ABSTRACT
The Hippo signaling pathway regulates cellular proliferation and survival, thus exerting profound effects on normal cell fate and tumorigenesis. The pivotal effector of this pathway is YAP1, a transcriptional co-activator amplified in mouse and human cancers where it promotes epithelial-to-mesenchymal transition (EMT) and malignant transformation. The Hippo tumor suppressor pathway has been suggested to inhibit the YAP1 function through serine phosphorylation-induced cytoplasmic retention and degradation. Here we report that the tyrosine188 (Y188) site of YAP1 isoform with 2 WW domains (known as YAP1-2) plays an important role in YAP1-induced cellular transformation. IP-Mass Spectrometry analysis of YAP1 identified the phosphorylation of Y188 but not other tyrosine residues. In contrast to the aberrant 3D acinus formation observed in YAP1-WT transduced cells, overexpression of YAP1-Y188F (non-phosphorylated mimic) displayed normal 3D structures. In addition, knockdown of the endogenous YAP1 in MDA-MB231 breast cancer cells inhibited cell proliferation and migration, which were then successfully rescued by the exogenous YAP1-WT and YAP1-Y188E but not Y188F. Mechanistically, we also demonstrated that YAP1-Y188F had a higher affinity to the upstream negative regulator PTPN14 and was extensively localized in the cytoplasm. Since the Y188 is located in the conserved aromatic core of the WW domain of YAP1, our finding has a wide implication for WW domain signaling in general, where Y phosphorylation may act as a common positive regulator of the complex formation via WW domains. In summary, our results indicate that tyrosine 188 plays an important role in the YAP1-induced cellular transformation and its phosphorylation may intriguingly serve as a positive indicator of YAP1 activation.
Introduction
The determination of organ size incorporates a delicate balance between growth, proliferation and apoptosis.Citation1 Initially discovered in Drosophila, the Hippo signaling pathway has been shown to be crucial in growth regulation, tissue regeneration and stem cell renewal in both Drosophila and mammalian cells.Citation2,3,4 The key downstream effectors of the Hippo pathway, YAP1 (Yes-associated protein 1) and TAZ (transcriptional co-activator with PDZ-binding motif), are tightly regulated by a number of upstream molecules, such as Mst1/2, Lats1/2 and RASSF family proteins.Citation4,5,6,7 Comprehensive survey of the most common solid cancer types revealed widespread and frequent YAP1 overexpression in lung, ovarian, pancreatic, colorectal, hepatocellular and prostate carcinomas.Citation2,8 We previously identified by the array-based comparative genomic hybridization (aCGH) that YAP, the pivotal effector of the Hippo pathway, is amplified in mouse and human breast tumors.Citation9 Of note, overexpression of YAP1 induces the epithelial-to-mesenchymal transition (EMT), suppression of apoptosis, growth factor-independent proliferation and anchorage-independent growth in soft agar.Citation9,10
Although YAP1 was originally identified as a binding partner for the Src family member, Yes protein-tyrosine kinase,Citation11,12 only recent studies raised the possibility that YAP1 could be phosphorylated at tyrosine sites in certain signaling scenarios.Citation13,14,15,16 In terms of context-dependent signaling it is important to note that the immunoprecipitated endogenous YAP1 protein from primary chicken embryo fibroblasts, which were labeled with radioactive orthophosphate, showed that only Serine was decorated by phosphate in the phospho-amino acid analysis.Citation12 The Stein laboratory was the first to report that tyrosine phosphorylation of YAP is necessary for its interaction with the Runx2 transcription factor, as well as for its subsequent nuclear trafficking.Citation13 Moreover, inhibition of the Src and Yes protein-tyrosine kinase activities reduced the tyrosine phosphorylation of YAP, resulting in dissociation of the endogenous Runx2-YAP complexes. It was also found that in response to DNA damage, YAP is phosphorylated by the c-Abl kinase at Y357 and this modification stabilizes the YAP protein. Phospho-Y357-YAP has increased affinity for the p73 transcription factor and this association results in elevated expression of the p73-dependent pro-apoptotic target genes.Citation14 In addition, active Yes kinase transiently binds and phosphorylates YAP at one or more tyrosine residues, which in turn recruits the TEAD2 complex to expression of key factors involved in the maintenance and self-renewal of embryonic stem (ES) cells.Citation15 Furthermore, the Hahn laboratory reported that in colon cancer, YAP and the transcription factor TBX5 form a complex with β-catenin and phosphorylation of YAP by the YES tyrosine kinase leads to localization of this complex to the promoter of anti-apoptotic genes, including BCL2L1 and BIRC5, defining a β-catenin-YAP-TBX5 complex essential for the transformation and survival of β-catenin-driven colon malignancy.Citation17 Most recently, using advanced tools of proteomics and proximity ligation, the O'Neill laboratory documented that Ras association domain family 1C (RASSF1C) was able to target Src and Yes kinases to epithelial cell to cell junctions to promote tyrosine phosphorylation of YAP along with β-catenin and E-cadherin and thereby promoting YAP-mediated invasion of carcinomas.Citation16 All these studies implicate tyrosine phosphorylation in the function of YAP. In this report we explore one mechanistic facet of tyrosine-phosphorylated YAP in breast cancer biology.
Here we identified for the first time the phosphorylation of YAP1-Y188 and demonstrated that overexpression of YAP1-Y188F had no effect on the 3D morphogenesis of MCF10A cells, in contrast to the invasive phenotype elicited by the YAP1-WT or YAP1 with mutations at other tyrosine sites. In breast MDA-MB231 cells, YAP1-WT or YAP1-Y188E rescued the phenotype associated with YAP1 knockdown, whereas YAP1-Y188F failed to do so. Last, immunostaining showed that YAP1-Y188F was extensively localized in the cytoplasm, in contrast to the nuclear localization of YAP1-WT. Notably, Y188 is located within the aromatic core of the first WW domain of YAP1 and Y188 phosphorylation acts as a positive regulator of complexes mediated by the YAP1 WW domain, and perhaps by other WW domains. Taken together, our data indicate that the phosphorylation of YAP1-Y188 plays an important role in its oncogenic functions in breast cancer.
Materials and methods
Cell culture and transfections
MCF10A and MCF12A cell cultures were performed as previously described.Citation18 MDA-MB-231, T47D and HCC1143 cells were cultivated in RPMI-1640 medium with 10% fetal calf serum (FBS); MCF7 and SK-BR3 cells were cultivated in DMEM medium with 10% FBS; SUM159 was cultivated in F12K medium plus 5% FBS, 5 µg/ml insulin and 1 µg/ml hydrocortisone. All media were supplemented with 100 units/ml penicillin, 100 µg/ml streptomycin and 2 mM glutamine. All cells were cultured in a humidified atmosphere of 95% air and 5% CO2 at 37°C. MDA-MB-231, T47D, HCC1143, MCF7, SK-BR3 and SUM159 cells were purchased from the American Type Culture Collection (ATCC, VA).
For knockdown experiments, shRNA hairpins targeting human YAP were obtained from the RNAi Consortium (The Broad Institute, Boston, MA). The target sequences are listed (in the 5′-3′ direction): shYAP: CCCAGTTAAATGTTCACCAA; and Control-shRNA: CAACAAGATGAAGAGCACCAA.
Retroviral or lentivirus packaging, MCF10A and MDA-MB231 cell transduction and drug selection were performed following standard protocols and was described previously.Citation19,20
Plasmid constructs
YAP1-2 gamma expression construct was described previously.Citation21 YAP1 tyrosine mutants and shYAP1 resistant constructs were established by PCR-based mutagenesis and mutant constructs were confirmed by DNA sequencing.
Antibodies and molecular biology analyses
Lats1 antibody was purchased from Cell Signaling Technology (Beverly, MA); YAP1 and PTPN14 antibodies from Santa Cruz biotechnology (Santa Cruz, CA); anti-phosphotyrosine antibody, clone 4G10 (EMD Millipore, MA); β-Actin antibody from Upstate (Lake Placid, NY); and Flag (M2) antibody from Sigma (St. Louis, MO). For protein extraction, cells were washed with phosphate-buffered saline and collected with IP buffer: 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 20% glycerol, 0.5% NP-40, 1x protease inhibitor cocktail (CompleteTM EDTA-free, Roche). Cell lysate was cleared by centrifugation at 14,000 rpm for 20 min at 4°C. Lysate was loaded onto 4–15% SDS-PAGE gel (ReadyGel, Bio-Rad) with SDS sample buffer. For immuno-blot, proteins were transferred onto Immobilon PVDF (Millipore), detected by various antibodies and visualized with Western Lightning Plus Chemiluminescence Kit (Perkin Elmer).
For RNA preparation and qRT-PCR detection, RNA was extracted using the Trizol reagent (Invitrogen). cDNA synthesis was performed using First-Strand cDNA Synthesis Kit (GE Healthcare) and quantitative real-time PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems).
Sequences of the qPCR primer pairs (in the 5′-3′ direction) are as follows:
GAPDH-F: GGTGAAGGTCGGAGTCAACGG;
GAPDH-R: GAGGTCAATGAAGGGGTCATTG;
FN1-F: GAAGCCGAGGTTTTAACTGC;
FN1-R: ACCCACTCGGTAAGTGTTCC;
CTGF-F: GCAGAGCCGCCTGTGCATGG;
CTGF-R: GGTATGTCTTCATGCTGG;
CYR61-F: CACACCAAGGGGCTGGAATG;
CYR61-R: CCCGTTTTGGTAGATTCTGG.
All measurements were performed in triplicate and standardized to the level of GAPDH.
Cell migration
Trans-well cell migration assay was performed as previously described.Citation9
Immunofluorescence microscopy
293 cells were cultured on coverslips to appropriate density. Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100 for 15 min. After blocking in 3% BSA for 30 min, slides were incubated with the primary antibody diluted in 1% BSA for 1 hr. After washing with PBS, slides were incubated with Alexa Fluor 488- or 594-conjugated secondary antibodies (1:1000 dilution) for 1 hr.
IP-nano-LC mass spec analysis
Flag-YAP1-WT transduced MCF10A lysates were harvested in IP buffer, immune-precipitated by the Anti-FLAG M2 Affinity Gel and eluted with the Flag peptide from Sigma (St. Louis, MO). The eluted products were run on a pre-cast 4-15% SDS-PAGE gel (ReadyGel, Bio-Rad) and were then stained with Coomassie blue. The band containing YAP1 were excised and digested with in-gel digestion protocol with trypsin only and solution samples were divided into 2 aliquots for dual enzyme digestion of trypsin and GluC according to the 2-step on pellet digestion protocol.Citation22,23 The digests were eluted on a 100 cm column with a 2-hr gradient with CID and ETD activation by Fusion Tribrid mass spectrometer (Thermo Scientific), using a previously described condition.Citation24 Raw files were searched with Proteome Discoverer (Thermo Scientific) against YAP1 sequence and Carbamidomethyl (57.021Da) was set as static modification on Cystine (C), and oxidation (15.995Da ) on methionine (M); phosphorylation (79.966 Da) on serine (S), threonine (T) and tyrosine (Y) were set as dynamic side chain modifications. The search results were merged with Scaffold (Proteome Software), and filtered to achieve a local peptide probability >95%.
Statistical analysis
Statistical analysis of data was performed using the SPSS statistics software package (SPSS, IL). All results are expressed as mean ± SD. *, P < .05; **, P < .001; ***, P < .0001.
Results
IP-Mass spectrometry analysis of YAP1 post-translational modifications
To examine the YAP1 post-translational modifications in cells, we overexpressed YAP1-wild-type (YAP1-WT), an isoform of YAP1 with 2 WW domains (YAP1-2 gama), in mammary epithelial MCF10A cells and performed a comprehensive IP-Mass spec analysis using a high-resolution nano-LC/MS system. As shown in , we confirmed 4 potential serine phosphorylation sites in YAP1 by Lats1/2 at 61, 109, 127 and 164;Citation25,26 4 potential phosphorylation sites by JNKs at T119, S138, T154 and S317;Citation27 and 5 potential phosphorylation sites at T119, S128, S138, S289 and S367 by CDK1.Citation28,29 More importantly, we identified several additional serine/threonine phosphorylation sites (). In particular, a phosphorylation modification of YAP1 at Y188 caught our attention. To confirm that such modification was not an artifact, we investigated the tyrosine phosphorylation status of endogenous YAP1 protein in a panel of breast cancer cell lines by immunoprecipitating YAP1 and then blotting with a pan-tyrosine phosphorylation antibody (4G10). The cell lines tested by us included: the immortalized non-transformed MCF10A and MCF12A cells; luminal type cells: T47D, SKBR3, MCF7; basal type cells: HCC1143, SUM159 and MDA-MB-231. Importantly, tyrosine phosphorylation of endogenous YAP1 was detected in the majority of the samples (), indicating the presence of and significance of this intrinsic modification in breast cancer cells.
Figure 1. Identification of post-translational modification sites of YAP1-2 gamma. Mass spectrometry analysis identified the potential YAP1 phosphorylation sites. The peptide sequences of YAP1 recovered by mass spectrometry are labeled in yellow; the serine and threonine phosphorylation sites are labeled in green; the tyrosine phosphorylation site was labeled by red.

Figure 2. Examination of endogenous YAP1 tyrosine phosphorylation in breast cancer cells. (A) Expression of tyrosine-phosphorylated YAP1 and total YAP1 in a panel of breast cancer cells as examined by immunoblot. β-actin was used as a loading control. (B) Tyrosine phosphorylation of- YAP1 induced by EGF treatment in MCF10A cells.
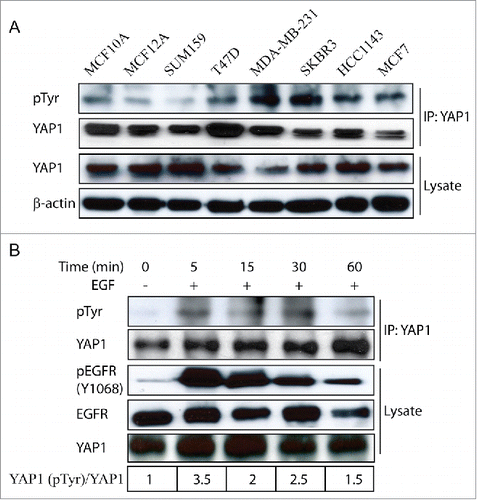
Recent studies have demonstrated that epidermal growth factor (EGF) stimulates YAP1 activity in Drosophila and MCF-10A cells, which contributes to cell proliferation.Citation30,31 Therefore, it would be of interest to know if there is any association between the growth stimulation and YAP1 modification. Accordingly, we treated the mammary epithelial MCF10A cells with EGF and found that YAP1 tyrosine phosphorylation was indeed increased (), indicating the dynamic changes of YAP1 tyrosine phosphorylation in response to growth factor stimulation.
YAP1-Y188F overexpression maintained the normal 3D morphogenesis
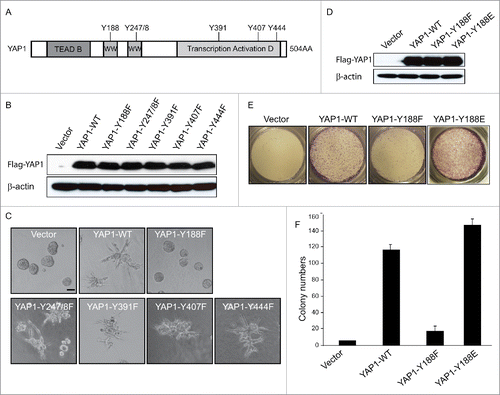
Altogether, there are 6 specific tyrosine (Y) residues within YAP1-2 gamma isoform,Citation32,33 i.e., Y-188, Y-247/248, Y-391, Y-407 and Y-444 (). An important question to ask then was whether the tyrosine phosphorylation of YAP1 has any functional role, especially in terms of its oncogenic properties. To do this, we generated retroviral expression constructs containing the specific tyrosine (Y) to phenylalanine (F) mutations, in the same YAP1 isoform with 2 WW domains (YAP1-2 gamma), which would mimic the non-phosphorylated-tyrosine. First, we showed that these Y-to-F mutations did not affect the YAP1 protein stability by Western blot (). Next, we took advantage of the 3D epithelial culture systems as it allowed epithelial cells to organize into structures resembling their in vivo architecture and thus made it possible to study the function of cancer genes and pathways in a biologically relevant context.Citation34 We had previously demonstrated that overexpression of YAP1-WT in MCF10A cells induced invasive 3D structures.Citation9 We then cultured MCF10A cells that were transduced with the different Y-F non-phosphorylated-tyrosine mimics of YAP1 in 3D. Importantly, only YAP1-Y188F but not any other Y-F mutations obliterated the invasive acinus formation in MCF10A cells (). This result suggested that Y188 of YAP1 plays a role in the abnormal 3D morphogenesis induced by YAP1.
Figure 3. YAP1-Y188E enhanced the YAP1 oncogenic functions. (A) Schematic representation of YAP1. The 6 tyrosine sites of YAP1 are indicated. (B) The Y-F mutations have no effect on the protein expression of YAP1, as shown by immunoblot in transduced MCF10A cells. β-Actin was used as a loading control. (C) Overexpression of YAP1-Y188F obliterates the aberrant acinar formation induced by YAP1 in 3D culture. Representative phase contrast images from 3 independent experiments are shown. (D) Expression of YAP1-WT, YAP1-Y188F and YAP1-Y188E at comparable levels in transduced MCF10A cells as revealed by immunoblot. β-Actin used as a loading control. (E) YAP1-Y188E overexpression increased cell migration. MCF 10A cells transduced with vector control or YAP1 variants were plated onto 8-µm transwell filters and allowed to migrate for 24 hrs. (***, p < 0.001) (F) YAP1-Y188E overexpression enhanced the colony formation in soft agar.
YAP1-Y188E overexpression enhanced the YAP1 oncogenic functions
To further confirm the effect of YAP1-Y188 modification on the YAP1 oncogenic function, we created a YAP1-Y188E mutant that mimicked the phosphorylated YAP1-Y188. MCF10A cells were then retro-virally transduced with the vector control, YAP1-WT, YAP1-Y188F and YAP1-Y188E constructs. Like Y188F, the Y188E mutation did not affect the protein expression of YAP1 (). However, YAP1-Y188E overexpression significantly enhanced cell migration in the Trans-well assay and colony formation in the soft agar assay, in striking contrast to YAP1-Y188F (). Our data showed that the phospho-YAP1-Y188 mimic strongly enhanced the YAP1 oncogenic functions.
YAP1-Y188F fails to rescue the loss-of-function of YAP1 in breast cancer cells
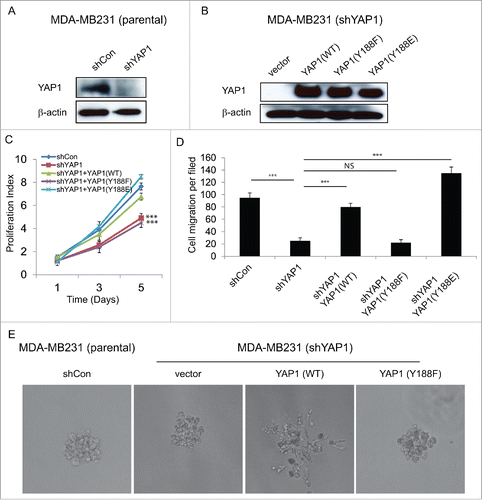
YAP1 is known to increase cell migration as well as promotion the EMT and breast epithelial cell transformation.Citation9,21,26 Our present results suggested that the modification of YAP1 at tyrosine 188 regulates its oncogenic functions (). We then elected to examine the effect of YAP1-Y188 modification on breast cancer malignancy and designed a rescue experiment using Flag-tagged YAP1 expression constructs that are resistant to shYAP1. As expected, knockdown of the endogenous YAP1 by shYAP1 in the MDA-MB-231 breast cancer cells () significantly decreased the cell proliferation (Figure S1A). However, when control and variant shYAP1-resistant constructs were used to rescue the phenotype observed for MDA-MB-231 cells with stable knockdown of endogenous YAP1 (), only the WT and Y188E but not Y188F construct restored the cell proliferation and migration (, S1B). In addition, we found that overexpression of YAP1-WT induced invasive 3D structure in MDA-MB231 cells, YAP1-Y188F failed to do so (). Taken together, these results indicated that YAP1-Y188F lost its ability to promote full malignant phenotype.
Figure 4. YAP1-Y188F fails to rescue the loss-of-function of YAP1 in MDA-MB-231 breast cancer cells. (A) Immunoblot demonstrates effective knockdown of YAP1 with the lentiviral shYAP1 construct in MDA-MB-231 cells. β-Actin was used as a loading control. (B) Expression of shYAP1-resistant WT-, Y188F- and Y188E-YAP1 variants in the MDA-MB-231 cells with stable knockdown of endogenous YAP1. (C) WT- and Y188E-YAP1, but not Y188F-YAP1, restores cell proliferation. (***, p < 0.001) (D) Quantifications of shControl or shYAP1 alone, and shYAP1 plus WT- , Y188F- or Y188E-YAP1 transduced MDA-MB-231 cell migration. (***, p < 0.001) (E) WT-YAP1, but not Y188F-YAP1, induced 3D invasive structure in MDA-MB231 cells.
Disrupted interaction between YAP1-Y188E and its upstream negative regulators
To investigate whether Y188F reduced the transcriptional co-activator activity of YAP1, we examined the effect of YAP1-Y188F on the expression of several, arbitrarily chosen, YAP1 target genes such as CTGF, Cyr61 and FN1.Citation21,35,36 Real-time qRT-PCR was performed using the RNA harvested from the vector control, YAP1-WT and YAP1-Y188F transduced MCF10A cells. The levels of CTGF, Cyr61 and FN1 were significantly reduced upon YAP1-Y188F expression as compared to the WT ().
Figure 5. Disrupted interaction between YAP1-Y188E and its upstream negative regulators. (A) qRT-PCR analysis of the YAP1 target genes in MCF10A cells. GAPDH was used as an internal control. Error bars equal ±SD. (** p < 0.01; * p < 0.05) (B) Disrupted interaction between YAP1-Y188E and Lats1. HEK293T cells were transfected with LATS1 alone or with LATS1 plus WT- , Y188F- or Y188E-YAP. YAP1 was immunoprecipitated and the co-immunoprecipitated Lats1 was detected by immunoblot. (C) Disrupted interaction between YAP1-Y188E with PTPN14. HEK293T cells were transfected with PTPN14 alone or with PTPN14 plus WT- , Y188F- or Y188E-YAP1. YAP1 was immunoprecipitated and the co-immunoprecipitated PTPN14 was detected by immunoblot. (D) Overexpression of YAP1-Y188F induces YAP1 nuclear exclusion. Immunofluorescence microscopy shows cytoplasmic localization of YAP1-Y188F in transduced-293 cells, in contrast to the nuclear localization of YAP-WT. (scale bar = 20µm). (E) Prediction of tyrosine kinases responsible for the phosphorylation of Y188. (F) Working model for the phosphorylation modification of YAP1-Y188. The in silico predicted kinases phosphorylate YAP1-Y188, the YAP1-Y188 phosphorylation enhances YAP1 oncogenic function through YAP1 nuclear activation.
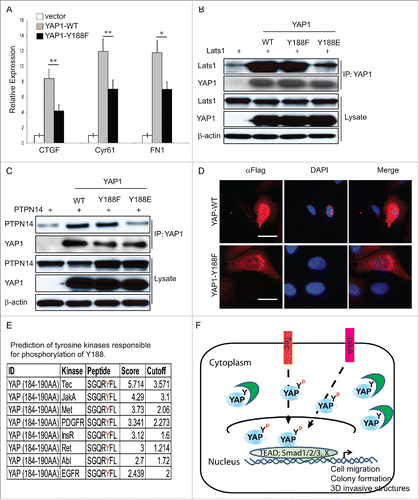
It is known that one of the YAP1 regulatory mechanisms in the Hippo signaling pathway is through YAP1 phosphorylation at Ser127 by Lats1/2 and subsequent cytoplasmic retention of YAP1.Citation5,25,26 Therefore, to investigate whether Y188F led to the YAP1 cytoplasmic sequestration, we set out to examine the interaction between YAP1-Y188F and Lats1 proteins. To do so, control vector, Flag-YAP1-WT, -Y188F or -Y188E and Lats1 was co-transfected into HEK293 cells and immunoprecipitation was performed. As suspected, YAP1-Y188E mutant lost its ability to interact with Lats1 (). Moreover, the Y188E mutation disrupted the interaction between YAP1 and its other upstream negative regulator: PTPN14Citation37,38,39,40 (). To confirm these biochemical findings, immunofluorescence microscopy was used to visualize the localization of non-phosphorylated mimic of YAP1-Y188F. Interestingly, YAP1-Y188F was extensively localized in the cytoplasm, in striking contrast to the exclusive localization of YAP1-WT in the nucleus (, S3).
Although Src, Yes, ErbB4 and EGFR are candidate kinases for Y188 phosphorylation of YAP1,Citation12,30,41,42 we decided to search for the potential tyrosine kinases responsible for the YAP1-Y188 phosphorylation in an unbiased way, via in silico analysis using the GPS 2.0 (Group-based Prediction System, version 2.0) software (http://gps.biocuckoo.org/).Citation43 We identified several intriguing candidates including Tec, JakA, PDGFR and Met kinases (). It remains to be determined how, which and how many of protein-tyrosine kinases regulate YAP1 via Y188 phosphorylation.
Our results are in agreement with a likely mechanism underlying the YAP1-Y188F inhibition of YAP1 oncogenic function, which might result from an increased affinity of the mutated WW domain toward LATS1/2, PTPN14 and other negative regulators of YAP1, including AMOTS, which all contain PPxY motif that is cognate for the WW domain.Citation33 Such scenario would result in enhanced sequestration of YAP1 in the cytoplasm ().
Discussion
We report here the identification of a novel tyrosine phosphorylation site Y188 in YAP1. We found that overexpression of YAP1-Y188E, the phosphorylated mimic, enhanced its oncogenic functions; whereas YAP1-Y188F obliterated the malignant potential of YAP1. Mechanistically, we showed that YAP1-Y188E disrupted the interaction between YAP1 and its upstream negative regulators, which might lead to its constitutive activation. On the other hand, YAP1-Y188F was extensively localized in the cytoplasm, which may partially explain for the inhibited YAP1 function. In summary, our present study demonstrates an important post-translational modification of YAP1 and provides a novel mechanism underlying the YAP1 oncogenic function in breast cancer ().
Post-translational modifications are widely seen in the regulation of multiple YAP functions such as oncogenesis as well as pro- and anti-apoptosis.Citation44 For example, phosphorylation by the LATS kinases leads to the YAP cytoplasmic retention and degradation.Citation25,26,45 In response to DNA damage, c-Abl kinase phosphorylates YAP1 (YAP1-1beta) at Y357 and enhances pro-apoptotic functions of p73.Citation14 Also, c-Jun N-terminal kinases (JNKs) phosphorylates YAP at T119, S138, T154, S317 and T362 and enhances its stabilization of ΔNp63.Citation27 Last, the CDK1 phosphorylates YAP at T119, S128, S138, S289 and S367, which promotes mitotic defects and decreases YAP mediated anti-apoptosis potential induced by the anti-tubulin drugs.Citation28,29 In the present study, we confirmed most of these post-translational modifications in YAP () and more interestingly, identified a novel Y188 phosphorylation site and showed that its phosphorylation played a critical role in the regulation of YAP oncogenic functions.
The Hippo pathway and its network of cross-talking proteins are enriched in WW domains and their ligands tend to contain PPxY motifs.Citation46,47 Consistently, YAP1-2 gamma contains 2 WW domains, which may interact with its ligands cooperatively.Citation48 Of note, Y188 is localized in the first WW domain of YAP and this WW domain has been reported to be important for the interactions with other binding partners.Citation46,49,50 We found that the phospho-YAP-Y188 mimic, YAP-Y188E, disrupted the interactions with its negative regulators Lats1 and PTPN14 (), providing a possible mechanism for the regulation of YAP function via Y188 phosphorylation. TAZ, the paralog of YAP, contains one WW domain and its tyrosine 141 equivalent to YAP-Y188. We predict that TAZ-Y141 phosphorylation may play a similar role like YAP-Y188. It will be of interest to investigate whether the phosphorylation of YAP-Y188 can actually serve as a positive indicator of the YAP activity. For future studies, we plan to identify the potential tyrosine kinase(s) that is responsible for the YAP-Y188 phosphorylation. Elucidation of the underlying mechanism and key players in YAP/TAZ regulation will contribute significantly to our understanding of the Hippo pathway in cancer biology and conceivably advance our therapeutic strategies.Citation51
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary files
Download Zip (1.9 MB)Funding
This work was supported by the Roswell Park Cancer Institute and National Cancer Institute (NCI) grant #P30 CA016056, the American Cancer Society Research Scholar Grant RSG-14-214-01-TBE and in part by the National Cancer Institute (NCI) R21 CA179693, (to J.Z).
References
- Irvine KD, Harvey KF. Control of organ growth by patterning and hippo signaling in Drosophila. Cold Spring Harb Perspect Biol 2015; 7(6):pii: a019224; PMID:26032720; http://dx.doi.org/10.1101/cshperspect.a019224
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell 2010; 19(4):491-505; PMID:20951342; http://dx.doi.org/10.1016/j.devcel.2010.09.011
- Barry ER, Camargo FD. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr Opin Cell Biol 2013; 25(2):247-53; PMID:23312716; http://dx.doi.org/10.1016/j.ceb.2012.12.006
- Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol 2011; 13(8):877-83; PMID:21808241; http://dx.doi.org/10.1038/ncb2303
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 2007; 130(6):1120-33; PMID:17889654; http://dx.doi.org/10.1016/j.cell.2007.07.019
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005; 122(3):421-34; PMID:16096061; http://dx.doi.org/10.1016/j.cell.2005.06.007
- Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015; 163(4):811-28; PMID:26544935; http://dx.doi.org/10.1016/j.cell.2015.10.044
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer 2013; 13(4):246-57; PMID:23467301; http://dx.doi.org/10.1038/nrc3458
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A 2006; 103(33):12405-10; PMID:16894141; http://dx.doi.org/10.1073/pnas.0605579103
- Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, Haber DA. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol 2009; 11(12):1444-50; PMID:19935651; http://dx.doi.org/10.1038/ncb1993
- Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module–the WW domain. FEBS Lett 1995; 369(1):67-71; PMID:7641887; http://dx.doi.org/10.1016/0014-5793(95)00550-S
- Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene 1994; 9(8):2145-52; PMID:8035999
- Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J 2004; 23(4):790-9; PMID:14765127; http://dx.doi.org/10.1038/sj.emboj.7600073
- Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell 2008; 29(3):350-61; PMID:18280240; http://dx.doi.org/10.1016/j.molcel.2007.12.022
- Tamm C, Bower N, Anneren C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J Cell Sci 2011; 124(Pt 7):1136-44; PMID:21385842; http://dx.doi.org/10.1242/jcs.075796
- Vlahov N, Scrace S, Soto MS, Grawenda AM, Bradley L, Pankova D, Papaspyropoulos A, Yee KS, Buffa F, Goding CR, et al. Alternate RASSF1 Transcripts Control SRC Activity, E-Cadherin Contacts, and YAP-Mediated Invasion. Curr Biol 2015; 25(23):3019-34; PMID:26549256; http://dx.doi.org/10.1016/j.cub.2015.09.072
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, et al. beta-Catenin-Driven Cancers Require a YAP1 Transcriptional Complex for Survival and Tumorigenesis. Cell 2012; 151(7):1457-73; PMID: 23245941; http://dx.doi.org/10.1016/j.cell.2012.11.026
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003; 30(3):256-68; PMID:12798140; http://dx.doi.org/10.1016/S1046-2023(03)00032-X
- Li YW, Shen H, Frangou C, Yang N, Guo J, Xu B, Bshara W, Shepherd L, Zhu Q, Wang J, et al. Characterization of TAZ domains important for the induction of breast cancer stem cell properties and tumorigenesis. Cell cycle 2015; 14(1):146-56; PMID:25602524; http://dx.doi.org/10.4161/15384101.2014.967106
- Frangou C, Li YW, Shen H, Yang N, Wilson KE, Blijlevens M, Guo J, Nowak NJ, Zhang J. Molecular profiling and computational network analysis of TAZ-mediated mammary tumorigenesis identifies actionable therapeutic targets. Oncotarget 2014; 5(23):12166-76; PMID:25361000; http://dx.doi.org/10.18632/oncotarget.2570
- Zhang J, Smolen GA, Haber DA. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res 2008; 68(8):2789-94; PMID:18413746; http://dx.doi.org/10.1158/0008-5472.CAN-07-6205
- Tu C, Li J, Young R, Page BJ, Engler F, Halfon MS, Canty JM Jr, Qu J. Combinatorial peptide ligand library treatment followed by a dual-enzyme, dual-activation approach on a nanoflow liquid chromatography/orbitrap/electron transfer dissociation system for comprehensive analysis of swine plasma proteome. Anal Chem 2011; 83(12):4802-13; PMID:21491903; http://dx.doi.org/10.1021/ac200376m
- Cao J, Gonzalez-Covarrubias V, Straubinger RM, Wang H, Duan X, Yu H, Qu J, Blanco JG. A rapid, reproducible, on-the-fly orthogonal array optimization method for targeted protein quantification by LC/MS and its application for accurate and sensitive quantification of carbonyl reductases in human liver. Anal Chem 2010; 82(7):2680-9; PMID:20218584; http://dx.doi.org/10.1021/ac902314m
- Nouri-Nigjeh E, Sukumaran S, Tu C, Li J, Shen X, Duan X, DuBois DC, Almon RR, Jusko WJ, Qu J. Highly multiplexed and reproducible ion-current-based strategy for large-scale quantitative proteomics and the application to protein expression dynamics induced by methylprednisolone in 60 rats. Anal Chem 2014; 86(16):8149-57; PMID:25072516; http://dx.doi.org/10.1021/ac501380s
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem 2008; 283(9):5496-509; PMID:18158288; http://dx.doi.org/10.1074/jbc.M709037200
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Gen Dev 2007; 21(21):2747-61; PMID:17974916; http://dx.doi.org/10.1101/gad.1602907
- Tomlinson V, Gudmundsdottir K, Luong P, Leung KY, Knebel A, Basu S. JNK phosphorylates Yes-associated protein (YAP) to regulate apoptosis. Cell Death Dis 2010; 1:e29; PMID:21364637; http://dx.doi.org/10.1038/cddis.2010.7
- Yang S, Zhang L, Liu M, Chong R, Ding SJ, Chen Y, Dong J. CDK1 phosphorylation of YAP promotes mitotic defects and cell motility and is essential for neoplastic transformation. Cancer Res 2013; 73(22):6722-33; PMID:24101154; http://dx.doi.org/10.1158/0008-5472.CAN-13-2049
- Zhao Y, Khanal P, Savage P, She YM, Cyr TD, Yang X. YAP-induced resistance of cancer cells to antitubulin drugs is modulated by a Hippo-independent pathway. Cancer Res 2014; 74(16):4493-503; PMID:24812269; http://dx.doi.org/10.1158/0008-5472.CAN-13-2712
- Fan R, Kim NG, Gumbiner BM. Regulation of Hippo pathway by mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. Proc Natl Acad Sci U S A 2013; 110(7):2569-74; PMID:23359693; http://dx.doi.org/10.1073/pnas.1216462110
- Reddy BV, Irvine KD. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev Cell 2013; 24(5):459-71; PMID:23484853; http://dx.doi.org/10.1016/j.devcel.2013.01.020
- Gaffney CJ, Oka T, Mazack V, Hilman D, Gat U, Muramatsu T, Inazawa J, Golden A, Carey DJ, Farooq A, et al. Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene 2012; 509(2):215-22; PMID:22939869; http://dx.doi.org/10.1016/j.gene.2012.08.025
- Sudol M. YAP1 oncogene and its eight isoforms. Oncogene 2013; 32(33):3922; PMID:23160371; http://dx.doi.org/10.1038/onc.2012.520
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer 2005; 5(9):675-88; PMID:16148884; http://dx.doi.org/10.1038/nrc1695
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Gen Dev 2008; 22(14):1962-71; PMID:18579750; http://dx.doi.org/10.1101/gad.1664408
- Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem 2011; 286(9):7018-26; PMID:21224387; http://dx.doi.org/10.1074/jbc.C110.212621
- Liu X, Yang N, Figel SA, Wilson KE, Morrison CD, Gelman IH, Zhang J. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene 2013; 32(10):1266-73; PMID:22525271; http://dx.doi.org/10.1038/onc.2012.147
- Wang W, Huang J, Wang X, Yuan J, Li X, Feng L, Park JI, Chen J. PTPN14 is required for the density-dependent control of YAP1. Gen Dev 2012; 26(17):1959-71; PMID:22948661; http://dx.doi.org/10.1101/gad.192955.112
- Michaloglou C, Lehmann W, Martin T, Delaunay C, Hueber A, Barys L, Niu H, Billy E, Wartmann M, Ito M, et al. The tyrosine phosphatase PTPN14 is a negative regulator of YAP activity. PloS one 2013; 8(4):e61916; PMID:23613971; http://dx.doi.org/10.1371/journal.pone.0061916
- Huang JM, Nagatomo I, Suzuki E, Mizuno T, Kumagai T, Berezov A, Zhang H, Karlan B, Greene MI, Wang Q. YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene 2013; 32(17):2220-9; PMID:22689061; http://dx.doi.org/10.1038/onc.2012.231
- Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem 2003; 278(35):33334-41; PMID:12807903; http://dx.doi.org/10.1074/jbc.M305597200
- Haskins JW, Nguyen DX, Stern DF. Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Sci Signal 2014; 7(355):ra116; PMID:25492965; http://dx.doi.org/10.1126/scisignal.2005770
- Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics 2008; 7(9):1598-608; PMID:18463090; http://dx.doi.org/10.1074/mcp.M700574-MCP200
- Kodaka M, Hata Y. The mammalian Hippo pathway: regulation and function of YAP1 and TAZ. Cell Mol Life Sci 2015; 72(2):285-306; PMID:25266986; http://dx.doi.org/10.1007/s00018-014-1742-9
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Gen Dev 2010; 24(1):72-85; PMID:20048001; http://dx.doi.org/10.1101/gad.1843810
- Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci 2010; 35(11):627-33; PMID:20598891; http://dx.doi.org/10.1016/j.tibs.2010.05.010
- Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A 1995; 92(17):7819-23; PMID:7644498; http://dx.doi.org/10.1073/pnas.92.17.7819
- Schuchardt BJ, Mikles DC, Hoang LM, Bhat V, McDonald CB, Sudol M, Farooq A. Ligand binding to WW tandem domains of YAP2 transcriptional regulator is under negative cooperativity. FEBS J 2014; 281(24):5532-51; PMID:25283809; http://dx.doi.org/10.1111/febs.13095
- Sudol M. Newcomers to the WW Domain-Mediated Network of the Hippo Tumor Suppressor Pathway. Gen Cancer 2010; 1(11):1115-8; PMID:21779434; http://dx.doi.org/10.1177/1947601911401911
- Sudol M, Hunter T. NeW wrinkles for an old domain. Cell 2000; 103(7):1001-4; PMID:11163176; http://dx.doi.org/10.1016/S0092-8674(00)00203-8
- Sudol M, Shields DC, Farooq A. Structures of YAP protein domains reveal promising targets for development of new cancer drugs. Semin Cell Dev Biol 2012; 23(7):827-33; PMID:22609812; http://dx.doi.org/10.1016/j.semcdb.2012.05.002
