ABSTRACT
The liver is the most important organ in cholesterol metabolism, which is instrumental in regulating cell proliferation and differentiation. The gene Tm7sf2 codifies for 3 β-hydroxysterol-Δ14-reductase (C14-SR), an endoplasmic reticulum resident protein catalyzing the reduction of C14-unsaturated sterols during cholesterol biosynthesis from lanosterol. In this study we analyzed the role of C14-SR in vivo during cell proliferation by evaluating liver regeneration in Tm7sf2 knockout (KO) and wild-type (WT) mice. Tm7sf2 KO mice showed no alteration in cholesterol content. However, accumulation and delayed catabolism of hepatic triglycerides was observed, resulting in persistent steatosis at all times post hepatectomy. Moreover, delayed cell cycle progression to the G1/S phase was observed in Tm7sf2 KO mice, resulting in reduced cell division at the time points examined. This was associated to abnormal ER stress response, leading to alteration in p53 content and, consequently, induction of p21 expression in Tm7sf2 KO mice. In conclusion, our results indicate that Tm7sf2 deficiency during liver regeneration alters lipid metabolism and generates a stress condition, which, in turn, transiently unbalances hepatocytes cell cycle progression.
Introduction
In the liver, cells are in a quiescent state and rarely divide. When the liver is injured or partially removed, residual hepatocytes manifest proliferative ability, and thus restore the original mass.Citation1 The most commonly used experimental method for studying cell proliferation in vivo is partial hepatectomy (PH).Citation2 Various components are called into play in liver regeneration.Citation1,3,4 The cytokines IL-6 and TNF-α, together with the second messenger cAMP, act on hepatocytes by adjusting their transition from the G0 to G1 phase, making them responsive to different growth factors, which are necessary for proliferation.Citation5-7 In this “priming phase,” transcription factors such as NF-κB, STAT3, AP-1, and C/EBPβ are also immediately activated.Citation1,4 These factors in turn regulate transcription and subsequent translation of immediate early genes required for the transition from G0 to G1, including c-fos, c-jun, c-myc, jun B. Then, other genes, defined as delayed, are translated and play a central role in the progression of hepatocytes through the cell cycle (i.e., those transcribing for Cyclin A, B, E, D, p53, p21, MDM2).Citation1 Furthermore, many other genes, some of which have been uncharacterized thus far, increase their expression or are modulated during liver regeneration. Among those are genes identified and/or studied by our group, such as Hops,Citation8-11 Lal-1,Citation12 Gucd-1,Citation13 Crem/Creb,Citation5-7 CD44,Citation14 and Tat.Citation15
An active metabolism of lipids occurs in the liver, whose correct functioning ensures normal liver regeneration.Citation16 In particular, among lipid species, cholesterol and triglycerides play the most important role in the regenerative process. Indeed, any alterations in triglycerides accumulation and catabolism or in cholesterol synthesis determine impaired hepatocellular proliferation.Citation17-19 In the first hours after PH, the regenerating liver accumulates fat consisting mainly of triglycerides, constituting the major energy source for cell metabolism. During liver regeneration, the peak of fat accumulation in mice tissue is between 12 and 24 h following PH, at a time when levels of cholesterol and triglycerides in the serum instead decrease.Citation16
Several data suggest that modifications in the cholesterol biosynthetic pathway would affect cell proliferation.Citation20,21 In this work, we analyzed the function of the gene Tm7sf2, encoding for 3β-hydroxysterol-Δ14-reductase (C14-SR), an enzyme that resides in the endoplasmic reticulum (ER) and catalyzes the reduction of the C14 double-bond during cholesterol synthesis.Citation22-25 The Tm7sf2 gene promoter contains Sterol Response Elements (SRE) along its sequence, indicating that its expression is controlled by cell's sterol levels through the transcription factor SREBP-2, which activates the Tm7sf2 promoter in response to low levels of cellular sterols.Citation25 Mice lacking Tm7sf2 (hereafter referred as Tm7sf2 KO) are apparently healthy, with normal cholesterol levels in liver membranes and in plasma.Citation23 This finding was explained by the compensatory action carried out by the inner nuclear membrane protein Lamin B receptor (LBR), characterized by a C-terminal sequence with sterol reductase activity,Citation26 which is responsible for catalyzing cholesterol biosynthesis in Tm7sf2 KO mice.Citation23 However, abnormal activation of the ER stress response was documented in quiescent livers of these mice.Citation27 Moreover, affymetrix microarray analysis of gene expression conducted in the liver of Tm7sf2 KO mice showed that many genes involved in cell-cycle progression are downregulated whereas genes involved in xenobiotic metabolism are upregulated.Citation23 Because of the importance of cholesterol in cell structure and functionality, we asked whether the absence of Tm7sf2 would affect the process of liver regeneration.Citation1-4 We performed PH experiments to understand the role of C14-SR in the process of cell proliferation in vivo. We documented triglycerides accumulation during liver regeneration in Tm7sf2 KO mice. This was associated to a delay in the proliferation of residual hepatocytes and increased ER stress, with altered level of p53, and abnormal activation of p21, which delayed cyclin expression.
Results
Expression of TM7SF2 and LBR during liver regeneration
We performed PH on Tm7sf2 KO and WT littermates and collected regenerating livers at different times following PH. We initially determined the temporal pattern of Tm7sf2 expression during liver regeneration. By analyzing Tm7sf2 mRNA expression level in WT mice by qPCR, we observed a decrease from 0 to 36 h following PH, time after which the levels are stabilized at approximately 50% of the initial value (). As expected, no transcription was observed in KO mice (not shown).
Figure 1. Expression of C14-SR and LBR during liver regeneration (A) Time course of Tm7sf2 mRNA expression in WT regenerating liver. The expression was normalized to mouse Gapdh. The amount of mRNA is expressed in relative fold of expression. Bars represent means ± SD (n = 3). (B) Lbr mRNA expression in regenerating liver in WT and Tm7sf2 KO mice. The expression was normalized to mouse Gapdh levels at each time point. The amount of mRNA is expressed in relative fold of expression. Bars represent means ± SD (n = 3). (C) Protein expression levels of C14-SR and LBR in regenerating livers from WT and Tm7sf2 KO mice, determined by Western Blotting (WB). Coomassie blue staining was used as loading control. Representative images are shown. Graphs represent the densitometric quantification of immunoblotting signal. Values are normalized to the level of histones staining with Coomassie Blue.
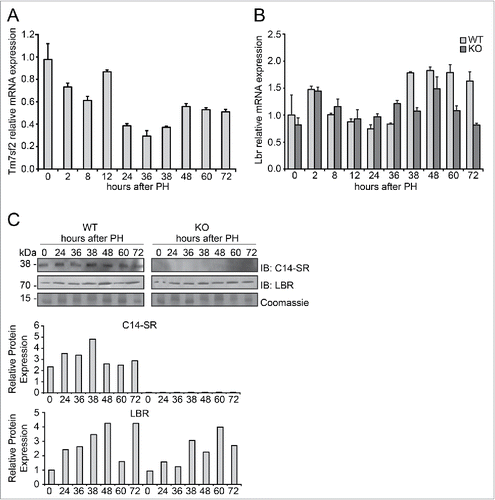
Given the importance of LBR in cholesterol biosynthesis in absence of Tm7sf2,Citation23 we monitored its expression during liver regeneration in both WT and Tm7sf2 KO mice. Gene expression analysis of Lbr showed an opposite trend compared to Tm7sf2, with an upregulation of almost 1.5-fold relative to normal liver starting from 38 h up to 72 h following PH in WT mice (), though the relative expression levels of Tm7sf2 were much higher than that of Lbr at all time points (Fig. S1), as previously described.Citation23 In Tm7sf2 KO mice, Lbr mRNA expression was only slightly induced at 38 h following PH compared to quiescent liver, thus remaining to levels significantly lower than in WT mice (). In order to confirm the reduced expression of LBR in Tm7sf2 KO mice compared to WT, we analyzed protein levels at different time points after PH. In WT mice, despite the reduced mRNA expression, C14-SR protein expression showed a mild increase in regenerating liver compared to normal liver (). Instead, in line with mRNA expression, we observed that LBR protein expression was induced during liver regeneration, with a peak at 48 h in WT mice, while its expression was reduced in KO compared to WT mice, as confirmed by densitometric analysis ().
To assess whether the absence of Tm7sf2 and the reduced expression of Lbr would affect cholesterol metabolism during liver regeneration in Tm7sf2 KO mice, cholesterol levels were measured in liver and plasma following PH. As previously observed in quiescent liver,Citation23 only mild differences were found in both liver and plasma of Tm7sf2 KO mice as compared to WT at all time points examined (). Moreover, we did not measure any accumulation of cholesterol biosynthesis intermediates in each time point analyzed (not shown).
Figure 2. Analysis of lipid content during liver regeneration in Tm7sf2 KO mice. (A) Cholesterol levels in liver and plasma during liver regeneration in WT and Tm7sf2 KO mice. Bars represent means ± SEM (n = 3–6). (B) Triglycerides levels in liver and plasma during liver regeneration in WT and Tm7sf2 KO mice. Bars represent means ± SEM (n = 3-6). Significant changes are shown. * P < 0.05, ** P < 0.01. (C) Lipid droplets formation in hepatocytes in WT and Tm7sf2 KO mice during liver regeneration, evaluated in paraffin-embedded liver sections harvested and stained with HE. Representative images are shown (magnification, 1000X). Bars indicate 10 µm.
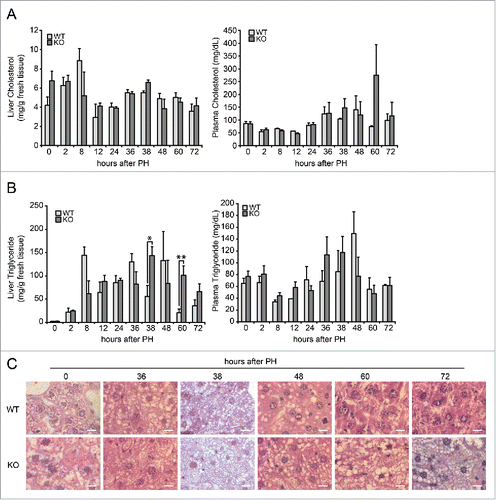
In conclusion, these results revealed that Tm7sf2 and Lbr are differently modulated in liver following PH, and that absence of Tm7sf2 does not impair cholesterol biosynthesis during liver regeneration.
Tm7sf2 KO mice exhibit a high-degree of transient hepatic steatosis during liver regeneration after PH
In the early stages of liver regeneration after PH, hepatocytes accumulate a significant amount of lipids in cellular structures called lipid droplets, which are mainly composed of triacylglycerol and cholesterol esters.Citation28 The functional meaning of this transient steatosis is not well defined as there are conflicting studies on how alteration of the hepatic lipid content affects the proliferative response.Citation29 We measured hepatic triglycerides content in WT and Tm7sf2 KO mice. In WT mice, levels of triglycerides – as an energy source for the process of cell proliferation – peak early at 8 h and later at 36 h following PH. In Tm7sf2 KO mice, we observed that levels of triglycerides progressively increase during regeneration, reaching significantly higher levels than in WT mice at 38 and 60 h following PH (). No significant variations were observed in plasma triglycerides levels in the same animals ().
In order to confirm the data obtained, we analyzed the presence of triglycerides, in the form of drops or vacuoles, in liver sections stained with hematoxylin-eosin (HE) in both WT and Tm7sf2 KO mice at different hours after PH. Lipid droplets were significantly increased in regenerating liver of both WT and Tm7sf2 KO mice compared to normal liver, starting from 38 h following PH. However, levels of hepatic steatosis was overall more prominent in Tm7sf2 KO, where lipid droplets were smaller but much more abundant than in WT mice (), a result in line with the observation of an increased hepatic triglycerides content in KO mice at 60 and 72 h after PH ().
In conclusion these results revealed a prolonged accumulation of triglycerides in livers of Tm7sf2 KO mice after PH, and point to a defective lipid metabolism that might affect hepatocytes proliferation.
Tm7sf2 deficiency leads to impaired hepatocyte proliferation
In order to evaluate hepatocytes proliferation, DNA synthesis was measured as bromodeoxyuridine (BrdU) incorporation in the nuclei of hepatocytes from WT and Tm7sf2 KO mice, injected 2 hours prior to the sacrifice. As previously observed,Citation5 2 peaks of BrdU incorporation in residual hepatocytes were found in WT mice at 38 and later at 60 h following PH, with 25% and 15% of cells involved in the G1/S transition, respectively in the first and second wave of cell division (). The BrdU analysis in KO mice revealed an early peak of about 12% BrdU-positive cells at 36 h post-PH. However, the peak remained unchanged at 38 h and reached significantly lower level than in WT mice. Furthermore, at 60 h following PH, only 6% of the cells were in G1/S phase, a percentage significantly lower than in the WT counterparts (). The total number of BrdU-positive nuclei at all time points analyzed was significantly reduced in Tm7sf2 KO compare to WT mice ().
Figure 3. Defective proliferation in Tm7sf2 KO regenerating livers (A) Representative micrographs of liver sections collected from WT and Tm7sf2 KO mice after PH, immunostained with anti-BrdU antibody (green). Cryosections were additionally stained with DAPI (blue). Representative merge images are shown. Bars indicate 10 µm. Magnification, 200X. (B) Quantitative analysis of BrdU labeling index (% of BrdU-positive hepatocytes) at each time of liver regeneration of WT and Tm7sf2 KO mice. Results represent means ± SD (n = 3). Significant changes are shown. *** P < 0.001 (C) Quantitative analysis of total BrdU labeled nuclei in the regenerating liver of WT and Tm7sf2 KO mice, obtained by summing the % of BrdU-positive cells at all experimental time points. Results represent means ± SD (n = 3). Significant changes are shown. ** P < 0.01. (D) Micrographs of liver sections, collected from WT and Tm7sf2 KO mice after PH, immunostained with monoclonal pH3 antibody (green) and DAPI (blue). Representative merge images are shown. Bars indicate 10 µm. Magnification, 400X. (E) Percentage of pH3-positive cells during liver regeneration in WT and Tm7sf2 KO mice. Results are means ± SD (n = 3). Significant changes are shown. ** P < 0.01. (F) Representative image of hepatocytes in G2 and M phase of the cell cycle in regenerating liver of WT and Tm7sf2 KO mice. Histological slides were immunostained with anti-pH3 antibody (red) and DAPI (blue). Merge images are shown. Bars indicate 10 µm. (G) Percentage of pH3-positive cells distinct from G2 and M phase at 48 hours PH in Tm7sf2 WT and Tm7sf2 KO mice. Results are means ± SD (n = 3). (H) Mitotic index determined by counting the percentage of mitotic figures on liver regenerating sections stained by H-E. Results are means ± SEM (n = 3-6). Significant changes are shown. ** P < 0.01, *** P < 0.001.
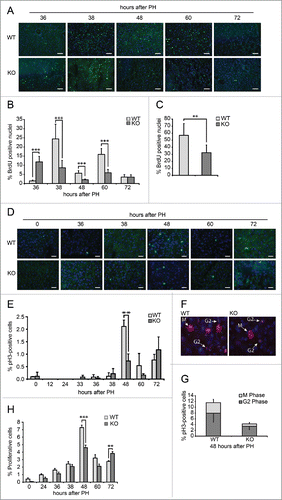
After analyzing G1/S progression, we evaluated the percentage of cells in G2/M phase. To this purpose, the expression of the phosphorylated form (Ser-10) of Histone H3 (pH3) was analyzed by immunohistochemistry. In both groups of mice, hepatocytes proliferate and divide at 48 h, even though in Tm7sf2 KO mice the percentage of cells expressing pH3 was remarkably lower with respect to WT mice (). However, fluorescence analysis allowing discriminating cells in the G2 versus M phases revealed that the proportion between cells in G2 and M phase was maintained in Tm7sf2 KO mice, where the percentage of cells in the mitotic phase amounted to about one-third of the total number of proliferating cells at 48 h, as in WT mice ().
In order to confirm the reduced number of mitotic cells in Tm7sf2 KO regenerating livers, mitotic index analysis was performed by counting the number of mitotic figures in HE-stained sections. Results showed a significant reduction in the percentage of proliferating cells in Tm7sf2 KO mice compared to WT at 48 h after PH, followed by an increase at 72 h, during the second wave of cell division, possibly as a compensatory mechanism ().
Overall, these results indicate a transient defect in the progression through the cell cycle of hepatocytes during liver regeneration in absence of Tm7sf2 gene.
Defective G1/S transition during liver regeneration in Tm7sf2 KO mice
To better understand whether Tm7sf2 has a role in triggering proliferation, we analyzed mRNA and protein expression of cyclin D1, cyclin A, cyclin-dependent kinase 4 (CDK4) and cyclin E1, important molecules related to the G1/S transition.
Cyclin D1 mRNA expression in WT mice showed an increase at 24 h following PH, followed by a progressive decrease until 38 h, down to the levels in normal liver. Then, it raised again at 48 h and remained constantly elevated at 60 and 72 h. The transcription level of cyclin D1 in Tm7sf2 KO mice, differently from the WT control, showed a lower but progressive increase starting at 24 h until 72 h after PH (). Because CDK4 interacts with cyclin D1 during the transition G1/S of the cell cycle, we analyzed its expression at the same time points. In WT livers Cdk4 mRNA showed a biphasic pattern, peaking at 36 and 48 h after PH, whereas, in KO livers its expression was reduced at all times examined (). Similarly, cyclin A mRNA expression in WT mice presented a peak at 48 h after PH, followed by a progressive reduction down to the level of normal liver, while in KO mice the peak of expression at 48 h was strongly reduced compared to WT and prolonged till 72 h (). Finally, the analysis of cyclin E1 mRNA showed 2 peaks of expression at 36 (about 5-fold increase) and 60 h (greater than 10-fold increase) in WT mice, whereas in Tm7sf2 KO mice the expression peaked at 36 h, with levels about 20-fold higher than in normal liver and, surprisingly, higher than in WT controls ().
Figure 4. Expression of markers of G1/S phase during liver regeneration in Tm7sf2 KO mice (A) mRNA levels of expression of cyclin D1, Cdk4, cyclin A and cyclin E1, detected by qPCR in WT and Tm7sf2 KO mice during liver regeneration at different times following PH. The expression of each gene was normalized to mouse Gapdh levels. The amount of mRNA is expressed in relative fold of expression. Bars represent means ± SD (n = 3). (B, C) Protein expression levels of cyclin D1, cyclin E (B), cyclin A and cdk4 (C) in regenerating livers from WT and Tm7sf2 KO mice, evaluated by WB. Coomassie Blue staining was used as loading control. Representative images are shown. Graphs represent the densitometric quantification of immunoblotting signal.
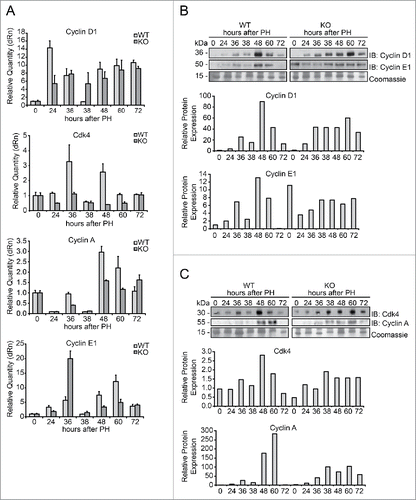
At the protein level, we observed a general trend characterized by reduced cyclins expression in Tm7sf2 KO regenerating livers. In particular, cyclin D1 protein expression in WT mice was upregulated starting from 36 h after PH, with the main peak of expression occurring at 48 h following PH (). Instead, in Tm7sf2 KO mice we observed a progressive increase in cyclin D1 levels which parallels that of mRNA, with reduced levels compared to WT mice, and a peak of expression at 60 h, as shown by densitometric analysis (). Similarly, cyclin E1 protein expression significantly increased at 36 h and peaked at 48 h after PH in WT mice, while in Tm7sf2 KO mice, despite high basal levels, cyclin E1 expression increased more gradually and remained at steady levels until 72 h after PH (). CDK4 protein expression also followed the same pattern of cyclin D1 expression, with Tm7sf2 KO exhibiting significantly reduced levels compared to WT mice (). Furthermore, cyclin A protein expression exhibited a peak at 48-60 h in WT, while in Tm7sf2 KO the increase was more gradual and the peak was significantly reduced compared to WT mice ().
Altogether, these data reveal that hepatocytes from Tm7sf2 KO mice have a defective progression through the G1/S phase, associated to delayed and/or reduced expression of central regulators of the cell cycle such as cyclin D1, cyclin A, cyclin E1 and CDK4.
Increased ER stress response during liver regeneration in Tm7sf2 KO mice
Recently it has been demonstrated that in Tm7sf2 KO mice the ER stress response to different stressors is impaired.Citation27 For this reason, we evaluated whether the adaptive program of the Unfolded Protein Response (UPR) triggered by ER stress was modified in Tm7sf2 KO mice in response to the stress that accompanies liver regeneration. The UPR and cellular stress regulators binding immunoglobulin protein/glucose-regulated protein 78 (Bip/Grp78), eukaryotic initiation factor 2A (eIF2a) and its phosphorylated form (p-eIF2a) were analyzed. In WT mice, Bip/Grp78 expression showed a progressive increase starting from 24 h until 72 h after PH. In Tm7sf2 KO mice, Bip/Grp78 expression at time 0 was slightly higher than in WT controls, and the increase during liver regeneration was more pronounced than in WT mice (), indicating an altered stress-induced response in KO with respect to WT mice. A mild increase was also found in eIF2a protein expression in Tm7sf2 KO mice, as revealed by densitometric analysis (). Moreover, the time course of eIF2a phosphorylation was fairly different between the WT and Tm7sf2 KO mice (). In WT mice, p-eIF2a appeared at 24 h and peaked at 38 h after PH, decreasing thereafter, while in Tm7sf2 KO mice, p-eIF2a raised at 38 h and remained elevated until 60 h after PH ().
Figure 5. Evaluation of the ER stress response in Tm7sf2 KO mice during liver regeneration (A) WB analysis of Bip/Grp78, eIF2a and p-eIF2a protein expression during liver regeneration in WT and Tm7sf2 KO mice. The control of the proteins amount is represented by Coomassie Blue staining. Representative images are shown. Graphs represent the densitometric quantification of immunoblotting signal. (B) Levels of expression of the proteins p53 and p21, evaluated by WB. Coomassie Blue staining was used as loading control. Representative images are shown. Graphs represent the densitometric quantification of immunoblotting signal.
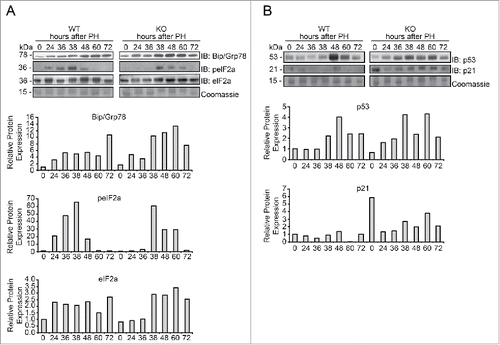
Given the role of the tumor suppressor p53 in stress sensing and in ER stress,Citation30 as well as in cell cycle gatekeeping through its target gene p21, we decided to explore whether p53 and p21 expression was different in Tm7sf2 WT and KO mice, and whether such a change could justify the data observed and be responsible for the impaired liver regeneration. We observed that in WT mice, a significant peak of p53 protein expression occurred at 48 h and declined at 72 h after PH, whereas in Tm7sf2 KO mice p53 expression was increased starting at 24 h and remained constantly high until 60 h after PH (). Furthermore, p53-target gene p21, one of the most important regulators of cell cycle arrest, was also highly expressed in Tm7sf2 KO livers, both in quiescent liver and at all time points of liver regeneration ().
In conclusion, these results revealed that during liver regeneration Tm7sf2 KO mice present an abnormal ER stress response, and increased p53-mediated p21 overexpression that might be responsible of the delayed hepatocyte replication.
Discussion
Tm7sf2 gene transcribes for C14-SR,Citation22-25 involved in cholesterol biosynthesis. It has been demonstrated that LBR has also sterol Δ14-reductase activity with redundant enzymatic activity with Tm7sf2.Citation22-24 For this reason, Tm7sf2 KO mouse is healthy, with no significant physiological alterations and normal cholesterol levels.Citation23
The main focus of our study was to investigate whether the loss of Tm7sf2 could affect hepatocyte proliferation. Indeed, the study of the proliferation kinetics of residual hepatocytes after PH showed a delay in Tm7sf2 KO mice. During the first mitotic wave, BrdU incorporation and pH3 expression are down-modulated and the transition from G1 to S-phase appears to be reduced and postponed. The expression of cyclins involved in G1/S transition is delayed both at the transcriptional and translational levels, indicating a difficulty to progress from G1 to S phase and, in turn, a delayed entry into G2/M phase of the cell cycle. Indeed, the percentage of cells in the mitotic phase at 48 h is lower in KO compared to WT mice.
In acute request of cholesterol, such as liver regeneration, we found an increased transcription of the Lbr gene in WT mice, thus suggesting that LBR might play the most relevant role in controlling cholesterol production during liver regeneration. What are the signals triggering the increase in LBR expression during liver regeneration and what is the reason for this redundancy in quiescent livers is unknown at this time. It is possible that LBR compensates for cholesterol production during all the period necessary to guarantee the appropriate amount to the residual hepatocytes and to the whole mouse. In Tm7sf2 deficient mice, Lbr gene expression is downregulated, thus suggesting that Lbr should not be able to compensate for the absence of Tm7sf2 in this condition. However, the analysis of total hepatic and plasma cholesterol levels did not reveal particular modifications between the 2 mouse strains during liver regeneration. In the light of these results, it appears clear that it is not cholesterol lack or reduction that affects the rate of proliferation of residual hepatocytes in KO mice. Other biological events involving Tm7sf2 are responsible for the delay in the cell cycle observed in Tm7sf2 KO mice. In particular, we found that hepatic triglycerides were differently accumulated during liver regeneration in WT and Tm7sf2 KO mice. Triglycerides are the lipids that meet the energy demand of the remaining hepatocytes, necessary to start and complete the intricate mechanism of reconstitution of the original liver mass.Citation31,32 In WT mice, triglycerides increase significantly at 8 h after PH with the peak in coincidence with the transition G1/S phase and with M-phase. This physiological increase in storage of triglycerides and lipids generally determines a state of transient steatosis. Animals showing an altered formation of lipid droplets regenerate liver with considerable difficulty.Citation17,19 Tm7sf2 KO mice lack the triglycerides peak at 8 h and accumulate lipids slowly, reaching at 38 and 60 h after PH higher values compared to WT mice. The steatotic condition is maintained longer in Tm7sf2 KO mice, in line with the delay observed in proliferation. Are triglycerides the cause or the effect in the delay of proliferation in Tm7sf2 KO mice? Can other factors affect the impairment in proliferation observed in absence of Tm7sf2 gene? Recently, it has been determined that Tm7sf2 KO mice present an altered ER stress response following different stress inducing treatmentsCitation27 and liver regeneration represents a strong stress condition in ER. Moreover, it has been demonstrated that lipids accumulated in hepatocytes after PH can trigger ER stress, which in turn may result in a delay in DNA replication and impairment of liver regeneration.Citation33 Our analysis of ER stress sensors showed differences between the 2 groups of mice during liver regeneration, indicating a variation in stress response. C14-SR resides in the endoplasmic reticulum, where synthesis, folding, assembly, and secretion of proteins, as well as lipid synthesis occur. Any disturbance of protein homeostasis and proper folding leads to endoplasmic reticulum stress and activation of ER stress signaling, including UPR that detects these alterations and brings the situation back to normality.Citation33,34 The analysis of ER stress sensors in Tm7sf2 KO during liver regeneration shows increased level of Bip/GRP78 and, to a lesser extent, of eIF2a respect to the WT, and an altered timing in the phosphorylation of eIF2a, whose role is to attenuate total protein synthesis, which is consistent with the delay in cyclins expression in Tm7sf2 KO.
It has been postulated that cell cycle arrest following activation of URP is linked to activation of p53, which in turn regulates p21 expression.Citation30,35,36 A sound difference between WT and Tm7sf2 KO mice has been detected in p53 expression. While in WT liver p53 shows the typical expression with a peak at 48 h that declines over the following hours, in Tm7sf2 KO mice p53 is intensely and constantly upregulated from 36 until 60 h after PH. This difference in p53 reflects the difference observed in p21 expression, on comparing WT and KO mice, that could be responsible of the delay disclosed in the start of the proliferation.
In conclusion, here we have described an altered progression of liver regeneration following PH in mice lacking the enzyme C14-SR. The defective proliferation was associated to prolonged lipid accumulation and consequent ER stress, inducing delayed cell cycle progression of hepatocytes. Future studies will shed light on the mechanism how absence of C14-SR alters triglycerides metabolism when hepatocytes are induced to proliferate, and will provide a better understanding of its role in the metabolism of regenerating liver.
Materials and methods
Animals
Adult C57BL/6 WT and Tm7sf2 KO male miceCitation23 were housed under standard conditions (12 h light/dark cycles) with food and water ad libitum before and after surgery. All experiments were performed using 8- to 12-week-old mice. A number of 3–4 mice each time point was used for most of the experiments, unless differently specified. All of the animal experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Protocols for the care and use of animals were approved by the University of Perugia Animal Care and Use Committee. PH experiments were conducted, according to Higgins and Anderson,Citation2 under deep anesthesia by inhalation of isoflurane (2-chloro-2-(difluoromethoxy)-1,1,1-trifluoro-ethane). Mice were subjected to removal of two-thirds of the liver mass and sacrificed at the indicated times post PH. At the time of sacrifice the regenerating liver was quickly extracted and tissue punches were used for RNA analysis, protein expression study and histological observation.
Liver histology and immunohistochemistry
Two hours before sacrifice, mice were injected with bromodeoxyuridine (BrdU, Sigma-Aldrich) in 0.9% NaCl (1 mg kg−1). Subsequently, the fragments of regenerated tissue were fixed in 10% formalin and embedded in paraffin after serial dehydration in alcohol for histology, or included in Tissue-Tek® O.C.T Compound (Optimum Cutting Temperature, Sakura) and stored at −80°C until use for immunohistochemistry. The paraffin-embedded livers were sectioned (5 µm) and stained by hematoxylin and eosin (HE) to study tissue structure and to calculate mitotic index by counting proliferating figures. Frozen sections were incubated with anti-BrdU antibody and anti-pH3 antibody. See Table S1 for primary and secondary antibodies used. Slides were examined with a Zeiss Axioplan fluorescence microscope. The images were acquired by using a Spot-2 cooled camera (Diagnostic Instruments). Images are representative of 3 independent experiments. Total BrdU and pH3 labeled hepatocytes were determined by counting positively stained and total cell nuclei in 10 microscope fields (200X and 400X magnification, respectively) per liver section at each experimental point. The hepatocyte proliferation index was calculated as percentage of BrdU positive cell nuclei.
Western blotting (WB) analysis
Fresh liver tissue was homogenized in Laemmli Buffer, boiled at 95°C, sonicated, and then centrifuged at maximum speed for 30 min. The extracts were normalized by Coomassie staining and subsequently separated by SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose or PVDF membrane. See Table S1 for antibodies used. WB were revealed using ECL (Amersham Biosciences). Protein loading was verified using Coomassie staining of gels. Protein band intensities were quantified by densitometric analysis using Image J software. Images are representative of 3 independent experiments.
RNA extraction and qPCR
Total RNA was extracted with TRIzol reagent (Invitrogen) and cDNAs were prepared using M-MLV Kit (Thermo Fisher Scientific Inc. Waltham, MA, USA) by using 25–50 ng of cDNA per sample. A mix of cDNA from 3–4 mice was used for qPCR experiments. qPCR was performed using Mx3000P qPCR System, Brilliant SYBR Green QPCR Master Mix (Agilent Technologies Inc., Stratagene) and ROX as the reference dye. See Table S2 for primer sequences. All values are relative to those of Gapdh mRNA levels at each time point. Bars represent the mean ± standard deviation (SD) (n = 3 technical replicates).
Liver lipid content
Lipids were extracted by Folch method,Citation37 and cholesterol and triglycerides were separated by thin-layer chromatography (n-hexane/diethyl ether/acetic acid, 70:30:1, v/v/v), visualized with Cu-acetate reagent,Citation38 and quantified. Purified cholesterol and triglycerides standards were run on the same plate as the samples to construct calibration curves. The images were acquired using the VersaDoc Imaging System and signals were quantified using Quantity One software (Bio-Rad Laboratories). Plasma cholesterol and triglycerides were determined enzymatically with commercial kits (Cholesterol Liquid and Triglyceride Liquid, Sentinel Diagnostic).
Statistical analysis
All data are presented as mean ± SD or SEM. Statistical significance was determined by the Student t test. Statistical significance was assumed when *P < 0.05.
Abbreviations
| C14-SR | = | 3β-hydroxysterol-Δ14-reductase |
| PH | = | Partial Hepatectomy |
| ER | = | Endoplasmic Reticulum |
| SRE | = | Sterol Response Elements |
| LBR | = | Lamin B Receptor |
| HE | = | hematoxylin-eosin |
| BrdU | = | bromodeoxyuridine |
| CDK4 | = | Cyclin-Dependent Kinase 4 |
| UPR | = | Unfolded Protein Response |
| Bip/Grp78 | = | Binding immunoglobulin protein/glucose-regulated protein 78 |
| eIF2a | = | eukaryotic Initiation Factor 2A |
| p-eIF2a | = | phospho-eIF2A |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1195939_Supplemental_Material.docx
Download MS Word (220 KB)Acknowledgment
We thank Paolo Puccetti for critical reading of the manuscript, Silvano Pagnotta and Maria Luisa Alunni for technical assistance, Bartolomeo Sebastiani for help with lipid analysis experiments, and all the member of G. Servillo's Lab for helpful discussion.
Funding
This study was supported by grants from Associazione Umbra Contro il Cancro (AUCC) and Fondazione Cassa di Risparmio di Perugia. G.S. is recipient of a PRIN Project n. 2010C2LKKJ_002. D.B. is recipient of AUCC fellowship program. M.M.B. was supported by a Fellowship from Fondazione Umberto Veronesi.
References
- Fausto N. Liver regeneration. J Hepatol 2000; 32:19-31; PMID:10728791; http://dx.doi.org/10.1016/S0168-8278(00)80412-2
- Higgins GM, Anderson RM. Experimental pathology of liver: restoration of liver in white rat following partial surgical removal. Arch Pathol (Chic) 1931; 12:186-202.
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science 1997; 276:60-6; PMID:9082986; http://dx.doi.org/10.1126/science.276.5309.60
- Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol 2004; 5:836-47; PMID:15459664; http://dx.doi.org/10.1038/nrm1489
- Servillo G, Della Fazia MA, Sassone-Corsi P. Transcription factor CREM coordinates the timing of hepatocyte proliferation in the regenerating liver. Genes Dev 1998; 12:3639-43; PMID:9851970; http://dx.doi.org/10.1101/gad.12.23.3639
- Servillo G, Della Fazia MA, Sassone-Corsi P. Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM. Exp Cell Res 2002; 275:143-54; PMID:11969286; http://dx.doi.org/10.1006/excr.2002.5491
- Servillo G, Penna L, Foulkes NS, Magni MV, Della Fazia MA, Sassone-Corsi P. Cyclic AMP signalling pathway and cellular proliferation: induction of CREM during liver regeneration. Oncogene 1997; 14:1601-6; PMID:9129151; http://dx.doi.org/10.1038/sj.onc.1200996
- Castelli M, Pieroni S, Brunacci C, Piobbico D, Bartoli D, Bellet MM, Colombo E, Pelicci PG, Della Fazia MA, Servillo G. Hepatocyte odd protein shuttling (HOPS) is a bridging protein in the nucleophosmin-p19(Arf) network. Oncogene 2013; 32:3350-8; PMID:22890319; http://dx.doi.org/10.1038/onc.2012.353
- Castelli M, Piobbico D, Bartoli D, Pieroni S, Brunacci C, Bellet MM, Chiacchiaretta M, Della Fazia MA, Servillo G. Different functions of HOPS isoforms in the cell: HOPS shuttling isoform is determined by RIP cleavage system. Cell Cycle 2014; 13:293-302; PMID:24240191; http://dx.doi.org/10.4161/cc.27054
- Della Fazia MA, Castelli M, Bartoli D, Pieroni S, Pettirossi V, Piobbico D, Viola-Magni M, Servillo G. HOPS: a novel cAMP-dependent shuttling protein involved in protein synthesis regulation. J Cell Sci 2005; 118:3185-94; PMID:16014383; http://dx.doi.org/10.1242/jcs.02452
- Pieroni S, Della Fazia MA, Castelli M, Piobbico D, Bartoli D, Brunacci C, Bellet MM, Viola-Magni M, Servillo G. HOPS is an essential constituent of centrosome assembly. Cell Cycle 2008; 7:1462-6; PMID:18418082; http://dx.doi.org/10.4161/cc.7.10.5882
- Della Fazia MA, Piobbico D, Bartoli D, Castelli M, Brancorsini S, Viola Magni M, Servillo G. lal-1: a differentially expressed novel gene during proliferation in liver regeneration and in hepatoma cells. Genes Cells 2002; 7:1183-90; PMID:12390252; http://dx.doi.org/10.1046/j.1365-2443.2002.00593.x
- Bellet MM, Piobbico D, Bartoli D, Castelli M, Pieroni S, Brunacci C, Chiacchiaretta M, Del Sordo R, Fallarino F, Sidoni A, et al. NEDD4 controls the expression of GUCD1, a protein upregulated in proliferating liver cells. Cell Cycle 2014; 13:1902-11; PMID:24743017; http://dx.doi.org/10.4161/cc.28760
- Della Fazia MA, Pettirossi V, Ayroldi E, Riccardi C, Magni MV, Servillo G. Differential expression of CD44 isoforms during liver regeneration in rats. J Hepatol 2001; 34:555-61; PMID:11394655; http://dx.doi.org/10.1016/S0168-8278(00)00065-9
- Della Fazia MA, Servillo G, Viola-Magni M. Different expression of tyrosine aminotransferase and serine deydratase in rat livers after partial hepatectomy. Biochem Biophys Res Comm 1992; 182:753-9; PMID:1370891; http://dx.doi.org/10.1016/0006-291X(92)91796-S
- Delgado-Coello B, Briones-Orta MA, Macias-Silva M, Mas-Oliva J. Cholesterol: recapitulation of its active role during liver regeneration. Liver Int 2011; 31:1271-84; PMID:21745289; http://dx.doi.org/10.1111/j.1478-3231.2011.02542.x
- Fernandez MA, Albor C, Ingelmo-Torres M, Nixon SJ, Ferguson C, Kurzchalia T, Tebar F, Enrich C, Parton RG, Pol A. Caveolin-1 is essential for liver regeneration. Science 2006; 313:1628-32; PMID:16973879; http://dx.doi.org/10.1126/science.1130773
- Lasuncion MA, Martin-Sanchez C, Canfran-Duque A, Busto R. Post-lanosterol biosynthesis of cholesterol and cancer. Curr Opin Pharmacol 2012; 12:717-23; PMID:22824432; http://dx.doi.org/10.1016/j.coph.2012.07.001
- Shteyer E, Liao Y, Muglia LJ, Hruz PW, Rudnick DA. Disruption of hepatic adipogenesis is associated with impaired liver regeneration in mice. Hepatology 2004; 40:1322-32; PMID:15565660; http://dx.doi.org/10.1002/hep.20462
- Fernandez C, Lobo Md Mdel V, Gomez-Coronado D, Lasuncion MA. Cholesterol is essential for mitosis progression and its deficiency induces polyploid cell formation. Exp Cell Res 2004; 300:109-20; PMID:15383319; http://dx.doi.org/10.1016/j.yexcr.2004.06.029
- Singh P, Saxena R, Srinivas G, Pande G, Chattopadhyay A. Cholesterol biosynthesis and homeostasis in regulation of the cell cycle. PloS One 2013; 8:e58833; PMID:23554937; http://dx.doi.org/10.1371/journal.pone.0058833
- Bennati AM, Castelli M, Della Fazia MA, Beccari T, Caruso D, Servillo G, Roberti R. Sterol dependent regulation of human TM7SF2 gene expression: role of the encoded 3beta-hydroxysterol Delta14-reductase in human cholesterol biosynthesis. Biochim Biophys Acta 2006; 1761:677-85; PMID:16784888; http://dx.doi.org/10.1016/j.bbalip.2006.05.004
- Bennati AM, Schiavoni G, Franken S, Piobbico D, Della Fazia MA, Caruso D, De Fabiani E, Benedetti L, Cusella De Angelis MG, Gieselmann V, et al. Disruption of the gene encoding 3beta-hydroxysterol Delta-reductase (Tm7sf2) in mice does not impair cholesterol biosynthesis. FEBS J 2008; 275:5034-47; PMID:18785926; http://dx.doi.org/10.1111/j.1742-4658.2008.06637.x
- Roberti R, Bennati AM, Galli G, Caruso D, Maras B, Aisa C, Beccari T, Della Fazia MA, Servillo G. Cloning and expression of sterol Delta 14-reductase from bovine liver. Eur J Biochem 2002; 269:283-90; PMID:11784322; http://dx.doi.org/10.1046/j.0014-2956.2001.02646.x
- Schiavoni G, Bennati AM, Castelli M, Fazia MA, Beccari T, Servillo G, Roberti R. Activation of TM7SF2 promoter by SREBP-2 depends on a new sterol regulatory element, a GC-box, and an inverted CCAAT-box. Biochim Biophys Acta 2010; 1801:587-92; PMID:20138239; http://dx.doi.org/10.1016/j.bbalip.2010.01.013
- Olins AL, Rhodes G, Welch DB, Zwerger M, Olins DE. Lamin B receptor: multi-tasking at the nuclear envelope. Nucleus 2010; 1:53-70; PMID:21327105; http://dx.doi.org/10.4161/nucl.1.1.10515
- Bellezza I, Roberti R, Gatticchi L, Del Sordo R, Rambotti MG, Marchetti MC, Sidoni A, Minelli A. A novel role for Tm7sf2 gene in regulating TNFalpha expression. PloS One 2013; 8:e68017; PMID:23935851; http://dx.doi.org/10.1371/journal.pone.0068017
- Garcia-Arcos I, Gonzalez-Kother P, Aspichueta P, Rueda Y, Ochoa B, Fresnedo O. Lipid analysis reveals quiescent and regenerating liver-specific populations of lipid droplets. Lipids 2010; 45:1101-8; PMID:21063798; http://dx.doi.org/10.1007/s11745-010-3492-2
- Newberry EP, Kennedy SM, Xie Y, Luo J, Stanley SE, Semenkovich CF, Crooke RM, Graham MJ, Davidson NO. Altered hepatic triglyceride content after partial hepatectomy without impaired liver regeneration in multiple murine genetic models. Hepatology 2008; 48:1097-105; PMID:18697204; http://dx.doi.org/10.1002/hep.22473
- Dioufa N, Chatzistamou I, Farmaki E, Papavassiliou AG, Kiaris H. p53 antagonizes the unfolded protein response and inhibits ground glass hepatocyte development during endoplasmic reticulum stress. Exp Biol Med (Maywood) 2012; 237:1173-80; PMID:23038705; http://dx.doi.org/10.1258/ebm.2012.012140
- Farrell GC. Probing Prometheus: fat fueling the fire? Hepatology 2004; 40:1252-5; PMID:15558710; http://dx.doi.org/10.1002/hep.20522
- Rudnick DA. Trimming the fat from liver regeneration. Hepatology 2005; 42:1001-3; PMID:16250044; http://dx.doi.org/10.1002/hep.20931
- Hamano M, Ezaki H, Kiso S, Furuta K, Egawa M, Kizu T, Chatani N, Kamada Y, Yoshida Y, Takehara T. Lipid overloading during liver regeneration causes delayed hepatocyte DNA replication by increasing ER stress in mice with simple hepatic steatosis. J Gastroenterol 2014; 49:305-16; PMID:23512345; http://dx.doi.org/10.1007/s00535-013-0780-7
- Fu S, Watkins SM, Hotamisligil GS. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab 2012; 15:623-34; PMID:22560215; http://dx.doi.org/10.1016/j.cmet.2012.03.007
- Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 2013; 32:805-18; PMID:22508478; http://dx.doi.org/10.1038/onc.2012.130
- Zhang F, Hamanaka RB, Bobrovnikova-Marjon E, Gordan JD, Dai MS, Lu H, Simon MC, Diehl JA. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. J Biol Chem 2006; 281:30036-45; PMID:16893887; http://dx.doi.org/10.1074/jbc.M604674200
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957; 226:497-509; PMID:13428781
- Macala LJ, Yu RK, Ando S. Analysis of brain lipids by high performance thin-layer chromatography and densitometry. J Lipid Res 1983; 24:1243-50; PMID:6631248
