ABSTRACT
Meiosis is an important process in sexual reproduction. Meiosis initiation has been found to be highly diverse among species. In yeast, it has been established that cyclin-dependent kinases (Cdks) and cyclins are essential components in the meiosis initiation pathway. In this study, we identified 4 Cdks in the model ciliate, Tetrahymena thermophila, and we found one of them, Cdk3, which is specifically expressed during early conjugation, to be essential for meiosis initiation. Cdk3 deletion led to arrest at the pair formation stage of conjugation. We then confirmed that Cdk3 acts upstream of double-strand break (DSB) formation. Moreover, we detected that Cdk3 is necessary for the expression of many genes involved in early meiotic events. Through proteomic quantification of phosphorylation, co-expression analysis and RNA-Seq analyses, we identified a conjugation-specific cyclin, Cyc2, which most likely partners with Cdk3 to initiate meiosis.
Introduction
Meiosis is an essential process in all sexually reproducing unicellular and multicellular eukaryotes, including animals, plants, yeasts, and protists. It involves a series of cellular events, including DNA double-strand break (DSB) formation, homologous chromosome recombination and chromosome segregation. Although all of these events are conserved among species, diverse mechanisms control meiosis initiation.Citation1,2 In Saccharomyces cerevisiae, starvation induces expression of IME1, which encodes a transcription factor that transactivates early meiotic genes.Citation3 One of the target genes, IME2, encodes the meiosis induction protein kinase (Ime2), which promotes meiotic DNA replication.Citation4,5 Sci1, an inhibitor of Cdc28 (a cyclin-dependent kinase, [Cdk]), is phosphorylated by Ime2 leading to its degradation. In conjunction with the cyclins Clb5 and Clb6, Cdk28 then triggers the initiation of pre-meiotic S phase.Citation6,7 In Schizosaccharomyces pombe, the meiosis initiation pathway consists of Ste11 (a transcription factor), Mei2 (an RNA-binding protein), Pat1 (a kinase), Mmi1 (another RNA-binding protein) and Cdc2 (a Cdk) bound to Cig2 (a cyclin). Here, responding to nutritional cues, Mei2 promotes exit from the mitotic cell cycle by antagonizing the elimination of meiotic mRNAs.Citation8-11 Thus, while the mechanisms of meiosis initiation are very different and the genes involved in these processes have little sequence homology, Cdks and cyclins play important roles in both S. cerevisiae and S. pombe.
Cdks, cyclins and their regulatory factors are key cell cycle regulators. Currently, more than 20 members of the Cdk family have been identified in humans.Citation12 These are characterized by a conserved structure comprising an ATP-binding domain, a PSTAIRE-like cyclin-binding domain and an activating T-loop domain.Citation13 Cdks are involved in 2 main processes: (1) integrating cellular signals to modulate gene transcription and (2) cell division.Citation12-16 In S. cerevisiae and S. pombe, all cell cycle events are controlled by a single essential Cdk (Cdc28 and Cdc2 respectively), assisted by Pho85.Citation17 In mammals, there are 3 subfamilies of cell cycle Cdks: Cdk1 (Cdk1–Cdk3), Cdk4 (Cdk4 and Cdk6) and Cdk5 (Cdk5 and Cdk14–Cdk18).Citation15 In yeast, Cdc28/Cdc2 is essential for meiosis initiation; further, a lack of Cdk2 or Cdk4 causes sterility in mice.Citation13,18-21 Therefore, Cdk involvement in meiosis may be conserved among species.
However, the molecular mechanism responsible for meiosis initiation in ciliates, an important branch of the eukaryotes, is unknown. The unicellular ciliate Tetrahymena thermophila is an excellent model eukaryote for studying meiosis. All ciliates, including T. thermophila possess 2 nuclei: a diploid germline micronucleus (MIC) and a polyploid somatic macronucleus (MAC).Citation22 When mixed together, cells of different mating types can conjugate under starvation conditions, which involves initiating synchronous meiosis of their MICs.Citation23 Meiosis has been well studied in T. thermophila in recent decades, thus providing a good model system for exploring the mechanism of ciliate meiosis initiation.Citation22-27
To determine whether Cdks are involved in meiosis initiation in ciliates, we screened T. thermophila for proteins containing the PSTAIRE-like cyclin-binding domain and identified Cdk3 (also named Tcdk3) as a candidate. Here we determine its role in meiosis and identify its likely cyclin partner.
Results and discussion
Cdk3 is a candidate meiotic regulator in T. thermophila
Cdks are a family of protein kinases that require cyclin binding for activation.Citation17 Therefore, to identify T. thermophila Cdks, we screened for genes that encode proteins that containing the PSTAIRE-like cyclin-binding domain. In total, 4 genes were identified: Cdk3, Cdc2, Cdk1 and Cdc28 (Table S1). Multiple sequence alignment showed that all 4 proteins have a conserved ATP-binding domain, and a T-loop (), indicating that they are cyclin-dependent kinases.
Figure 1. Cdk3 is a T. thermophila conjugation-specific Cdk with maximal expression at the start of conjugation. (A) Multiple sequences alignment of proteins containing a cyclin-binding domain (PSTAIRE) in T. thermophila and the Cdk1 subfamily of Homo sapiens and S. cerevisiae. (B) Phylogenetic analysis of Cdks in T. thermophila, H. sapiens and S. cerevisiae. Scale bar represents 10 nucleotide substitutions per 100 nucleotides. (C) Expression of CDKs during the T. thermophila life cycle. Red box, CDK3 expression during exponential growth (E) and starvation (S) conditions, and at 0–18 h post mixing. Relative expression values were based on microarray data retrieved from TetraFGD. (D) CDK3 expression profile based on microarray and RNA-Seq data.
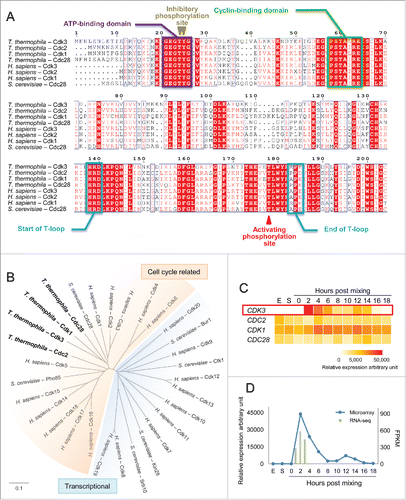
To determine whether these 4 Cdks could be cell cycle Cdks, we constructed a phylogenetic tree together with human Cdk1–Cdk20 and 6 well characterized S. cerevisiae Cdks ().Citation15 Cdks are classified into 2 groups: cell cycle (; shown in red) and transcriptional Cdks (; shown in blue).Citation15 In humans, only 4 Cdks are directly involved in cell cycle control: Cdk1, Cdk2, Cdk4 and Cdk6.Citation17 Our phylogenetic analysis showed that all 4 T. thermophila Cdks we found are cell cycle related: 3 (Cdk3, Cdk1 and Cdc28) are members of the Cdk1 subfamily, and Cdc2 is in the Cdk5 subfamily ().
To further investigate the function of all 4 Cdks, their gene expression profiles were determined by microarray analysis under different conditions ().Citation28,29 CDC2, CDK1 and CDC28 were expressed under growth, starvation and conjugation conditions; in contrast, CDK3 was specifically expressed during conjugation, with maximal expression at 2 h after we had triggered the meiotic program by mixing cells. RNA-Seq data on CDK3 expression was consistent with the microarray data, confirming that maximal expression occurs at 2 h post mixing the cells (). Since meiosis initiation occurs about 2 h after the trigger, Cdk3 may play an important role in meiosis initiation. And if meiosis initiation is promoted by a Cdk in T. thermophila, Cdk3 is the most likely candidate.
Meiosis is arrested at the pair formation stage in cdk3Δ
In T. thermophila, meiosis can be initiated by mixing starved cells of different mating types. After meiosis initiation, programmed DSBs induced by Spo11 induce MIC elongation during meiotic prophase.Citation24,30 Homologous chromosome pairing occurs during maximal MIC elongation and DSBs are repaired by homologous recombination. The MIC then shortens; homologous chromosomes are segregated at anaphase I and sister-chromatids at anaphase II.Citation25,31 During the first 3 hours post mixing, DSB formation occurs, as a response to which the MIC elongation and DSB repair are initiated.
To investigate whether Cdk3 is involved in meiosis, Cdk3 knockout strains (cdk3Δ, 2 mating types) were constructed. RNA-Seq analysis showed that CDK3 expression in these strains was totally abolished during meiosis (Fig. S1). MIC development was monitored in wild-type (WT) and cdk3Δ strains. In WT cells, MICs started to elongate at meiosis initiation; in contrast, MIC elongation did not occur in cdk3Δ cells (). A time course of meiosis showed that most WT pairs had completed meiosis by 8 h post mixing; whereas cdk3Δ paired cells were arrested at the pair formation stage ().
Figure 2. Meiosis is arrested at the pair formation stage in cdk3Δ cells. (A) DAPI staining shows progression through early conjugation in WT and cdk3Δ strains. (B) Time course of conjugation stages in WT and cdk3Δ strains. (C) r values for genes expressions of whole transcriptome of each samples. Higher r value shows samples are more similar and lower r value shows samples are more different. (D) Venn diagram shows overlapping DEGs at 1, 2, and 3 h post mixing. Numbers, gene number of each subset. (E) Numbers of up- and down-regulated genes at 1, 2, and 3 h post mixing.
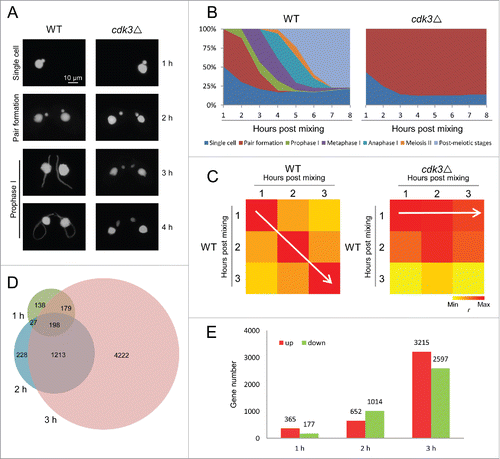
We also compared gene expression in WT and cdk3Δ cells at 1–3 h post mixing. Pearson correlation coefficient (r) values for gene expression had a diagonal distribution in WT cells (; white arrow in left panel); in contrast, r values in cdk3Δ cells at all 3 time points showed the greatest correlation with those seen at 1 h post mixing in WT cells (; white arrow in right panel). These data suggest that meiosis progressing in WT but arrested at the pair formation stage in cdk3Δ cells.
Next, differentially expressed genes (DEGs) were identified at each time point using a twofold cutoff between WT and cdk3Δ. Overlapping analysis of DEGs at the 1, 2, and 3 h time points showed that most genes with altered expression at early time points were also affected at later time points (). This means that the DEGs expression is not delayed but prevented altogether. (If a gene expression is delayed, this gene with altered expression at early time points but may not affected at later time points.) An analysis of DEGs in cdk3Δ showed an increase in the number of up- and down-regulated genes from 1 to 2 h and from 2 to 3 h post mixing (). This suggests that a subset of genes is differently regulated early and that more genes are differentially regulated later due to the early arrest.
Cdk3 acts upstream of DSB formation in T. thermophila
Since both cytological and gene expression analyses showed arrested meiosis at the pair formation stage in cdk3Δ cells, we next confirmed whether Cdk3 acts upstream of meiosis. There are 3 important events in the prophase stage of T. thermophila meiosis: DSB formation, MIC elongation and DSB repair. Neither MIC elongation nor DSB repair occur in spo11Δ cells which cannot form DSBs, indicating that MIC elongation and DSB repair are initiated by DSB formation.Citation24 Since DAPI staining showed that MIC elongation does not occur in cdk3Δ cells (see above) (), Cdk3 must act upstream of this process.
Therefore, we tested whether DSBs are formed in cdk3Δ cells. To this end, we analyzed the mobility of meiotic DNA species using pulsed-field gel electrophoresis. Chromosomes of generative nuclei are too big to enter a pulsed-field gel, and only fragmented meiotic DNA can do so.Citation30 In the WT control, a DNA band of DSB-generated fragment was present 3 h post mixing, and nearly absent after DSB repair, i.e. 5 h post mixing (). A similar band was seen in the mre11Δ control (which forms DSBs but cannot repair them), at 3h and 5 h post mixing (). In contrast, only a background signal (of similar strength to the bands at 0 h post mixing in the controls) was seen in the cdk3Δ samples, indicating that DSBs were not formed (). This result was consistent with down-regulation of SPO11 in cdk3Δ cells (). Additionally, in WT cells, γ-H2A.X signals, in response to DSB formation, were observed in MIC, while there were no signals in cdk3Δ cells (). Therefore, it is likely that Cdk3 acts upstream of DSB formation and is necessary for MIC elongation and DSBs repair.
Figure 3. DSB formation and repair are inhibited in Cdk3 deficient cells. (A) Detection of DSBs. Pulse field gel electrophoresis was used to separate the genomic DNA and a MIC specific southern probe was used to detect DSBs. (B) Downregulation of genes related to DSB formation and repair in cdk3Δ cells. (C) γ-H2A.X staining of WT and cdk3Δ cells. (D) GO enrichment of downregulated DEGs at 3 h post mixing. Color bar, corrected probability value (corr-p) of significance of enrichment.Citation49
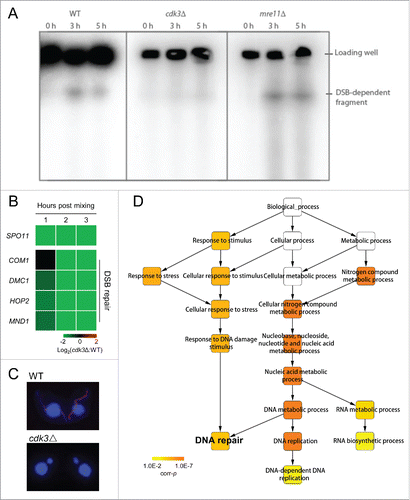
Next, we studied the expression of DSB repair genes in cdk3Δ cells. Gene Ontology (GO) enrichment analysis of downregulated DEGs at 2 h and 3 h post mixing showed significant enrichment of genes involved in DNA repair (Table S2; , 3 h post mixing, corrected probability value [corr-p] = 4.7×10−5), suggesting that this process is not triggered or may be impaired when Cdk3 is absent. To confirm that DSB repair is abnormal in cdk3Δ cells, a series of genes were selected for further investigation (Table S1). After DSB formation in T. thermophila, Com1 interacts with DSBs to produce 3′ single-stranded DNA molecules.Citation30 Dmc1 then binds to the 3′ ends of these DNA molecules and the Hop2–Mnd1 complex stabilizes Dmc1-ssDNA binding to promote homology search DNA strand invasion, an important stage in DSB repair.Citation24,32 Analysis of fold changes in gene expression between cdk3Δ and WT showed that genes related to DSB repair were down-regulated in cdk3Δ cells (). These results support our finding that DSB repair is not triggered or is abnormal in cdk3Δ cells.
Cdk3 may initiate meiosis together with Cyc2
To understand how Cdk3 acts on meiotic events, quantitative phosphorylome analysis of WT and cdk3Δ cells was undertaken at the pair formation stage of conjugation using tandem mass tag labeling and affinity enrichment followed by high-resolution liquid chromatography tandem mass spectrometry analysis. Altogether, 5272 phosphorylation sites in 2710 protein groups were identified, of which, 2109 sites in 1340 proteins were quantified. The ratios of unlabeled WT and cdk3Δ to 15N-labeled protein internal controls were determined first and the base 2 logarithms of the cdk3Δ:WT ratios (log2[cdk3Δ:WT]) were then calculated as described in the Material and Methods (also see Fig. S2). Of the total quantified sites, 87 sites in 77 proteins possess an S/T*-P-x-K/R motif (where x represents any amino acid and the asterisk indicates the phosphorylation site), which has been proven to be the full Cdk consensus site in several organisms.Citation17,33 Therefore, we focused on these sites and proteins for further analysis.
To determine whether the Cdk consensus site is conserved in T. thermophila, we assessed the distribution of the absolute value of log2(cdk3Δ:WT). If the Cdk3 consensus site resembles S/T*-P-x-K/R, the mean abundance of phosphopeptides containing this consensus site should be decreased in cdk3Δ cells due to the decrease of the phospho-Cdk3 targets. Consistent with this, ANOVA revealed that phosphopeptides containing the full consensus site were significantly less abundant in the absence of Cdk3 (). Therefore, we defined 3 criteria for a Cdk3 substrate. First, its phosphorylation site must be identical to the full Cdk consensus sequence. Second, the amount of phospho-peptide must be significantly reduced in cdk3Δ cells. Third, its expression pattern should resemble that of Cdk3.
Figure 4. Cdk3 may initiate meiosis together with Cyc2. (A) Distribution of abundances changes of phosphorylated peptides between cdk3Δ and WT cells (log2[Lcdk3Δ/LWT], details see Fig. S2). (B) Relative abundance (log2[LSample/HWT], details see Fig. S2) of phosphorylated Cyc2 in WT and cdk3Δ cells. (C) The phosphorylation site in Cyc2. (D) CDK3 and CYC2 are co-expressed in WT cells. (E) Expression profiles of core meiotic genes in cdk3Δ, cyc2Δ and spo11Δ cells. Values for cyc2Δ expression were retrieved from NCBI (GSE79286) and those for spo11Δ expression from TetraFGD. (F) Relationship among Cdk3, Cyc2, and proteins that function in early meiosis. Green arrow, promote genes function at their expression level; orange arrow, promote an event by a phosphorylation pathway; gray arrow, perform the function in the meiosis events.
![Figure 4. Cdk3 may initiate meiosis together with Cyc2. (A) Distribution of abundances changes of phosphorylated peptides between cdk3Δ and WT cells (log2[Lcdk3Δ/LWT], details see Fig. S2). (B) Relative abundance (log2[LSample/HWT], details see Fig. S2) of phosphorylated Cyc2 in WT and cdk3Δ cells. (C) The phosphorylation site in Cyc2. (D) CDK3 and CYC2 are co-expressed in WT cells. (E) Expression profiles of core meiotic genes in cdk3Δ, cyc2Δ and spo11Δ cells. Values for cyc2Δ expression were retrieved from NCBI (GSE79286) and those for spo11Δ expression from TetraFGD. (F) Relationship among Cdk3, Cyc2, and proteins that function in early meiosis. Green arrow, promote genes function at their expression level; orange arrow, promote an event by a phosphorylation pathway; gray arrow, perform the function in the meiosis events.](/cms/asset/617e4455-5d8f-421a-b990-6f13d1ceac1d/kccy_a_1207838_f0004_oc.gif)
This triple filtering approach identified a phosphorylation site within the conjugation-specific cyclin, Cyc2, as the most likely Cdk3 target. In the absence of Cdk3, the abundance of the specific phosphopeptide was strongly decreased (; log2[cdk3Δ:WT] = −5.2, corr-p = 4.0×10−37). Moreover, the sequence of the phosphorylation site is SPQK, which matches the full Cdk consensus (). Additionally, Cyc2 is co-expressed with Cdk3 (; r = 0.944, p = 4×10−6).
A recent study shed light on the function of T. thermophila Cyc2.Citation34 Similar to cdk3Δ cells, CYC2 knockout results in arrest of the meiotic conjugation process before the start of meiotic divisions. MIC elongation (which is dependent on DSBs) does not occur in cyc2Δ cells. Also γ-H2A-X, which is a marker of DNA damage, does not localize to MICs. These results strongly suggest that Cdk3 initiates meiosis by acting together with Cyc2. Further, the Cyc2 phosphorylation site might be necessary for downstream signaling or may be a target for phosphorylation-dependent ubiquitination which is important for meiotic progression, in view of previous studies in human.Citation34-36
We next compared expression of meiosis-related genes (Table S1) in cdk3Δ, cyc2Δ and spo11Δ cells. The effects of Cdk3 deficiency and Cyc2 deficiency appeared to be similar, while those of Spo11 deficiency were different (). In both cdk3Δ and cyc2Δ cells, most meiosis-related genes were downregulated; however, in spo11Δ, most of these genes did not show differential expression (only a few genes were slightly up-regulated) (). Heat map analysis of DEGs suggested that Cdk3 and Cyc2 might belong to the same meiosis initiation pathway.
The heat map also revealed that some DEGs, down-regulated in cdk3Δ and cyc2Δ cells, are involved in DSB formation and processing, which are among the earliest events in meiosis (). Especially, SPO11, some DSB repair genes (EXO1, COM1, DMC1, HOP2, and MND1) and some genes related to chromosomal crossover (MSH4, MSH5 and SGS1) were downregulated in both cdk3Δ and cyc2Δ cells (), suggesting that both proteins regulate the expression of several important meiotic genes (). Studies of Cdks and cyclins in other organisms suggest that a transcription factor might link Cdk3 and Cyc2 to these meiotic genes.Citation13 On the other hand, regulation of some other genes that are not under strict meiotic control like MRE11, RAD50 and RAD51 (involved in DNA repair) as well as MUS81 and EME1 (also named MMS4) (involved in chromosomal crossover) were not regulated by Cdk3 and Cyc2 (). In addition, the expression of most genes involved in chromosome condensation (SMC2 and SMC4) and cohesion (SMC1, SMC3 and REC8), which are important events in late prophase, were not regulated by Cdk3 and Cyc2. This suggests that Cdk3 and Cyc2 regulate gene expression at early prophase during conjugation but (an)other signaling pathway(s) may also be actively regulating prophase mechanisms by controlling another set of genes involved in these pathways. Notably, while Spo11 triggers meiotic MIC elongation and homologous pairing via a kinase signaling pathway involving Atr1 (), it has no influence on the progression of later meiotic events, hence, in spo11Δ meiosis, meiotic divisions take place.Citation24 This DSB-independence of downstream events was confirmed by our observation of practically unchanged expression of meiosis-related genes in spo11Δ cells ().
Our results suggest that Cdk3 and Cyc2 together have essential function in meiosis initiation in T. thermophila. They promote the expression of genes important for early prophase. In the absence of either of these proteins, conjugation was arrested at the pair formation stage and early meiotic events (DSB formation, MIC elongation, and DSB repair) did not occur. These results are consistent with previous studies in yeast,Citation1,17 suggesting that the mechanism controlling Cdk and cyclin function in meiosis initiation may be conserved among species.
Materials and methods
Cell culture and conjugation induction
T. thermophila WT strains CU427 (mating type VI) and CU428 (mating type VII) (Tetrahymena Stock Center [http://tetrahymena.vet.cornell.edu/]) were grown in Super Proteose Peptone (SPP) medium (1 % Proteose Peptone, 0.2 % glucose, 0.1 % yeast extract, 0.003 % Sequestrene).Citation26 Cells of 2 different mating types (at ∼2 ×105 cells/ml) were starved in 10 mM Tris-Cl (pH 7.4) for 12–16 h, and then mixed in equal proportions for conjugation induction.
Gene identification, phylogenetic analysis, and gene expression analysis
Since Cdks bind cyclin via the cyclin-binding PSTAIRE-like motif, we first screened for proteins containing this motif based on the gene predictions of T. thermophila (version 2014: http://ciliate.org/index.php/home/downloads) using FIMO.Citation37 Domain composition was then examined using InterproScan.Citation38
For phylogenetic analysis, multiple sequences alignment were first aligned with clustalW.Citation39 Then, the alignment was used to construct phylogenetic tree by MEGA6 with the NJ method and 1500 bootstrap replicates.Citation40 Multiple sequence alignment result was visualized by ESPript 3.Citation41
CDK3 knockout
To construct the CDK3 knockout strains (cdk3Δ), one DNA fragment upstream of the CDK3 open reading frame and 2 downstream fragments were amplified using the following primers: CDK3-up1f-NotI, CDK3-up1r-HA and CDK3-down1f-HA, CDK3-down1r-N4 and CDK3-down2f-N4, CDK3-down2r-NotI, respectively (Table S3). Using fusion PCR, the 2 downstream fragments were joined to the NEO4 cassette which contains a neomycin resistance gene driven by a Cd2+-inducible MTT1 metallothionein promoter. To construct the knockout plasmid, the upstream fragment and fusion fragment were then cloned into the pBlueScript SK (+) backbone. To obtain CDK3 knockout strain of 2 mating types, the knockout construct was obtained by NotI digestion and shot into starved WT CU427 and CU428 cells by biolistic transformation, respectively. Transformants were cultured in SPP containing decreasing CdCl2 (from 1 μg/ml to 0.05 μg/ml) and increasing paromomycin concentrations (from 120 μg/ml to 40 mg/ml) until all WT chromosomes in the MAC were completely replaced by knockout chromosomes.Citation42
Detection of DSBs by pulsed-field gel electrophoresis
Detection of DSB-generated fragments followed a previously described method.Citation30 In short, intact DNA was extracted in agarose plugs. DNA of intact MIC chromosomes cannot enter the gel, while fragmented DNA migrates as a single band. For detecting DSB-generated signal, MIC-borne DNA fragments were then highlighted using Southern detection of a MIC-specific DNA (Tlr1). The Tlr sequence was excised from the pMBR2 vector (NCBI accession number AF451863),Citation43 gel-isolated and radioactively labeled by random priming with 32P-dATP, and hybridized to MIC DNA on the membrane.Citation30
RNA-seq analysis
At 1, 2, and 3 h post mixing, total RNA was extracted from WT and cdk3Δ pairs using the RNeasy Protect Cell Mini Kit (Qiagen), as described (TetraFGD: http://tfgd.ihb.ac.cn/index/smphelp).Citation44 mRNA with Poly-A tail were then enriched by the Sera-Mag magnetic oligo (dT) beads (GE). Illumina sequencing libraries were then constructed based on manufacturer's recommendations. Paired-end (150 bp×2) sequencing has been made for all the samples with an Illumina Hiseq4000 sequencer. All sequence data have been submitted to GenBank databases under accession number GSE80977. Adaptors of raw reads were trimmed using Trim-Galore (version 0.4.0).Citation45 Trimmed-reads were mapped to the T. thermophila MAC genome (version 2014: http://ciliate.org/index.php/home/downloads) using TopHat (version 2.0.9).Citation46 Genes expression values were quantify to the number of fragments per kilobase of exon per million fragments mapped (FPKM) using Cuffdiff (version 2.1.1).Citation47
r values between each samples was calculated using R software on the basis of genes expression values (FPKM).Citation48 DEGs were screened using a cutoff with 2 fold changes between cdk3Δ and WT cells for each same time point, and for control the false positives, very low expressed genes (FPKM < 10) were excluded. And then, GO enrichment analyses were performed using BiNGO (version 3.0.2).Citation49,50 Hypergeometric test was used for statistical test, the familywise error rate (FWER) was used for multiple testing corrections to control the false positives, and the criteria of significant enrichment were defined as corr-p < 0.01.
Proteomic quantification of phosphorylation
Proteomic quantification of phosphorylation was down as previously described.Citation51 Briefly, bacteria were grown in M9 minimal medium containing 15N-labeled ammonium sulfate as the sole nitrogen source. For the internal control, both CU427 and CU428 T. thermophila cells were added to a stationary phase bacteria population to convert all proteins from the light to heavy forms. Meanwhile, both mating types of WT and cdk3Δ strains were cultured in standard SPP medium (not 15N-enriched). For all labeled WT, unlabeled WT and unlabeled cdk3Δ cells, the 2 mating types were mixed together after starvation to induce conjugation. Proteins were extracted from all 3 samples at the pair formation stage and equal proportions of labeled and unlabeled proteins were mixed. To quantify protein phosphorylation, the ratios of unlabeled samples to spiked labeled protein internal controls were first determined and the base 2 logarithm of final ratios between cdk3Δ and WT was calculated from the relative ratios of light vs heavy forms in cdk3Δ cells divided by those from WT cells (log2[Lcdk3Δ:HWT/LWT:HWT] = log2[Lcdk3Δ/LWT]; illustrated in Fig. S2). Differentially phosphorylated proteins were identified using DESeq2.Citation52
Abbreviations
| Cdks | = | cyclin-dependent kinases |
| cdk3Δ | = | CDK3 knockout strains |
| DEGs | = | differentially expressed genes |
| DSBs | = | double-strand breaks |
| FPKM | = | fragments per kilobase of exon per million fragments mapped |
| GO | = | gene ontology |
| MAC | = | macronucleus |
| MIC | = | micronucleus |
| WT | = | wild-type |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary files
Download Zip (608.7 KB)Acknowledgments
We thank Prof. Josef Loidl, Center for Molecular Biology, University of Vienna, for his critical comments.
Funding
This work was supported by grants from the International Science & Technology Cooperation Program of China (No. 2013DFG32390) and by Project CN10/2013 from the OEAD (Austria). Work by M.T. and A.S. was supported by grants P27313-B20 and W1238-B20 from the Austrian Science Fund (FWF) to J. Loidl.
References
- Pawlowski WP, Sheehan MJ, Ronceret A. In the beginning: the initiation of meiosis. Bio Essays 2007; 29:511-4
- Wang CJ, Tseng CC. Recent advances in understanding of meiosis initiation and the apomictic pathway in plants. Front Plant Sci 2014; 5:497; PMID:25295051
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science 1998; 282:699-705; PMID:9784122; http://dx.doi.org/10.1126/science.282.5389.699
- Clifford DM, Stark KE, Gardner KE, Hoffmann-Benning S, Brush GS. Mechanistic insight into the Cdc28-related protein kinase Ime2 through analysis of replication protein A phosphorylation. Cell Cycle 2014; 4:1826-33; http://dx.doi.org/10.4161/cc.4.12.2214
- Foiani M, Nadjar-Boger E, Capone R, Sagee S, Hashimshoni T, Kassir Y. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol Gen Genet 1996; 253:278-88; PMID:9003314; http://dx.doi.org/10.1007/s004380050323
- Dirick L, Goetsch L, Ammerer G, Byers B. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 1998; 281:1854-7; PMID:9743499; http://dx.doi.org/10.1126/science.281.5384.1854
- Stuart D, Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev 1998; 12:2698-710; PMID:9732268; http://dx.doi.org/10.1101/gad.12.17.2698
- Borgne A, Murakami H, Ayte J, Nurse P. The G1/S cyclin Cig2p during meiosis in fission yeast. Mol Biol Cell 2002; 13:2080-90; PMID:12058071
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 2006; 442:45-50; PMID:16823445; http://dx.doi.org/10.1038/nature04881
- Kitamura K, Katayama S, Dhut S, Sato M, Watanabe Y, Yamamoto M, Toda T. Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev Cell 2001; 1:389-99; PMID:11702950; http://dx.doi.org/10.1016/S1534-5807(01)00037-5
- Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev 1991; 5:1990-9; PMID:1657709; http://dx.doi.org/10.1101/gad.5.11.1990
- Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, Morgan DO, Tsai LH, Wolgemuth DJ. Cyclin-dependent kinases: a family portrait. Nat Cell Biol 2009; 11:1275-6; PMID:19884882; http://dx.doi.org/10.1038/ncb1109-1275
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 2013; 140:3079-93; PMID:23861057; http://dx.doi.org/10.1242/dev.091744
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009; 9:153-66; PMID:19238148; http://dx.doi.org/10.1038/nrc2602
- Malumbres M. Cyclin-dependent kinases. Genome Biol 2014; 15:122; PMID:25180339; http://dx.doi.org/10.1186/gb4184
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 1997; 13:261-91; PMID:9442875; http://dx.doi.org/10.1146/annurev.cellbio.13.1.261
- Morgan DO. The cell cycle: Principles of control. London: New Science Press Ltd, 2007
- Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 Knockout Mice Are Viable. Curr Biol 2003; 13:1775-85; PMID:14561402; http://dx.doi.org/10.1016/j.cub.2003.09.024
- Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet 2003; 35:25-31; PMID:12923533; http://dx.doi.org/10.1038/ng1232
- Rane SG, Dubus P, Mettus RV, Galbreath EJ, Boden G, Reddy EP, Barbacid M. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in β-islet cell hyperplasia. Nat Genet 1999; 22:44-52; PMID:10319860; http://dx.doi.org/10.1038/8751
- Tsutsui T, Hesabi B, Moons DS, Pandolfi PP, Hansel KS, Koff A, Kiyokawa H. Targeted disruption of CDK4 delays cell cycle entry with enhanced p27(Kip1) activity. Mol Cell Biol 1999; 19:7011-9; PMID:10490638; http://dx.doi.org/10.1128/MCB.19.10.7011
- Orias E, Cervantes MD, Hamilton EP. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res Microbiol 2011; 162:578-86; PMID:21624459; http://dx.doi.org/10.1016/j.resmic.2011.05.001
- Martindale DW, Allis CD, Bruns PJ. Conjugation in Tetrahymena thermophila: A temporal analysis of cytological stages. Exp Cell Res 1982; 140:227-36; PMID:7106201; http://dx.doi.org/10.1016/0014-4827(82)90172-0
- Mochizuki K, Novatchkova M, Loidl J. DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. J Cell Sci 2008; 121:2148-58; PMID:18522989; http://dx.doi.org/10.1242/jcs.031799
- Loidl J, Scherthan H. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J Cell Sci 2004; 117:5791-801; PMID:15522890; http://dx.doi.org/10.1242/jcs.01504
- Orias E, Hamilton EP, Orias JD. Tetrahymena as a laboratory organism: Useful strains, cell culture, and cell line maintenance. Methods Cell Biol 2000; 62:189-211; PMID:10503191; http://dx.doi.org/10.1016/S0091-679X(08)61530-7
- Yan GX, Dang H, Tian M, Zhang J, Shodhan A, Ning YZ, Xiong J, Miao W. Cyc17, a meiosis-specific cyclin, is essential for anaphase initiation and chromosome segregation in Tetrahymena thermophila. Cell Cycle 2016; 14:1-10.
- Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One 2009; 4:e4429; PMID:19204800; http://dx.doi.org/10.1371/journal.pone.0004429
- Xiong J, Lu X, Lu Y, Zeng H, Yuan D, Feng L, Chang Y, Bowen J, Gorovsky M, Fu C, et al. Tetrahymena Gene Expression Database (TGED): a resource of microarray data and co-expression analyses for Tetrahymena. Sci China Life Sci 2011; 54:65-7; PMID:21253873; http://dx.doi.org/10.1007/s11427-010-4114-1
- Lukaszewicz A, Howard-Till RA, Novatchkova M, Mochizuki K, Loidl J. MRE11 and COM1/SAE2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena. Chromosoma 2010; 119:505-18; PMID:20422424; http://dx.doi.org/10.1007/s00412-010-0274-9
- Shodhan A, Lukaszewicz A, Novatchkova M, Loidl J. Msh4 and Msh5 function in SC-independent chiasma formation during the streamlined meiosis of Tetrahymena. Genetics 2014; 198:983-93; PMID:25217051; http://dx.doi.org/10.1534/genetics.114.169698
- Howard-Till RA, Lukaszewicz A, Loidl J. The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS Genetics 2011; 7:193-202; http://dx.doi.org/10.1371/journal.pgen.1001359
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature 2003; 425:859-64; PMID:14574415; http://dx.doi.org/10.1038/nature02062
- Xu Q, Wang R, Ghanam AR, Yan G, Miao W, Song X. The key role of CYC2 during meiosis in Tetrahymena thermophila. Protein Cell 2016; 7(4):236-49
- Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev 1997; 11:957-72; PMID:9136925; http://dx.doi.org/10.1101/gad.11.8.957
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 2001; 294:173-7; PMID:11533444; http://dx.doi.org/10.1126/science.1065203
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics 2011; 27:1017-8; PMID:21330290; http://dx.doi.org/10.1093/bioinformatics/btr064
- Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 2015; 43:D213-21; PMID:25428371; http://dx.doi.org/10.1093/nar/gku1243
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947-8; PMID:17846036; http://dx.doi.org/10.1093/bioinformatics/btm404
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/10.1093/molbev/mst197
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 2014; 42:W320-4; PMID:24753421; http://dx.doi.org/10.1093/nar/gku316
- Mochizuki K. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene 2008; 425:79-83; PMID:18775482; http://dx.doi.org/10.1016/j.gene.2008.08.007
- Wuitschick JD, Gershan JA, Lochowicz AJ, Li S, Karrer KM. A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic Acids Res 2002; 30:2524-37; PMID:12034842; http://dx.doi.org/10.1093/nar/30.11.2524
- Xiong J, Lu X, Zhou Z, Chang Y, Yuan D, Tian M, Zhou Z, Wang L, Fu C, Orias E, et al. Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using Deep RNA Sequencing. PLoS One 2012; 7:e30630; PMID:22347391; http://dx.doi.org/10.1371/journal.pone.0030630
- Wu Z, Wang X, Zhang X. Using non-uniform read distribution models to improve isoform expression inference in RNA-Seq. Bioinformatics 2011; 27:502-8; PMID:21169371; http://dx.doi.org/10.1093/bioinformatics/btq696
- Kim D, Salzberg SL. TopHat-Fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol 2011; 12:R72; PMID:21835007; http://dx.doi.org/10.1186/gb-2011-12-8-r72
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7:562-78; PMID:22383036; http://dx.doi.org/10.1038/nprot.2012.016
- Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2015
- Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005; 21:3448-9; PMID:15972284; http://dx.doi.org/10.1093/bioinformatics/bti551
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. Nat Genet 2000; 25:25-9; PMID:10802651; http://dx.doi.org/10.1038/75556
- Zhang C, Liu Y, Andrews PC. Quantification of histone modifications using 15N metabolic labeling. Methods 2013; 61:236-43; PMID:23454290; http://dx.doi.org/10.1016/j.ymeth.2013.02.004
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550; PMID:25516281; http://dx.doi.org/10.1186/s13059-014-0550-8
