function is consistent with the localization at the mid-zone and mid-body of the mitotic and meiotic spindle. To the best of our knowledge this is the first study that reveals a role of PGRMC1 during late mitotic and meiotic events and represents an advancement of the state of the art in the field by giving information of a precise stage of cell division in which PGRMC1 exerts a function. Furthermore, PGRMC1's action possibly involves a direct interaction with AURKB, as revealed by PLA studies. Importantly this function seems to be conserved in mammalian cell mitosis and oocyte meiosis. Thus this observations provides a strong rationale for future studies on the precise mechanism of PGRMC1s action in the female gamete, in which technical limitation do not allow mechanistic conclusions, especially in large animal species. For example, since PGRMC1 exists in multiple forms (monomer and specific higher molecular weight forms and post translational modifications,Citation15,40) in future studies it will be important to assess which of these modifications have a predominant role in regulating PGRMC1 function during final karyo/cytokinesis and are important in targeting PGRMC1 to specific sites within the different subcellular domains. Moreover, since PGRMC1 is a mediator of P4 actions, further studies would be needed to address whether and how PGRMC1 function at this particular site of action is mediated by P4.
Materials and methods
Reagents
All the chemicals were purchased from Sigma-Aldrich (St. Louis, MO) except for those specifically mentioned. Gene silencing was performed by Stealth RNAi™ siRNA technology from Life Technologies (https://www.lifetechnologies.com/it/en/home/life-science/rnai/synthetic-rnai-analysis/stealth-rnai-technology.html?icid=cvc-invivo-sirna-c2t2). The ‘BLOCK-iT™ RNAi Designer’ tool from Life Technologies (https://rnaidesigner.lifetechnologies.com/rnaiexpress/) was used to design PGRMC1 Stealth RNAi (PGRMC1 RNAi) within the coding region of the bovine PGRMC1 sequence (NM_001075133). Sequence of the PGRMC1 RNAi used was: (RNA)-GAG UUG UAG UCA AGU GUC UUG GUC U. Negative control (cat n. 12935-200) was chosen among the Stealth RNAi negative control (CTRL RNAi) duplexes available from Life Technologies,. Stock solution of AG205 (16 mM) was prepared in Dimethyl sulfoxide (DMSO) and stored at −20C°. Primary antibodies were: rabbit polyclonal anti-PGRMC1 (Sigma, cat. HPA002877, lot number: A01099 and A27579), mouse monoclonal anti β tubulin (Sigma, cat. T8328, clone AA2) and mouse monoclonal anti AURKB (BD transduction Laboratories, cat. 611082, clone 6/AIM-1). Primer pair sequences were synthetized from Primm s.r.l. (Milan, Italy).
Sample collection
COCs were collected from pubertal Holstein dairy cows recovered at the abattoir (INALCA S.p.A., Ospedaletto Lodigiano, LO, IT 2270M CE, Italy) as previously described from 2–6 mm ovarian antral follicles.Citation5 Only medium-brown in color COCs, with five or more complete layers of cumulus cells and oocytes with finely granulated homogenous ooplasm were used.
After COC retrieval, the bGC were collected, washed and plated. Cells were cultured in growth medium, which was Dulbecco's modified medium supplemented with 10% bovine calf serum, 100 U/ml of penicillin G, 100 µg/ml of streptomycin and 1U/ml Glutamax (Gibco,Thermo Scientific), in humidified air at 37°C with 5% CO2. After 24 h cells were washed with PBS, cultured in growth medium for 6–7 d until confluent, and then used according to the experimental design.
RNAi treatment
bGC RNAi treatment
2 × 105 bGC were plated in 35-mm culture dishes. For immunofluorescence studies cells were plated on cover glasses, while for time-lapse experiments, glass bottom dishes (CELLview, Greiner bio-one) were used. After 24 h of culture (50–70% confluence), bGC were transfected with 6 µl of 20 µM PGRMC1 Stealth RNAi or CTRL RNAi combined with 10 µl of Lipofectamine® RNAiMAX Transfection Reagent (Thermo Fisher Scientific) in a final volume of 2 ml Opti-MEM® Reduced Serum Medium (Gibco, Thermo Fisher Scientific). Transfection efficiency, evaluated using the BLOCK-iT™ Alexa Fluor® Red Fluorescent Control (Invitrogen, Thermo Fisher Scientific) and calculated as the number of red fluorescent cells on the total number of DAPI stained nuclei, was 80% after 24 h treatment and remained constant up to 72 h.
Oocyte RNAi treatment
COCs were collected in medium supplemented with the 3-isobutyl-1-methyl-xanthine (IBMX) at the final concentration of 0.5 mM, as previously described.Citation47 Groups of 20–30 COCs were maintained meiotically arrested by adding 10 μM cilostamide as before describedCitation48,49 until microinjection (minimum 30 minutes). A microinjection apparatus (Narishige Co. Ltd.) mounted on an inverted microscope (Nikon Diaphot; Nikon Corp.) and a Femtojet microinjector (Eppendorf, Hamburg, Germany) were used to inject 10 pl of 20 µM PGRCM1 RNAi or CTRL RNAi into the ooplasm of COC. After injection, COCs were cultured with cilostamide overnight (the total treatment with cilostamide lasted 18 hours). COCs were then washed and in vitro matured for 24 h as previously described.Citation50
Treatment with AG205
Groups of 15–20 COCs or denuded oocytes (DOs) were in vitro matured for 24 h with or without 10, 20 or 40 µM AG 205.Citation19 To obtain DOs, oocytes were mechanically separated from cumulus cells as previously described.Citation50,51 The control group (0 µM AG205) was cultured with an equivalent amount of DMSO that was used to dissolve AG205.
Quantitative reverse Transcriptase-polymerase chain reaction (qRT-PCR)
Oocyte's total RNA was extracted with the Pico-Pure RNA Isolation Kit (Applied Biosystems) following the manufacturer's protocol and including DNase treatment (Qiagen) on the purification column, while bGC total RNA was extracted with the RNeasy Mini Kit (Qiagen).
Total RNA was retro-transcribed (RT) with random hexamers using the First-strand Synthesis System for RT-PCR System (Invitrogen, Life Technologies). 1 µg of bGC RNA or cDNA equivalent to 0.3 oocytes per reaction were used. Primer pairs are shown in Table S1. SYBR green (Bio-Rad) was used according to the manufactures instructions and reactions were developed in an iQ5 PCR Thermal Cycler (Bio-Rad). Primer pairs specificity was assessed using standard RT-PCR and sequencing analysis. PGRMC1 relative expression level was analyzed with the ΔΔ-ct methodCitation52 using the HIS1H2ACitation53,54 and/or GAPDHCitation55 as reference genes with the Biorad iQ5 Software. Data from 3 independent experiments were imported and grouped into a single ‘Gene Study’.
Western blot analysis
SDS-PAGE of bGC lysates was conducted as previously described.Citation15 Briefly, PGRMC1 or CTRL RNAi treated bGC were lysed in radioimmunoprecipitation assay (RIPA) buffer Total amount of protein was determined using the Qubit® Protein Assay Kit and Qubit® fluorometer (Thermo Fisher Scientific). Twenty µg of total protein/lane were used for Western blotting.
SDS-PAGE of oocytes was optimized to detect protein expression in small samples as previously described.Citation56,57 RNAi treated oocytes were denuded, washed and collected in 2 µl of PBS supplemented with proteases and phosphatase inhibitor cocktails. Samples were than mixed with 2 µl of 2X SDS–Laemmli loading buffer (Bio-Rad), boiled for 5 min and stored at –80°C until essayed. After thawing, samples were boiled for additional 5 minutes and resolved on a 10% SDS-PAGE with a 4% stacking gel. Micro-wells in the stacking gel were 1mm in width, 0.5 mm in thickness and 10 mm height, contains a maximum volume of 5 µl. The comb for these micro wells was made with a 3-D printer in William Kinsey laboratory and kindly provided by Lynda McGinnis, University of Kansas Medical Center.
Run transfer and immunoblotting were performed as previously describedCitation11 using the anti PGRMC1 rabbit polyclonal antibody (1:200). PGRMC1 was revealed using a goat anti rabbit IgG peroxidase conjugated antibody (1:1000, Thermo scientific, cat.32460) and detected using the Super Signal West Dura Extended Duration Substrate (Thermo scientific, cat.37071). The nitrocellulose membrane was stripped and re-probed with the anti β tubulin antibody (1:1000) as described,Citation11 which was revealed using a stabilized goat anti mouse IgG peroxidase conjugated antibody (1:1000, Thermo scientific, cat.32430). Relative amount of protein was quantified on scanned filmsCitation58 using Image J software. The intensity of each PGRMC1 band was firstly normalized for the corresponding β tubulin band and than the ratio between PGRMC1 RNAi and CTRL RNAi corresponding bands were calculated. PGRMC1 expression in PGRMC1 RNAi treated oocytes was expressed as a percentage of the corresponding CTRL RNAi treated group. These values were pooled for statistical analysis.
Assessment of bGC growth, effect on cell cycle and mitosis
After treatments, cells were collected after trypsinization. Care was taken to avoid loss of cells during collection and to ensure complete detachment of the cells from the plate by looking at the culture dishes under the microscope after trypsinization. Total cell number was counted with a Neubauer chamber. Cell growth rate was calculated as the ratio of the total number at each time point on the total cell number at the time of plating. Flow cytometry analysis was conducted in order to evaluate the percentage of cells at each cell cycle phase as previously described.Citation59 Cells were collected, fixed in 70% cold EtOH and kept at +4°C until essayed.
For time-lapse analysis CTRL and PGRMC1 RNAi treated bGC were stained with 0.05 µg/ml of Hoechst 33342 for 30 minutes and then substituted with fresh medium and imaged on a biostation IM (Nikon). Images were captured every 5 minutes for 6 h after 30 h from treatment. Each time 6 different fields were captured at a 20X magnification.
Assessment of PBI extrusion and meiotic progression of bovine oocytes
After IVM, oocytes were denuded and examined under the stereomicroscope at the highest magnification (50 X) to assess complete extrusion of the PBI in the perivitelline space. Then, oocytes were fixed in 4% paraformaldehyde in PBS, permeabilizaed in 0.1% Triton-X 100 in PBS for 10 minutes and washed in 0.1% PVA in PBS (PBS/PVA). Samples were mounted on slides with the anti-fade medium Vectashield (Vector Laboratories) supplemented with 1 µg/ml 40,6-diamidino-2-phenylindole (DAPI) to visualize DNA. Samples were analyzed on an epifluorescence microscope (Eclipse E600; Nikon Corp.) equipped with a digital camera (DS-Fi2; Nikon Corp.) to assess the stage of meiotic progression as previously described.Citation18
Immunofluorescence
Immunofluorescence analysis of PGRMC1 and AURKB localization were conducted as previously described on bGC grown on cover glassesCitation11 or denuded oocytesCitation5,18 with minor modification. Briefly, after culture, washing and fixation procedures were conducted with pre-warmed media. Paraformaldehyde fixed samples were incubated overnight at 4°C with a combination of the rabbit anti-PGRMC1 (dilution 1:50) and anti AURK B (dilution 1:50) antibodies. Secondary antibodies used were: TRITC-labeled donkey anti-rabbit antibody (dilution 1:100; Vector Laboratories, Inc.) and Alexa Fluor 488-labeled donkey anti-mouse antibody (dilution 1:500; Invitrogen, Life Technologies) for 1 h at room temperature. The samples were mounted on slides in the antifade medium Vecta Shield (Vector Laboratories) supplemented with 1 µg/ml 40,6-diamidino-2-phenylindole (DAPI). Samples were analyzed on an epifluorescence microscope (Eclipse E600; Nikon) equipped with a 60X objective, a digital camera, and software (NIS elements Imaging Software; Nikon). Immunofluorescence controls, which were performed by omitting one of the 2 primary antibodies while both fluorescently labeled secondary antibodies were present in all reactions, did not show any staining.
In situ proximity ligation Assay (PLA)
The interaction between PGRMC1 and AURKB was assessed using in situ PLA (Duolink II; OLINK Bioscience, Uppsala, Sweden http://www.olink.com/) in in vitro cultured bGC following the manufacturer protocol. Primary antibodies for PGRMC1 and AURKB were the same used for immunofluorescence while anti-rabbit PLUS and anti-mouse MINUS PLA probes were used as secondary antibodies. Negative controls were performed omitting one of the two primary antibodies. Cells were mounted with Duolink mounting medium (OLINK Bioscience). Samples were analyzed as described for immunofluorescence and ImageJ software was used to calculate the total area of the fluorescent signal corresponding to proteins interaction during the different mitotic phases.
Statistical analysis
Experiments were run in triplicates, unless otherwise specified. All statistical analysis was done using GraphPad Prism software (GraphPad Prism v. 6.0e, La Jolla, CA, USA). Data from the replicate experiments were pooled and the data expressed as a mean ± SEM. Student's t test was used to determine differences between two groups. When more than two groups were compared, one-way ANOVA followed by Tukey's Multiple Comparison test or two way ANOVA followed by Bonferroni post test were used. Fisher's exact test was used to analyze percentage data. Details on the statistical analysis are indicated for each experiment in the figure caption.
Abbreviations
| AG205 | = | cis-2-[[1-(4-Chlorophenyl)-1H-tetrazol-5-yl]thio]-1-(1,2,3,4,4a,9b-hexahydro-2,8-dimethyl-5H-pyrido[4,3-b]indol-5-yl)-ethanone, cis-5-({[1-(4-chlorophenyl)-1H-tetraazol-5-yl]sulfanyl}acetyl)-2,8-dimethyl-2,3,4,4a,5,9b-hexahydro-1H-pyrido[4,3-b]indole |
| Ana/Telo I | = | Anaphase/Telophase I |
| ANOVA | = | ANalysis Of Variance |
| AURKB | = | Aurora Kinase B |
| Bgc | = | bovine granulosa cells |
| betaTUB | = | β tubulin |
| COCs | = | cumulus cell-oocyte complexes |
| CTRL | = | control |
| DAPI | = | 40,6-diamidino-2-phenylindole |
| DEG: | = | degenerated |
| DOs | = | denuded oocytes |
| DMSO | = | Dimethyl sulfoxide |
| DNA | = | DNA |
| G0/G1 | = | Gap 0/Gap1 phases of the cell cycle |
| G2/M | = | Gap 2/ Mitosis phases of the cell cycle |
| GAPDH | = | glyceraldehyde-3-phosphate dehydrogenase |
| GV | = | germinal vesicle |
| HIST1H2A | = | histone cluster 1, H2ah |
| IVA | = | in vitro meiotic arrest |
| IVM | = | in vitro maturation |
| MI and II | = | Metaphase I and II |
| mRNA | = | Messenger Ribonucleic acid |
| PBI | = | first polar body |
| PBS/PVA | = | Phosphate-buffered saline/ Polivinilalcool |
| PGRMC1 | = | Progesterone Receptor Membrane Component 1 |
| PLA | = | Proximity Ligation Assay |
| S | = | Synthesys phase of the cell cycle |
| SDS-PAGE | = | Sodium Dodecyl Sulfate - PolyAcrylamide Gel Electrophoresis |
| SEM | = | standard error of the mean |
| SIGCs | = | spontaneously immortalized granulosa cells |
| RNAi | = | small interfering RNA |
| qRT-PCR | = | quantitative reverse transcriptase-polymerase chain reaction |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Notes on contributors
L.T performed all experiments related to bovine granulosa cells, participated in oocyte collection and RNAi procedures and contributed to the writing of the manuscript. I.T. carried out RNAi microinjections in COC and contributed to the writing of the manuscript; F.R. performed the Western blot analysis; F.F. contributed to microinjections procedures optimization, to data analysis and interpretation and contributed to the writing of the manuscript; V.M., S.G., M.Z contributed in time-lapse and fluocytometry analysis; G.M contributed to flow cytometry analysis. V.L performed AG205 experiments, qRT-PCR, WB and immunofluorescence analysis; V.L conceived the project, designed the experiments, analyzed the data and wrote the paper. All authors read and approved the final manuscript.
1192731_Supplemental_Material.docx
Download MS Word (5 MB)Supplemental_Movie_1.mov
Download QuickTime Video (1.1 MB)Supplemental_Movie_2.m4v
Download (2.3 MB)Supplemental_Movie_3.mov
Download QuickTime Video (3.6 MB)Supplemental_Movie_4.mov
Download QuickTime Video (2.7 MB)Acknowledgments
Authors thank L. K. McGinnis, (University of Southern California, Los Angeles CA) for providing the comb for micro-Western blotting; Prof C. Bandi, Dr C. Bazzocchi and Dr. S. Epis (University of Milan) for technical support in RT-qPCR study; G. Sivelli for helping with oocyte manipulation. Authors are thankful to A.M. Luciano (University of Milan), S.C. Modina (University of Milan), and J.J. Peluso (University of Connecticut Health Center) for thoughtful discussions.
Funding
This work was supported by CIG - Marie Curie Actions FP7-Reintegration-Grants within the 7th European Community Framework Program (Contract: 303640,"Pro-Ovum") awarded to V.L., and ‘Fondo Piano di sviuppo UNIMI linea B – Giovani ricercatori’ - Grant n.:15-6-3027000-54, awarded to V.L.F.F. is supported by FP7-PEOPLE-2013-IOF GA 624874 MateRNA
References
- Cahill MA. Progesterone receptor membrane component 1: an integrative review. J Steroid Biochem Mol Biol 2007; 105:16-36; PMID:17583495; http://dx.doi.org/10.1016/j.jsbmb.2007.02.002
- Peluso JJ. Progesterone receptor membrane component 1 and its role in ovarian follicle growth. Front Neurosci 2013; 7:99; PMID:23781168; http://dx.doi.org/10.3389/fnins.2013.00099
- Peluso JJ, Pappalardo A, Losel R, Wehling M. Progesterone membrane receptor component 1 expression in the immature rat ovary and its role in mediating progesterone's antiapoptotic action. Endocrinology 2006; 147:3133-40; PMID:16513825; http://dx.doi.org/10.1210/en.2006-0114
- Luciano AM, Corbani D, Lodde V, Tessaro I, Franciosi F, Peluso JJ, Modina S. Expression of progesterone receptor membrane component-1 in bovine reproductive system during estrous cycle. European J Histochem 2011; 55:e27; http://dx.doi.org/10.4081/ejh.2011.e27
- Luciano AM, Lodde V, Franciosi F, Ceciliani F, Peluso JJ. Progesterone receptor membrane component 1 expression and putative function in bovine oocyte maturation, fertilization, and early embryonic development. Reproduction 2010; 140:663-72; PMID:20739377; http://dx.doi.org/10.1530/REP-10-0218
- Mansouri MR, Schuster J, Badhai J, Stattin EL, Losel R, Wehling M, Carlsson B, Hovatta O, Karlstrom PO, Golovleva I, et al. Alterations in the expression, structure and function of progesterone receptor membrane component-1 (PGRMC1) in premature ovarian failure. Hum Mol Genetics 2008; 17:3776-83; http://dx.doi.org/10.1093/hmg/ddn274
- Schuster J, Karlsson T, Karlstrom PO, Poromaa IS, Dahl N. Down-regulation of progesterone receptor membrane component 1 (PGRMC1) in peripheral nucleated blood cells associated with premature ovarian failure (POF) and polycystic ovary syndrome (PCOS). Reprod Biol Endocrinol 2010; 8:58; PMID:20537145; http://dx.doi.org/10.1186/1477-7827-8-58
- Elassar A, Liu X, Scranton V, Wu CA, Peluso JJ. The relationship between follicle development and progesterone receptor membrane component-1 expression in women undergoing in vitro fertilization. Fertility Sterility 2012; 97:572-8; PMID:22245528; http://dx.doi.org/10.1016/j.fertnstert.2011.12.026
- Peluso JJ, Pru JK. Non-canonical progesterone signaling in granulosa cell function. Reprod 2014; 147:R169-78; http://dx.doi.org/10.1530/REP-13-0582
- Peluso JJ, Gawkowska A, Liu X, Shioda T, Pru JK. Progesterone receptor membrane component-1 regulates the development and Cisplatin sensitivity of human ovarian tumors in athymic nude mice. Endocrinol 2009; 150:4846-54; http://dx.doi.org/10.1210/en.2009-0730
- Lodde V, Peluso JJ. A novel role for progesterone and progesterone receptor membrane component 1 in regulating spindle microtubule stability during rat and human ovarian cell mitosis. Biol Reproduction 2011; 84:715-22; http://dx.doi.org/10.1095/biolreprod.110.088385
- Peluso JJ, Griffin D, Liu X, Horne M. Progesterone receptor membrane component-1 (PGRMC1) and PGRMC-2 interact to suppress entry into the cell cycle in spontaneously immortalized rat granulosa cells. Biol Reprod 2014; 91:104; PMID:25253729; http://dx.doi.org/10.1095/biolreprod.114.122986
- Peluso JJ, Liu X, Gawkowska A, Lodde V, Wu CA. Progesterone inhibits apoptosis in part by PGRMC1-regulated gene expression. Mol Cell Endocrinol 2010; 320:153-61; PMID:20144686; http://dx.doi.org/10.1016/j.mce.2010.02.005
- Peluso JJ, Romak J, Liu X. Progesterone receptor membrane component-1 (PGRMC1) is the mediator of progesterone's antiapoptotic action in spontaneously immortalized granulosa cells as revealed by PGRMC1 small interfering ribonucleic acid treatment and functional analysis of PGRMC1 mutations. Endocrinol 2008; 149:534-43; http://dx.doi.org/10.1210/en.2007-1050
- Peluso JJ, Lodde V, Liu X. Progesterone regulation of progesterone receptor membrane component 1 (PGRMC1) sumoylation and transcriptional activity in spontaneously immortalized granulosa cells. Endocrinol 2012; 153:3929-39; http://dx.doi.org/10.1210/en.2011-2096
- Bonner MK, Poole DS, Xu T, Sarkeshik A, Yates JR, 3rd, Skop AR. Mitotic spindle proteomics in Chinese hamster ovary cells. PloS one 2011; 6:e20489; PMID:21647379; http://dx.doi.org/10.1371/journal.pone.0020489
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A 2006; 103:5391-6; PMID:16565220; http://dx.doi.org/10.1073/pnas.0507066103
- Luciano AM, Franciosi F, Lodde V, Tessaro I, Corbani D, Modina SC, Peluso JJ. Oocytes isolated from dairy cows with reduced ovarian reserve have a high frequency of aneuploidy and alterations in the localization of progesterone receptor membrane component 1 and aurora kinase B. Biol Reprod 2013; 88:58; PMID:23325810; http://dx.doi.org/10.1095/biolreprod.112.106856
- Ahmed IS, Rohe HJ, Twist KE, Mattingly MN, Craven RJ. Progesterone receptor membrane component 1 (Pgrmc1): a heme-1 domain protein that promotes tumorigenesis and is inhibited by a small molecule. J Pharmacol Exp Ther 2010; 333:564-73; PMID:20164297; http://dx.doi.org/10.1124/jpet.109.164210
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 2007; 8:798-812; PMID:17848966; http://dx.doi.org/10.1038/nrm2257
- Kitagawa M, Lee SH. The chromosomal passenger complex (CPC) as a key orchestrator of orderly mitotic exit and cytokinesis. Front Cell Dev Biol 2015; 3:14; PMID:25798441; http://dx.doi.org/10.3389/fcell.2015.00014
- Vogt E, Kipp A, Eichenlaub-Ritter U. Aurora kinase B, epigenetic state of centromeric heterochromatin and chiasma resolution in oocytes. Reprod Biomed Online 2009; 19:352-68; http://dx.doi.org/10.1016/S1472-6483(10)60169-1
- Maddox AS, Azoury J, Dumont J. Polar body cytokinesis. Cytoskeleton (Hoboken) 2012; 69:855-68; PMID:22927361; http://dx.doi.org/10.1002/cm.21064
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol 2012; 28:29-58; PMID:22804577; http://dx.doi.org/10.1146/annurev-cellbio-101011-155718
- Evans JP, Robinson DN. The spatial and mechanical challenges of female meiosis. Mol Reprod Dev 2011; 78:769-77; PMID:21774026; http://dx.doi.org/10.1002/mrd.21358
- Kaitna S, Pasierbek P, Jantsch M, Loidl J, Glotzer M. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous Chromosomes during meiosis. Curr Biol 2002; 12:798-812; PMID:12015116; http://dx.doi.org/10.1016/S0960-9822(02)00820-5
- Rogers E, Bishop JD, Waddle JA, Schumacher JM, Lin R. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J Cell Biol 2002; 157:219-29; PMID:11940606; http://dx.doi.org/10.1083/jcb.200110045
- Sharif B, Na J, Lykke-Hartmann K, McLaughlin SH, Laue E, Glover DM, Zernicka-Goetz M. The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J Cell Sci 2010; 123:4292-300; PMID:21123620; http://dx.doi.org/10.1242/jcs.067447
- Yang KT, Li SK, Chang CC, Tang CJ, Lin YN, Lee SC, Tang TK. Aurora-C kinase deficiency causes cytokinesis failure in meiosis I and production of large polyploid oocytes in mice. Mol Biol Cell 2010; 21:2371-83; PMID:20484572; http://dx.doi.org/10.1091/mbc.E10-02-0170
- Normand G, King RW. Understanding cytokinesis failure. Adv Exp Med Biol 2010; 676:27-55; PMID:20687468; http://dx.doi.org/10.1007/978-1-4419-6199-0_3
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 2003; 299:1743-7; PMID:12637748; http://dx.doi.org/10.1126/science.1081412
- Aizen J, Thomas P. Role of Pgrmc1 in estrogen maintenance of meiotic arrest in zebrafish oocytes through Gper/Egfr. J Endocrinol 2015; 225:59-68; PMID:25720537; http://dx.doi.org/10.1530/JOE-14-0576
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ 2009; 16:3-11; PMID:18846107; http://dx.doi.org/10.1038/cdd.2008.150
- Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol 2011; 12:385-92; PMID:21527953; http://dx.doi.org/10.1038/nrm3115
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ 2012; 19:107-20; PMID:21760595; http://dx.doi.org/10.1038/cdd.2011.96
- Peluso JJ, Liu X, Saunders MM, Claffey KP, Phoenix K. Regulation of ovarian cancer cell viability and sensitivity to cisplatin by progesterone receptor membrane component-1. J Clin Endocrinol Metab 2008; 93:1592-9; PMID:18319313; http://dx.doi.org/10.1210/jc.2007-2771
- Clark NC, Friel AM, Pru CA, Zhang L, Shioda T, Rueda BR, Peluso JJ, Pru JK. Progesterone receptor membrane component 1 promotes survival of human breast cancer cells and the growth of xenograft tumors. Cancer Biol Ther 2016; 17(3):262-71; PMID:26785864; http://dx.doi.org/10.1080/15384047.2016.1139240
- Friel AM, Zhang L, Pru CA, Clark NC, McCallum ML, Blok LJ, Shioda T, Peluso JJ, Rueda BR, Pru JK. Progesterone receptor membrane component 1 deficiency attenuates growth while promoting chemosensitivity of human endometrial xenograft tumors. Cancer Lett 2015; 356:434-42; PMID:25304370; http://dx.doi.org/10.1016/j.canlet.2014.09.036
- Neubauer H, Ma Q, Zhou J, Yu Q, Ruan X, Seeger H, Fehm T, Mueck AO. Possible role of PGRMC1 in breast cancer development. Climacteric 2013; 16:509-13; PMID:23758160; http://dx.doi.org/10.3109/13697137.2013.800038
- Neubauer H, Clare SE, Wozny W, Schwall GP, Poznanovic S, Stegmann W, Vogel U, Sotlar K, Wallwiener D, Kurek R, et al. Breast cancer proteomics reveals correlation between estrogen receptor status and differential phosphorylation of PGRMC1. Breast Cancer Res 2008; 10:R85; PMID:18922159; http://dx.doi.org/10.1186/bcr2155
- Chen WS, Chen PL, Li J, Lind AC, Lu D. Lipid synthesis and processing proteins ABHD5, PGRMC1 and squalene synthase can serve as novel immunohistochemical markers for sebaceous neoplasms and differentiate sebaceous carcinoma from sebaceoma and basal cell carcinoma with clear cell features. J Cutan Pathol 2013; 40:631-8; PMID:23557589; http://dx.doi.org/10.1111/cup.12147
- Mir SU, Ahmed IS, Arnold S, Craven RJ. Elevated progesterone receptor membrane component 1/sigma-2 receptor levels in lung tumors and plasma from lung cancer patients. Int J Cancer 2012; 131:E1-9; PMID:21918976; http://dx.doi.org/10.1002/ijc.26432
- Aparicio IM, Garcia-Herreros M, O'Shea LC, Hensey C, Lonergan P, Fair T. Expression, regulation, and function of progesterone receptors in bovine cumulus oocyte complexes during in vitro maturation. Biol Reprod 2011; 84:910-21; PMID:21228216; http://dx.doi.org/10.1095/biolreprod.110.087411
- Assou S, Anahory T, Pantesco V, Le Carrour T, Pellestor F, Klein B, Reyftmann L, Dechaud H, De Vos J, Hamamah S. The human cumulus–oocyte complex gene-expression profile. Hum Reprod 2006; 21:1705-19; PMID:16571642; http://dx.doi.org/10.1093/humrep/del065
- Xu J, Zeng C, Chu W, Pan F, Rothfuss JM, Zhang F, Tu Z, Zhou D, Zeng D, Vangveravong S, et al. Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nat Commun 2011; 2:380; PMID:21730960; http://dx.doi.org/10.1038/ncomms1386
- Yoshitani N, Satou K, Saito K, Suzuki S, Hatanaka H, Seki M, Shinozaki K, Hirota H, Yokoyama S. A structure-based strategy for discovery of small ligands binding to functionally unknown proteins: combination of in silico screening and surface plasmon resonance measurements. Proteomics 2005; 5:1472-80; PMID:15798990; http://dx.doi.org/10.1002/pmic.200401032
- Lodde V, Franciosi F, Tessaro I, Modina SC, Luciano AM. Role of gap junction-mediated communications in regulating large-scale chromatin configuration remodeling and embryonic developmental competence acquisition in fully grown bovine oocyte. J Assisted Reprod Genetics 2013; 30:1219-26; http://dx.doi.org/10.1007/s10815-013-0061-7
- Luciano AM, Franciosi F, Modina SC, Lodde V. Gap junction-mediated communications regulate chromatin remodeling during bovine oocyte growth and differentiation through cAMP-dependent mechanism(s). Biol Reprod 2011; 85:1252-9; PMID:21816847; http://dx.doi.org/10.1095/biolreprod.111.092858
- Franciosi F, Coticchio G, Lodde V, Tessaro I, Modina SC, Fadini R, Dal Canto M, Renzini MM, Albertini DF, Luciano AM. Natriuretic peptide precursor C delays meiotic resumption and sustains gap junction-mediated communication in bovine cumulus-enclosed oocytes. Biol Reprod 2014; 91:61; PMID:25078681; http://dx.doi.org/10.1095/biolreprod.114.118869
- Luciano AM, Lodde V, Beretta MS, Colleoni S, Lauria A, Modina S. Developmental capability of denuded bovine oocyte in a co-culture system with intact cumulus-oocyte complexes: role of cumulus cells, cyclic adenosine 3',5'-monophosphate, and glutathione. Mol Reprod Dev 2005; 71:389-97; PMID:15803456; http://dx.doi.org/10.1002/mrd.20304
- Modina S, Beretta M, Lodde V, Lauria A, Luciano AM. Cytoplasmic changes and developmental competence of bovine oocytes cryopreserved without cumulus cells. Eur J Histochem 2004; 48:337-46; PMID:15718199
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/10.1006/meth.2001.1262
- Robert C, McGraw S, Massicotte L, Pravetoni M, Gandolfi F, Sirard MA. Quantification of housekeeping transcript levels during the development of bovine preimplantation embryos. Biol Reprod 2002; 67:1465-72; PMID:12390877; http://dx.doi.org/10.1095/biolreprod.102.006320
- Vigneault C, Gilbert I, Sirard MA, Robert C. Using the histone H2a transcript as an endogenous standard to study relative transcript abundance during bovine early development. Mol Reprod Dev 2007; 74:703-15; PMID:17133602; http://dx.doi.org/10.1002/mrd.20665
- Assidi M, Dufort I, Ali A, Hamel M, Algriany O, Dielemann S, Sirard MA. Identification of potential markers of oocyte competence expressed in bovine cumulus cells matured with follicle-stimulating hormone and/or phorbol myristate acetate in vitro. Biol Reprod 2008; 79:209-22; PMID:18417710; http://dx.doi.org/10.1095/biolreprod.108.067686
- Naito K, Kagii H, Iwamori N, Sugiura K, Yamanouchi K, Tojo H. Establishment of a Small-scale Western Blotting System Named as "Micro-Western Blotting" for Mammalian Ova Analysis. J Mamm Ova Res 1999; 16:154-7; http://dx.doi.org/10.1274/jmor.16.154
- McGinnis LK, Albertini DF, Kinsey WH. Localized activation of Src-family protein kinases in the mouse egg. Dev Biol 2007; 306:241-54; PMID:17449027; http://dx.doi.org/10.1016/j.ydbio.2007.03.024
- Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying Western blots: pitfalls of densitometry. Electrophoresis 2009; 30:1845-55; PMID:19517440; http://dx.doi.org/10.1002/elps.200800720
- Vigone G, Merico V, Redi CA, Mazzini G, Garagna S, Zuccotti M. FSH and LH receptors are differentially expressed in cumulus cells surrounding developmentally competent and incompetent mouse fully grown antral oocytes. Reprod Fertil Dev 2015; 27(3):497-503; http://dx.doi.org./10.1071/RD13251.
