One of the greatest challenges to maintaining genome integrity is the equal segregation of sister chromatids during mitosis. This process requires the assembly and coordination of over 200 proteins, including molecular motors and kinases, at centromeres and kinetochores. One centromere/kinetochore-associated factor critical for accurate chromosomal segregation is the Polo-like kinase 1-interacting checkpoint helicase (PICH). PICH is a substrate for the mitosis-specific Polo-like kinase 1, and is an ATP-dependent helicase.Citation1 During mitosis, PICH localizes to centromeres and kinetochores where it is required for resolution of chromatin bridges and completion of chromosome segregation. Loss of PICH or its ATPase activity results in the formation of anaphase bridges and micronuclei following cytokinesis.Citation2,3 How PICH is appropriately localized to centromeres and kinetochores, and its precise molecular role in preventing chromatin bridges, are not fully understood. A recent study from Sridharan and Azuma provides new insights by identifying small ubiquitin-related modifier (SUMO)-interacting motifs (SIMs) in PICH that are essential for both its localization and function in preventing chromatin bridges.Citation4
SUMOs function as posttranslational protein modifications and are essential during mitosis. Sumoylation regulates the localization and activities of a variety of centromere and kinetochore-associated proteins, including CENP-E and Topoisomerase IIα (TopoIIα). Sumoylation functions in part by promoting interactions between SUMO-modified proteins and downstream effector proteins containing SIMs. The most common and frequently studied SIMs contain a hydrophobic core, Val/Ile-X-Val/Ile-Val/Ile (V/I-X-V/I-V/I), flanked on either side by acidic residues. Although individual sumoylated proteins must associate with specific SIM-containing effectors, how specificity is achieved is largely unexplored and unknown. Whether effector proteins containing multiple SIMs may interact with multiple, different sumoylated proteins is also unclear.
Sridharan and Azuma have identified 3 SIMs (SIM1-SIM3) in PICH that confer distinct effects on PICH localization and activity. Using inducible human cell lines expressing specific SIM mutants, the authors demonstrate that the C-terminal SIM3 is required for PICH localization to centromeres, whereas the more centrally located SIM1 and SIM2 are uniquely required for PICH function in preventing chromatin bridges. Surprisingly, SIM3-mediated localization at centromeres is not required for PICH function in chromatin bridge resolution, a finding that widens the current view of the localization-dependent functions of PICH activity. Taken together, the authors present the model that the individual SIMs in PICH interact with different SUMO-modified proteins.
PICH was previously found by the Azuma lab to bind to SUMO-modified PARP1 and TopoIIαCitation5. Both PARP1 and TopoIIα localize to centromeres and kinetochores during mitosis and also function in resolving chromatin bridges. Another sumoylated factor involved in the resolution of chromatin bridges is the BLM helicase, although direct binding interactions with PICH have not been described. The SIMs in PICH could coordinate interactions with and between SUMO-modified forms of PARP1, TopoIIα, or BLM to mediate appropriate localization and function. In this context, PICH and SUMO can be likened to nucleation factors that facilitate assembly of proteins at sites of catenated DNA. Similar nucleating functions have been described for PML and SUMO in the formation of PML nuclear bodies,Citation6 and multiple factors are also recruited to sites of DNA damage through SUMO-SIM mediated assembly.Citation7 Determining whether sumoylated forms of PARP1, TopoIIα, or BLM bind preferentially to one or the other functionally unique SIMs in PICH could clarify specific roles in affecting PICH localization and function.
More generally, the identification and characterization of functionally distinct SIMs in PICH provides unique opportunities to explore SUMO-SIM interactions and the spatial and temporal requirements for sumoylation during mitosis. Whether the SIMs present in PICH affect interactions with unique sumoylated proteins in a temporally defined manner or mediate simultaneous assembly of multiple different sumoylated proteins will be interesting to investigate. The contributions that the SUMO substrate and the attached SUMO signals make in determining interactions with functionally distinct SIMs in PICH will also be an important question for future exploration ().
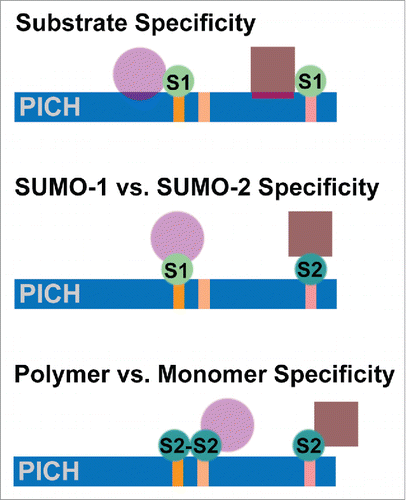
Figure 1. Alternative mechanisms for selective interactions between individual SIMs in PICH (indicated by vertical lines) and distinct SUMO-modified proteins. Specificity may be mediated by a bipartite interaction with PICH involving SUMO and the SUMO-modified protein (top). Alternatively, specificity may be determined by paralog-specific modification of PICH interacting proteins and paralog-specific SIM binding (middle). Finally, specificity may be determined by monomeric or polymeric modification of PICH interacting proteins and selective binding of monomeric or polymeric SUMO to the SIMs in PICH (bottom). Key: PICH = Polo-like kinase interacting protein-1; S1 = SUMO-1, S2 = SUMO-2.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Baumann C, Körner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 2007; 128:101-14; PMID:17218258; http://dx.doi.org/10.1016/j.cell.2006.11.041
- Kaulich M, Cubizolles F, Nigg EA. On the regulation, function, and localization of the DNA-dependent ATPase PICH. Chromosoma 2012; 121:395-408; PMID:22527115; http://dx.doi.org/10.1007/s00412-012-0370-0
- Nielsen CF, Huttner D, Bizard AH, Hirano S, Li T-N, Palmai-Pallag T, Bjerregaard VA, Liu Y, Nigg EA, Wang H-C, Hickson ID. PICH promotes sister chromatid disjunction and co-operates with topoisomerase II in mitosis. Nat Commun 2015; 6:8962; PMID:26643143; http://dx.doi.org/10.1038/ncomms9962
- Sridharan W, Azuma Y. SUMO-interacting motifs (SIMs) in Polo-like kinase 1-interacting checkpoint helicase (PICH) ensure proper chromosome segregation during mitosis. Cell Cycle 2016; 15(16):2135-44; PMID:27230136; http://dx.doi.org/10.1080/15384101.2016.1191713
- Sridharan V, Park H, Ryu H, Azuma Y. SUMOylation regulates polo-like Kinase 1-interacting checkpoint helicase (PICH) during mitosis. J Biol Chem 2015; 290:3269-76; PMID:25564610; http://dx.doi.org/10.1074/jbc.C114.601906
- Zhong S, Müller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood 2000; 95:2748-52; PMID:10779416
- Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 2012; 151:807-20; PMID:23122649; http://dx.doi.org/10.1016/j.cell.2012.10.021
