ABSTRACT
Induction of cell apoptosis is one of the major host defense mechanisms through which macrophages control Mycobacterium tuberculosis (Mtb) infection. However, the mechanisms underlying macrophage apoptosis triggered by Mtb infection are still largely unknown. In this study, a microarray profiling survey revealed 14 miRNAs were down-regulated in CD14+ monocytes from active pulmonary tuberculosis patients, and only the reduction of miR-20a-5p could be reversed after successful anti-tuberculosis treatment. Validation of miR-20a-5p expression was confirmed using real time qPCR. Moreover, miR-20a-5p expression also decreased in differentiated THP-1 macrophages after mycobacterial infection in vitro. Functional assays through forced or inhibited expression of miR-20a-5p in THP-1 macrophages demonstrated that miR-20a-5p functioned as a negative regulator of mycobacterial-triggered apoptosis. Importantly, inhibition of miR-20a-5p expression resulted in more efficient mycobacterial clearance from infected THP-1 macrophages while miR-20a-5p overexpression promoted mycobacterial survival. Mechanistically, miR-20a-5p was demonstrated to regulate Bim expression in a JNK2-dependent manner, unlike Bcl2, and luciferase assay showed JNK2 was a novel direct target of miR-20a-5p. Together, our findings indicate that downregulation of miR-20a-5p triggers macrophage apoptosis as a novel mechanism for host defense against mycobacterial infection.
Introduction
Tuberculosis (TB), a chronic infectious disease caused by Mycobacterium tuberculosis (Mtb), is a major global health threat responsible for almost 9.6 million new cases and 1.5 million deaths in 2014.Citation1 Macrophages are the first line of host defense against Mtb infection.Citation2 When appropriately activated, macrophages can effectively clear intracellular Mtb through multiple mechanisms such as autophagy, apoptosis, etc.Citation3,4 Apoptosis can serve as a defense mechanism for cells confronted with intracellular Mtb and reduce the viability of various mycobacterial species. TNF is required for induction of apoptosis in response to Mtb infection, and activates caspase-8-mediated extrinsic death domain pathway involving the kinases ASK1, p38 and c-Abl, which lowers the spread of mycobacteria.Citation4,5 In addition, mitochondrial outer membrane permeabilization (MOMP) leading to activation of the intrinsic apoptotic pathway is required. MOMP and apoptosis can occur independently of mitochondrial permeability transition (MPT), when MOMP is induced by members of the Bcl-2 family of apoptosis inducing proteins.Citation6 Among them, Bim is a crucial regulator of apoptosis induced by Mtb, which is mediated by the MAPK-dependent pathway.Citation7,8 Furthermore, FasL-induced apoptosis of infected human macrophages could be an immune effector mechanism not only depriving mycobacteria from their growth environment but also reducing viable bacterial counts.Citation9 Conversely, to facilitate the persistent infection, Mtb develops multiple strategies to avoid being killed by macrophages through inhibiting apoptosis. For example, virulent strains such as H37Rv inhibit macrophage apoptosis by activating the release of membrane-bound TNFR2 as the soluble form to evade TNF-dependent apoptosis.Citation10 The battle between Mtb and macrophages to control cell apoptosis is critical in determining the outcome of infection.
miRNAs are ∼22 nucleotide long, non-coding, highly-conserved, single-stranded RNAs, which play critical roles in regulating gene expression by targeting the mRNAs of protein-coding genes.Citation11 These miRNA targeted proteins can be directly or indirectly involved in cell cycle progression, cellular proliferation and apoptosis.Citation12 Individual miRNAs or miRNAs families can regulate the expression of hundreds of genes and, conversely, gene targets can be regulated by different miRNAs. Several previous studies revealed differential expression of miRNAs in TB patients (both active and latent infection) versus healthy donors,Citation13-16 but with little agreement between the reports and these miRNAs signatures should be validated in more studies. Recent studies demonstrated that Mtb can modulate miRNAs regulating a variety of mechanisms to escape immune control, such as TLR signaling pathways,Citation17 apoptosis regulation,Citation18 and cytokine production.Citation19 Although the role of miRNAs in regulation of the immune system and response to infection is becoming clearer, we do not yet have a clear picture of the global impact of miRNAs on the immune response to Mtb.
The miR-17∼92 cluster is a prototypical example of a polycistronic miRNA gene. In the human genome, the miR-17∼92 cluster encodes 6 miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1), which are tightly grouped within an 800 base-pair region of human chromosome 13.Citation20 This cluster is frequently overexpressed in human cancers and has been shown to promote several aspects of oncogenic transformation, including proliferation and evasion of apoptosis.Citation21,22 Among the members of miR-17 family, miR-20a has been extensively studied on its role in tumor development and progression. It was reported that miR-20a was up-regulated in anaplastic thyroid cancer,Citation23 and it could promote gastric cancer cell proliferation and inhibit cell apoptosis via post-transcriptional modulation of p21 and TP53INP1.Citation24 In addition, miR-20a also plays an important role in the regulation of pro-inflammatory cytokine release, such as controlling ASK1 expression, in inflammatory disease.Citation25 Furthermore, miR-20a was proved to downregulate Fas expression in osteosarcoma, thus increasing the metastatic potential of osteosarcoma cells by allowing survival in the FasL+ lung microenvironment.Citation26
Alternation of miRNA expression profiles has been observed in serum and sputum from TB patients,Citation14,16 but it still remains unknown in monocytes from peripheral blood. In this study, miRNA microarray was used to generate miRNA expression profiles in CD14+ monocytes, and we found that miR-20a-5p expression was downregulated in pulmonary TB patients and recovered after successful anti-TB treatment. Results were validated using real time qPCR. In view of the role of apoptosis in controlling mycobacterial infection and the role of miR-20a in regulating cancer cell apoptosis, we sought to further explore the role of miR-20a-5p on apoptosis regulation during mycobacterial infection. Here we showed that inhibition of miR-20a-5p expression significantly increased apoptosis to facilitate mycobacterial clearance, which was corroborated by the finding that over-expression of miR-20a-5p significantly reduced apoptosis to promote mycobacterial survival. Mechanistically, miR-20a-5p was demonstrated to regulate Bim expression in a JNK2-dependent manner, and luciferase assay showed JNK2 was a novel direct target of miR-20a-5p. In vivo data indicated that the expression of miR-20a-5p was negatively correlated with JNK2 and Bim in CD14+ monocytes. Our results uncover an additional layer of regulation of apoptosis during mycobacterial infection, dependent on the down-regulation of miR-20a-5p.
Results
MiR-20a-5p is reduced after mycobacterial infection in vivo and in vitro
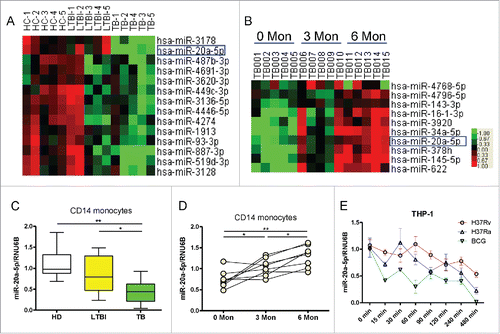
In an effort to identify candidate miRNAs that are critical for immune regulation, we performed microRNA profiling analysis in CD14+ monocytes isolated from 5 pulmonary tuberculosis (PTB) patients, 5 latent tuberculosis infections (LTBI), and 5 healthy donors (HD). We observed that miR-20a-5p was significantly down-regulated in CD14+ monocytes from PTB patients (), interestingly, among the 14 downregulated miRNAs in PTB patients, only miR-20a-5p expression increased after 3 months and 6 months of anti-TB treatment (). To validate these results, we measured miR-20a-5p expression in CD14+ monocytes using a conventional TaqMan real-time qPCR assay in an additional cohort. In accordance with miRNA arrays, miR-20a-5p was significantly down-regulated in active PTB patients compared with HD and LTBI individuals (P < 0.01, 0.05, ), such reduction could be reversed after successful anti-TB treatment (P < 0.05, ). Moreover, we used the live virulent strain of Mtb (H37Rv), attenuated strain of Mtb (H37Ra) or BCG to infect the differentiated THP-1 macrophages in vitro, and found that all mycobacterial strains could significantly reduce miR-20a-5p expression in a time-dependent manner (). These results indicate that miR-20a-5p expression is reduced after mycobacterial infection in vivo and in vitro, and recovers after successful anti-Mtb treatment.
Figure 1. MiR-20a-5p is reduced after mycobacterial infection in vivo and in vitro. (A) Comparison of the miRNA expression profiles determined by miRNA array between HD, LTBI, and TB CD14+ monocytes samples. Samples were clustered according to the expression profiles of differentially expressed miRNAs. Data from each miRNA was median centered. Heatmap analysis showed down-regulated (green) and upregulated (red) miRNAs. (B) Dynamic changes in the miRNA expression profiles of CD14+ monocytes from active PTB patients at 0, 3, and 6 months after anti-TB treatment. (C) Expression of miR-20a-5p in CD14+ monocytes from validation cohort consisting of HD (n = 10), LTBI (n = 10), and TB (n = 10) was determined by real time qPCR assay. Relative gene expression was normalized to RNU6B. (D) Validation of miR-20a-5p expression after anti-TB treatment using qPCR assay. (E) Differentiated THP-1 cells were infected with H37Rv, H37Ra or BCG at a MOI of 10, and miR-20a-5p expression was determined at different time points. * P < 0.05, ** P < 0.01.
MiR-20a-5p regulates cell apoptosis to control mycobacterial infection
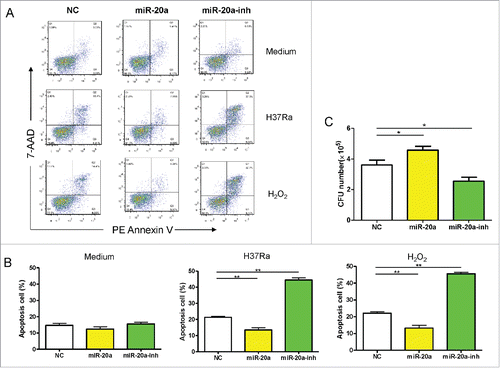
To investigate whether miR-20a-5p can influence mycobacterial-induced cell apoptosis, the lentivirus expressing miR-20a-5p and miR-20a-5p-inh was transduced into THP-1 cells respectively. GFP+ cells were isolated and infected with H37Ra for 8 hours or as a positive control of apoptosis, treated with H2O2 for 30 minutes. We found that at the basal level, no significant differences were observed in the apoptosis rate of THP-1 cells among all groups (), however, after stimulation with H37Ra or H2O2, the apoptosis rate of THP-1 cells infected with negative control was significantly higher than those infected with miR-20a-5p, and significantly lower than those infected with miR-20a-5p-inh (). In order to unambiguously determine the role of miR-20a-5p in bacterial clearance, we directly scored for bacterial CFUs in H37Ra infected THP-1 cells after up- or downregulation of miR-20a-5p. Compared with the negative control, survival of Mtb was augmented in the presence of miR-20a-5p overexpressing cells, and compromised in cells treated with miR-20a-5p-inh (). These data support the hypothesis that down-regulation of miR-20a-5p promote cell apoptosis to facilitate Mtb clearance.
Figure 2. MiR-20a-5p regulates cell apoptosis to control mycobacterial infection. (A) THP-1 cells were infected with miR-20a-5p, miR-20a-5p-inh, and NC lentivirus, GFP+ cells were sorted and differentiated into macrophages, which were then incubated without (upper panel) or with H37Ra for 8 hs (middle panel) or H2O2 for 30 mins (lower panel), and cells were stained with PE-labeled Annexin V and 7-AAD. Flow cytometric plots demonstrate that no difference was observed in the rate of apoptotic cells at basal level, but after H37Ra or H2O2 stimulation, a lower percentage of apoptotic cells were observed in THP-1 cells overexpressing miR-20a-5p, and higher percentage when miR-20a-5p was inhibited. (B) ANOVA/Newman-Keuls multiple comparison test was used to compare the difference of apoptosis rate among various groups. (C) GFP+ THP-1 cells were sorted after lentivirus infection and differentiated into macrophages, then cells were infected with H37Ra at a MOI of 10, and 3 d later, the cell lysates and culture supernatants were collected to determine CFU. ANOVA/Newman-Keuls multiple comparison test was used to compare the difference among all groups. * P < 0.05, ** P < 0.01.
MiR-20a-5p affects apoptosis related genes expression in THP-1 cells
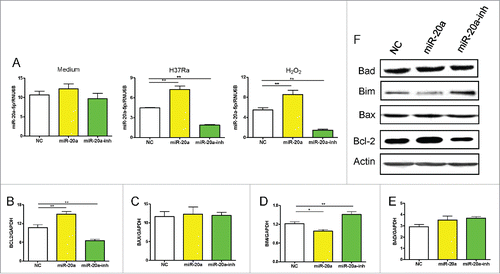
In order to determine how miR-20a-5p inhibited apoptosis in THP-1 cells, we investigated the underlying molecular mechanisms. Apoptosis can be triggered in a cell through 2 major pathways, an intrinsic pathway and extrinsic pathway. Firstly, we measured mitochondria-related anti-apoptotic gene transcriptional levels by real-time qPCR, such as Bcl-2, as well as pro-apoptotic gene transcriptional levels, including Bax, Bim, and Bad, in THP-1 cells infected with miR-20a-5p, miR-20a-5p-inh, or negative control lentivirus. After lentivirus infection, no differences were observed in miR-20a-5p expression in THP-1 cells without stimulation due to higher endogenous miR-20a-5p, but miR-20a-5p could be up- or down- regulated as expected after H37Ra or H2O2 stimulation (). Meanwhile, we found H37Ra-induced Bcl-2 expression was increased in THP-1 cells infected with miR-20a-5p lentivirus, and decreased when the miR-20a-5p was blocked (). However, the transcript levels of the pro-apoptotic gene, Bim, was decreased in miR-20a-5p overexpressed THP-1 cells, and increased when miR-20a-5p was inhibited (). However, there were no significant differences on the transcriptional levels of Bax and Bad (). We confirmed the transcript data at the protein level using western-blot assay (). These results clearly demonstrate that miR-20a-5p influences apoptosis through regulating Bcl-2 and Bim expression in THP-1 cells.
Figure 3. The level of MiR-20a-5p influences apoptosis related genes expression. (A) MiR-20a-5p expression in GFP+ THP-1 cells after lentivirus infection was detected without or with H37Ra or H2O2 stimulation. (B-E) Real-time qPCR assay was used to measure mitochondria-related anti/pro-apoptotic gene expression after up- and down-regulation of miR-20a-5p. The Bcl-2 gene expression increased in THP-1 cells infected with miR-20a-5p lentivirus, and decreased in THP-1 cells infected with miR-20a-5p-inh lentivirus (B), Bim gene presented an inverse result (D). No differences were observed in Bax and Bad gene expression (C, E). (F) Western-blot assay was used to detect anti/pro-apoptotic protein expression in H37Ra infected GFP+ THP-1 cells after up- and downregulation of miR-20a-5p. *P < 0.05, ** P < 0.01.
MiR-20a-5p regulates Bim expression via JNK2
Previous studies reported that miR-20a-5p dysfunction occurs in a range of cancers and inflammatory diseases.Citation22,25,27,28 However, there has been limited research to define the underlying mechanism. The Bim gene expression was inversely associated with miR-20a-5p, suggesting it could be a target gene of miR-20a-5p. However, we analyzed the whole Bim gene sequence without detecting an obvious seed region. Thus we predict that miR-20a-5p targets the upstream signaling genes to regulate Bim expression indirectly. It was proven in previous studies that AKT/JNK pathway could activate Bim gene expression to promote apoptosis,Citation29-32 so we measured AKT1, AKT2, JNK1 and JNK2 gene expression after up- or downregulation of miR-20a-5p. The level of JNK2 decreased following miR-20a-5p overexpression in THP-1 cells, and increased when miR-20a-5p was inhibited (). The transcript levels of AKT1 was increased in miR-20a-5p overexpressing THP-1 cells, but no significant difference was observed between miR-20a-inh and negative control (). However, there were no differences in AKT2 and JNK1 gene expression between experimental groups (). In order to validate the transcript data, we detected total JNK2 and p-JNK2 (p54) expression at the protein level using western-blot assay, which showed a similar pattern like mRNA level ().
Figure 4. MiR-20a-5p regulates Bim expression via JNK2. (A-D) Real-time qPCR assay was used to measure AKT/JNK signaling pathways genes expression in H37Ra infected GFP+ THP-1 cells after up- or down-regulation of miR-20a-5p: AKT1 (A), AKT2 (B), JNK1 (C), JNK2 (D). (E) Expression of total JNK2 and p-JNK was detected by western-blot assay in H37Ra infected GFP+ THP-1 cells. (F) THP-1 cells were infected with miR-20a-5p-inh or NC lentivirus after JNK2 siRNA-carrying lentivirus infection, and 3 d later, GFP+ THP-1 cells were isolated and differentiated followed by stimulation with H37Ra at a MOI of 10 for 8 hs, Bim and Bcl2 gene expression was measured using real time qPCR assay. (G) After silencing of JNK2, THP-1 cells were infected with miR-20a-5p-inh or NC lentivirus, GFP+ cells were dealt with as above and apoptosis cell rate was determined using flow cytometry. ANOVA/Newman-Keuls multiple comparison test was used to compare the difference among all groups. *P < 0.05, ** P < 0.01.
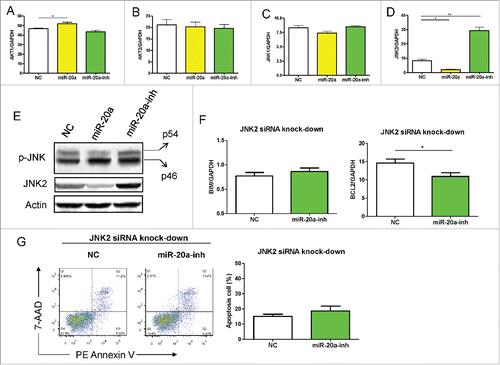
Next we investigated whether miR-20a-5p affects Bim expression via JNK2, we firstly used lentiviral vectors expressing gene-specific small interfering RNA (siRNA) to specifically block JNK2 expression, then Bim and Bcl2 expression was measured after inhibition of miR-20a-5p in THP-1 cells with H37Ra infection. Knockdown of JNK2 abrogated the regulation of miR-20a-5p on Bim expression, while Bcl2 was regulated independently of JNK2 (). Furthermore, there was no difference in the rate of apoptosis cells after silencing JNK2 between NC and miR-20a-inh group (). These results indicate that miR-20a-5p regulates Bim expression and mycobacterial induced apoptosis in a JNK2-dependent manner.
JNK2 is a direct target of MiR-20a-5p
Analysis of the JNK2 gene sequence identified 8 nucleotides at the 5′ end of miR-20a-5p matching the 3′UTR of wild JNK2 (). Therefore we made mutant JNK2 3′UTR in the predicted seed region of miR-20a-5p (). In order to investigate whether miR-20a-5p could directly target JNK2 3′UTR, we cloned wild or mutant JNK2 3′UTR into pmirGLO vector, then co-transfected into 293T and Hela cells with miR-20a-5p, and miR-20a-5p-inh. 48 hours post transfection, the intensity of Firefly and Renilla luciferase was assayed representing the transcriptional levels of JNK2 and internal reference, respectively. The relative transcript levels of JNK2 (F/R value) was decreased in 293T and Hela cells overexpressing miR-20a-5p co-transfected with wild type (WT) JNK2 3′UTR, and increased when miR-20a-5p was inhibited. However, no significant differences were observed for the JNK2 Mutant after miR-20a-5p up- or down-regulation (). These results suggest that miR-20a-5p directly block the transcription of wild type JNK2, by binding its 3′UTR.
Figure 5. MiR-20a-5p directly targets JNK2 gene. (A) The target region of miR-20a-5p in JNK2 3′UTR was determined and corresponding mutant was designed. (B) The wild type or mutant JNK2 3′UTR reporter vector was co-transfected into 293T or Hela cells with miR-20a-5p or miR-20a-5p-inh, demonstrating that significant differences were observed in WT reporter vector while up- or downregulation of miR-20a-5p, however, no differences were observed in Mut vector. The relative transcriptional level of JNK2 was presented as F/R value. (C-E) Comparison of expression of JNK2 (C), Bim (D), Bcl2 (E) determined by real time qPCR assay between HD (n = 10), LTBI (n = 10), and PTB (n = 10) CD14+ monocytes samples. Expression of JNK2 and Bim increased in PTB patients compared to HD. (F-H) The correlation analysis between miR-20a-5p and apoptosis related genes (F: JNK2; G: Bim; H: Bcl2) in CD14+ monocytes. * P < 0.05, ** P < 0.01.
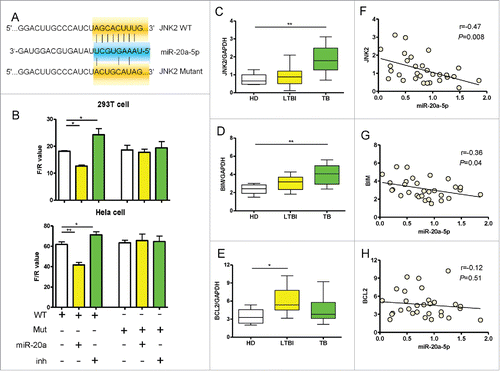
In order to further validate the above results, we measured JNK2, Bim and Bcl-2 transcript levels by real-time qPCR in CD14+ monocytes from clinical samples. The levels of JNK2 and Bim in CD14+ monocytes of PTB patients were significantly higher than healthy donors (). However, the level of Bcl-2 in CD14+ monocytes of LTBI was significantly higher than healthy donors, and there was no significant difference between PTB patients and healthy donors (). Both the levels of JNK2 and Bim were negatively correlated with miR-20a-5p expression in CD14+ monocytes of these 30 subjects (P < 0.05, ), the levels of Bcl-2 showed no correlation with miR-20a-5p (). Taken together, these results confirm that miR-20a-5p regulates Bim gene expression indirectly to promote apoptosis through its direct target JNK2.
Discussion
Mtb is spread by airborne droplet nuclei, transmitted when an infected individual coughs and disperses these droplets, which can then be inhaled into the airways and alveoli of a new host.Citation33 The interactions between Mtb and macrophages are more complex. While macrophages function as the first line of defense against Mtb infection, they also serve as the primary niche for Mtb replication. Triggered upon mycobacterial infection, multiple molecules and their associated signaling pathways in macrophages are involved in the regulation of protective immune responses. Mtb can be recognized by numerous receptors and induces a network of signaling pathways to drive inflammatory responses, involving toll-like receptors (TLRs), C-type lectins, Aryl hydrocarbon receptor (AhR), and Cyclic GMP-AMP synthase (cGAS).Citation34-36 Studies on gene expression profiles of Mtb-infected human macrophages provided the evidence for the importance of IL-1β and IL-12 in combating Mtb.Citation37 Importantly, one study identified CCL1, produced from macrophages of TB patients after Mtb stimulation, was involved in host susceptibility to TB.Citation38 Here, we demonstrated that miR-20a-5p was reduced after mycobacterial infection in vitro and in vivo, which implicates this pathway maybe a potential defense mechanism to promote an effective immune response against infection.
Apoptosis is an important defense immune response after the encounter of intracellular mycobacterial infection. Although virulent species of mycobacterial clearly induced less apoptosis than the attenuated species, it still induced a twofold increase in apoptosis compared to apoptosis levels in uninfected primary human alveolar macrophages.Citation4 Consistently, mycobacterial could induce apoptosis in vitro infection assays using monocyte derived macrophages obtained from human blood and human macrophage-like cell lines.Citation39,40 The importance of apoptosis during mycobacterial infection was suggested by the demonstration that apoptosis of infected cells reduced viability of intracellular mycobacterial, while necrosis of infected cells does not.Citation41,42 Remarkably, it was shown that phagocytosis of apoptotic bodies derived from Mtb-infected macrophages by dendritic cells contributed to the presentation of mycobacterial protein and glycolipid antigens and subsequent activation of specific CD8 T cells,Citation43 and apoptotic bodies containing mycobacterial antigens could crossprime CD8 T cells and protect against Mtb.Citation44 Taken together, induction of apoptosis is critical for the mycobacterial infected macrophages to facilitate bacterial clearance.
During Mtb infection, multiple molecules and their associated signaling pathways in macrophages are involved in the regulation of macrophage apoptosis. The extrinsic pathway is triggered by TNF-α in an autocrine/paracrine manner and requires infection-induced priming for TNF-receptor death signals since uninfected cells were resistant to exogenous TNF-α.Citation9 Phagosomal ROS production is clearly associated with TNF-dependent apoptosis.Citation45 In macrophages infected with attenuated Mtb, apoptosis is also associated with MOMP.Citation6,46 Silencing of the gene for the pro-apoptotic Bcl-2 like protein BAX, which is required for the release of cytochrome c and apoptosis inducing factor from the mitochondrial intermembrane space, abrogating Mtb-induced apoptosis.Citation46 Infection with Mtb significantly increases Bim expression, and downregulation of Bim levels markedly abrogates clinical isolate MT103-induced apoptosis.Citation7 An MEK/ERK inhibitor could rescue BCG-infected macrophages from apoptosis, which is accompanied by the complete suppression of Bim phosphorylation.Citation8 Other factors, such as eicosanoids, are involved in the regulation of apoptosis in Mtb-infected macrophages.Citation47 Avirulent Mtb strains such as H37Ra induce PGE2 production, which initiates protection of the mitochondria from bacteria-induced damage and promotes apoptosis, while virulent Mtb inhibits plasma membrane repair based on its ability to block PGE2 synthesis.Citation48,49 The elucidation of molecular mechanisms by which macrophages regulate apoptosis during mycobacterial infection will contribute to the development of new drug targets and/or vaccine candidates.
MiR-20a is a member of the miR-17∼92 cluster, which is the most extensively studied cluster that has an oncogenic function. MiR-20a has been reported to contribute to cancer pathogenesis by directly regulating genes associated with cell apoptosis and proliferation. The oncogenic roles of miR-20a were confirmed by the findings that inhibition of miR-20a by antisense oligonucleotides induced apoptosis in a human lung cancer cell line, Calu6 overexpressing miR-17∼92.Citation50 Interestingly, miR-20a expression in human leukemia cells was inhibited by oridonin, a natural agent with potent anticancer activity, which triggered apoptosis and reversed chemoresistance.Citation51 Mechanically, miR-20a could down-regulate Fas expression to allow osteosarcoma cells to circumvent FasL mediated apoptosis.Citation26 However, the effect of miR-20a on apoptosis regulation in human macrophages during mycobacterial infection still remains unknown. In this study, our findings reveal that miR-20a-5p functioned as a negative regulator in mycobacterial-triggered cell apoptosis, and inhibition of miR-20a-5p expression promoted cell apoptosis to facilitate efficient mycobacterial clearance.
Furthermore, we investigated the underlying mechanisms by which miR-20a-5p regulates apoptosis in mycobacterial infected macrophages. Here, we found Bim was increased in both miR-20a-5p inhibited THP-1 cells and CD14+ monocytes of PTB patients. In accordance with our results, previous studies also showed absence of miR-17∼92 leads to increased levels of the pro-apoptotic protein Bim.Citation52 Moreover, we investigated the upstream signaling genes regulating Bim gene expression. Previous study showed that a JNK inhibitor failed to increase the number of viable BCG-infected macrophages.Citation8 However, JNK1 and JNK2, belonging to the JNK kinase family, have distinct or even opposing functions in different diseases. They phosphorylate and activate a number of nuclear and non-nuclear proteins, including the transcription factor AP-1, ATF-2, c-Myc, as well as cell apoptotic proteins (Fas, Bim, Bcl-xl).Citation53,54 These proteins control a diversity of cellular responses, such as proliferation, differentiation, cell death and survival.Citation55 Our data shows that miR-20a-5p regulates Bim expression and mycobacterial induced apoptosis in a JNK2-dependent manner. We further demonstrated that up-regulation of miR-20a-5p inhibited JNK2 expression and miR-20a-5p could directly target JNK2 3′UTR. In our study, Bcl-2 was only decreased in THP-1 cells infected with miR-20a-5p-inh lentivirus, but not in monocytes of PTB patients. In consideration of the complicated mechanism in PTB patients, we predicted that Bcl-2 might be concurrently regulated by several factors. Although, TB patients presented with a higher percentage of CD14+ cells than health control,Citation56,57 it was confirmed that the apoptosis of macrophages was triggered after Mtb infection,Citation48 and the percentage of apoptotic monocytes dramatically decreased after Mtb treatment.Citation58
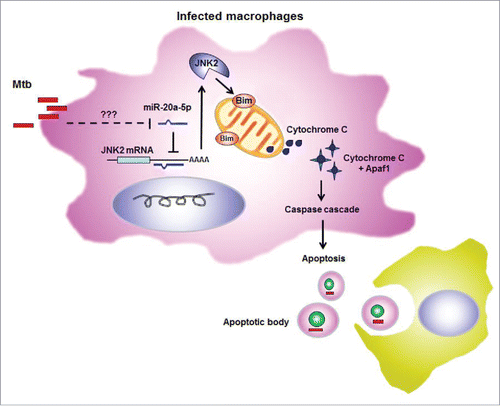
In conclusion, we elucidate a novel mechanism of host response against mycobacterial in humans. We show that miR-20a-5p is downregulated after mycobacterial infection and indirectly increased Bim expression, although the molecular mechanisms required for the reduction of miR-20a-5p by mycobacterial have yet to be fully elucidated, which promotes macrophage apoptosis to facilitate mycobacterial clearance. Furthermore, we report for the first time that JNK2 is the direct target of miR-20a-5p, through which miR-20a-5p performs the regulation of Bim expression and apoptosis after mycobacterial infection (). Taken together, our findings reveal a novel pathway through which host can control mycobacterial infection by reducing miR-20a-5p. Since, the percentage of apoptotic monocytes dramatically decreased after treatment,Citation58 miR-20a-5p has the potential to be a biomarker to diagnose tuberculosis and evaluate treatment outcomes.
Figure 6. Schematic diagram illustrating apoptosis regulated by miR-20a-5p in mycobacterial infected macrophages. Expression of miR-20a-5p was downregulated in macrophages after mycobacterial infection, but it remains unknown about the mechanisms by which miR-20a-5p is reduced. MiR-20a-5p can directly target JNK2 3′UTR and inhibit JNK2 expression, so JNK2 expression increases significantly when miR-20a-5p is inhibited, and therefore induces Bim expression. Then apoptosis is triggered and apoptotic body is formed to facilitate mycobacterial clearance.
Materials and methods
Ethics statement
All protocols for this study were reviewed and approved by the Research Ethics Committee of Shenzhen Third People's Hospital (No. 2012–007), and conducted according to the Declaration of Helsinki. The Research Ethics Committee approved the collection of peripheral blood exclusively for research purposes with the written informed consent of all participants.
Human subjects and samples collection
Healthy donors and patients were recruited and enrolled at Shenzhen Third People's Hospital, Guangdong, China. The diagnosis of active pulmonary TB was based on clinical presentation, chest radiography, and Mtb bacteriological confirmation. Patients infected with hepatitis B virus (HBV), hepatitis C virus (HCV) or human immunodeficiency virus (HIV) were excluded. Patients with a history of diabetes were also excluded. All of the pulmonary TB patients accepted standard anti-TB treatment, and underwent regular follow-up for at least 6 months. LTBI was indicated by asymptomatic individuals with a positive result from the interferon gamma release assays (IGRAs). Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood through density gradient centrifugation over Ficoll-Hypaque, and CD14+ monocytes were isolated by positive magnetic selection and stored in liquid nitrogen.
MiRNAs array
Total RNA of CD14+ monocytes was extracted and labeled with a miRCURY Hy3/Hy5 labeling kit (Exiqon, Vedbaek, Denmark). Then the labeled samples were hybridized to the miRCURY LNA array version 8.0 (Exiqon). The hybridization was performed following the miRCURY LNA array manual. Following hybridization, the slides were washed by Wash buffer kit (Exiqon), dried and scanned on a GenePix 4000B array scanner (Molecular Devices Co., Sunnyvale, CA, USA). The data was analyzed using unsupervised hierarchical clustering (Cluster 3.0) and TreeView analysis (Stanford University, Stanford, CA, USA).
Real-time quantitative PCR
Total RNA was extracted from CD14+ monocytes or cultured cells using the miRNeasy Mini Kit (Qiagen, Hilden, Germany). The TaqMan™ Advanced miRNA cDNA Synthesis Kit (Applied biosystems, Waltham, MA) was used for qPCR assay of miR-20a-5p as described by the manufacturer. Total 2 μL of extracted RNA was used for a single RT reaction, and the cDNA was used as template for real-time qPCR reactions using Applied Biosystems Vii7 Sequence Detection System (Applied biosystems, Waltham, MA) in 20 μl PCR mixtures containing TaqMan™ master mix and specific miRNA primers from the manufacturer. The apoptosis related genes expression was measured by real-time qPCR using cDNA synthesized with an oligo-dT primer and SuperScript II reverse transcriptase (Takara, Japan), SYBR-Green real-time PCR Master Mix (Takara, Japan), and a set of gene-specific oligonucleotide primers. The primers were designed as follows: Bcl2, 5′-GGTGGGGTCATGTGTGTGG-3′ (forward) and 5′-CGGTTCAGGTACTCA GTCATCC-3′ (reverse); Bax, 5′-CCCGAGAGGTCTTTTTCCGAG-3′ (forward) and 5′-CCAGCCCATGATGGTTCTGAT-3′ (reverse); Bim, 5′-TAAGTTCTGAGT GTGACCGAGA-3′ (forward) and 5′-GCTCTGTCTGTAGGGAGGTAGG-3′ (reverse); Bad,5′-CCCAGAGTTTGAGCCGAGTG-3′ (forward) and 5′-CCCATCCCTTCGTCGTCCT-3′ (reverse); AKT1, 5′-AGCGACGTGGCT ATTGTGAAG-3′ (forward) and 5′-GCCATCATTCTTGAGGAGGAAGT-3′ (reverse); AKT2, 5′-ACCACAGTCATCGAGAGGACC-3′ (forward) and 5′-GGAGCCACACTTGTAGTCCA-3′ (reverse); JNK1, 5′-TGTGTGGAATC AAGCACCTTC-3′ (forward) and 5′-AGGCGTCATCATAAAACTCGTTC-3′ (reverse); JNK2, 5′-GAAACTAAGCCGTCCTTTTCAGA-3′ (forward) and 5′-TCCAGCTCCATGTGAATAACCT-3′ (reverse); GAPDH, 5′- GCACCGTCA AGGCTGAGAAC-3′ (forward) and 5′-TGGTGAAGACGCCAGTGGA-3′ (reverse). All samples were assayed in triplicate. The U6 snRNA (RNU6B) and GAPDH levels were used to normalize the relative abundance of miRNA and mRNA, respectively. Relative expression levels were calculated via the 2−ΔΔCt method.
Lentiviral-mediated miR-20a-5p up- or down- regulation and RNA interference
For up- and down- regulation of miR-20a-5p, we used lentiviral vectors expressing miR-20a-5p or miR-20a-5p inhibitor. Oligonucleotide sequences were as follows: miR-20a-5p, 5′-TAAAGTGCTTATAGTGCAG GTAG-3′, miR-20a-5p inhibitor, 5′-GATGGTGCAGATATTGCACTTTT-3′. The stem-loop oligonucleotides were synthesized and cloned into a lentivirus-based vector carrying the green fluorescent protein (GFP) gene (pGLV3-GFP, Genechem, Shanghai, China). A universal sequence (LV1-NC: 5′-TTCTCCGAACGTGTCACGT-3′) was used as a negative control for lentiviral vector. For knockdown of JNK2, lentiviral vector expressing gene-specific small interfering RNA (siRNA) was used to block gene expression. Oligonucleotide sequences of JNK2 specific siRNAs were as follows: 5′-AAGAGAGCTTATCGTGAACTT-3′. The packaging of lentivirus was performed by Genechem co. ltd. For infection with lentiviral vector constructs, the viruses were diluted in serum-free OptiMEM (Life Technology) and cells were infected at a multiplicity of infection (MOI) of 10 for 3 h in the presence of 5 ug/mL polybrene. After 72h of infection in serum-containing medium, GFP+ cells were isolated with fluorescence activated cell sorting (FACS) for further analysis.
Vector construction
A DNA fragment of JNK2 3′UTR region that contains a potential target sequence of miR-20a-5p was annealed using the following complementary oligonucleotides: 5′-TCGAACAAATGCTTGCTTGGACTTGCCCATCTAGCAC TTTGG-3′ (forward, underlined letters showed XhoI site) and 5′-CTAGCCAAAGTGCTAGATGGGCAAGTCCAAGCAAGCATTTGT-3′ (reverse, underlined letters showed XbaI site). The annealed products were inserted into the downstream of luciferase during pmirGLO-report vector (Promega) and the resulting vector was designated pmirGLO-JNK2-WT. Mutated JNK2 3′UTR bases in the potential target sequence of miR-20a-5p are indicated in . The complementary oligonucleotides were annealed and inserted into pmirGLO-report vector, which was designated pmirGLO-JNK2-Mut.
Cells transfection and dual-luciferase assay
293T and Hela cells were cultured in RPMI 1640 supplemented with 10% (v/v) FCS (Gibco), 1% (v/v) antibiotic/antimycotic solution. The cells were plated in 24-well plates 24 h before transfection. A total of 0.8 ug mixture including 0.6 ug of either JNK2 WT or Mut reporter vector, and 0.2 ug of miR-20a-5p or inhibitor, were co-transfected into the cells using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Transfected cells were lysed in 150 μL passive lysis buffer (Promega) following 24h incubation. The lysates were assayed for both the Firefly and Renilla luciferase activities using the dual-luciferase reporter assay system (Promega). Promoter activity was measured as the ratio between firefly and Renilla luciferase (F/R value). The transfection for each construct was performed 3 times and each construct was assayed for promoter activity in duplicate.
Apoptosis assays
Annexin V PE/7AAD apoptosis detection kit I (BD PharmingenTM, San Diego, CA, USA) was used to determine cell apoptosis. In brief, THP-1 cells were infected with miR-20a-5p, miR-20a-5p-inh or NC lentivirus, GFP+ cell were isolated with FACS after 72 hours incubation. For knockdown of JNK2, gene-specific siRNA lentivirus was used to infect THP-1 cells. After differentiation in the presence of phorbol12-myristate13-acetate (PMA, 20 ng/mL) for 24 hours, the cells were stimulated by the live attenuated strain H37Ra for 8 hours or treated with 0.4 mM hydrogen peroxide (H2O2) for 30 minutes. Then the cells were resuspended in binding buffer, at a density of 1 × 106 cells/ml, and incubated with annexin V-PE and 7AAD for 15 min. The annexin V(+) and annexin V(+) 7AAD(+) cells were defined as the total number of apoptotic cells, determined by flow cytometric analysis.
Mycobacterial CFU determinations
After infection with miR-20a-5p, miR-20a-5p-inh or NC lentivirus, GFP+ THP-1 cells were isolated by FACS and differentiated into macrophages in 24-well plates at 5 × 105 cells per well, then cells were infected by H37Ra at a MOI of 10. The cells were washed 3 times with PBS after 6 hours incubation, and 3 d later, the cell lysates and culture supernatants of the infected cells were collected, serial dilutions of lysates and supernatants were plated on 7H11 agar plates supplemented with 10% OADC and incubated at 37°C for 3–4 weeks. CFUs were calculated in triplicate using standard procedures.
Western blot analysis
Cellular proteins were prepared using cell lysis buffer (50 mM Tris-HCl, pH 8.0, 1% NP-40, 2 mM EDTA, 10 mM NaCl, 2 mg/ml aprotinin, 5 mg/ml leupeptin, 2 mg/ml pepstatin, 1 mM DTT, 0.1% SDS and 1 mM phenylmethylsulfonyl fluoride). Equal amounts of protein (50 μg) were separated by 12% SDS-PAGE and then transferred to nitrocellulose membranes (NY, USA) by electroblotting. The membranes were blocked with 5% BSA in TBST (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.05% Tween 20) for 1 h, and then incubated with primary antibodies overnight at 4°C before subsequent incubation with HRP-labeled secondary antibody (Promega) for 1 h at 37°C. The primary antibodies were from the following sources: Bad antibody (Cell Signaling Technology, #9292), Bim antibody (Cell Signaling Technology, #2819), Bax antibody (Cell Signaling Technology, #2772), Bcl2 antibody (Cell Signaling Technology, #2876), JNK2 antibody (N-18, Santa Cruz, sc-827), p-JNK Antibody (29.Thr 183/Tyr 185, Santa Cruz, sc-293137), β-Actin Antibody (Cell Signaling Technology, #4967). Protein was visualized by exposure to X-ray films using enhanced chemiluminescence reagent (Thermo Fisher).
Statistical analysis
The one-way analysis of variance (ANOVA)/Newman-Keuls multiple comparison test was used for statistical analyses to compare the differences among multiple groups. The paired t test was used to compare the miR-20a-5p expression with or without treatment of anti-TB. Correlations were assessed using Spearman's rank correlation, and Pearson's t test was used to analyze the correlation. GraphPad Prism software (version 5.0) was used for all the statistical analysis. A P value of less than 0.05 was considered to indicate a statistically significant difference.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank staffs, doctors and nurses of Shenzhen Third People's Hospital for patient management, sample collection and analysis in this work.
Funding
This study was supported by Twelfth-Fifth and Thirteenth-Fifth Mega-Scientific Projects on “prevention and treatment of AIDS, viral hepatitis and other infectious diseases” (No. 2014ZX10004001, 2016ZX10004206), Natural Science Foundation of China (No. 81471913, 81501714, 81500004); Natural Science Foundation of Guangdong (No. 2014A030313789); Shenzhen Scientific and Technological Foundation (No. JCYJ20140411111718166).
References
- WHO. Global tuberculosis report 2015. 2015
- Hmama Z, Pena-Diaz S, Joseph S, Av-Gay Y. Immunoevasion and immunosuppression of the macrophage by Mycobacterium tuberculosis. Immunol Rev 2015; 264:220-32; PMID:25703562; http://dx.doi.org/10.1111/imr.12268
- Bradfute SB, Castillo EF, Arko-Mensah J, Chauhan S, Jiang S, Mandell M, Deretic V. Autophagy as an immune effector against tuberculosis. Curr Opin Microbiol 2013; 16:355-65; PMID:23790398; http://dx.doi.org/10.1016/j.mib.2013.05.003
- Behar SM, Martin CJ, Booty MG, Nishimura T, Zhao X, Gan HX, Divangahi M, Remold HG. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol 2011; 4:279-87; PMID:21307848; http://dx.doi.org/10.1038/mi.2011.3
- Kundu M, Pathak SK, Kumawat K, Basu S, Chatterjee G, Pathak S, Noguchi T, Takeda K, Ichijo H, Thien CB, et al. A TNF- and c-Cbl-dependent FLIP(S)-degradation pathway and its function in Mycobacterium tuberculosis-induced macrophage apoptosis. Nat Immunol 2009; 10:918-26; PMID:19597496; http://dx.doi.org/10.1038/ni.1754
- Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol 2008; 18:157-64; PMID:18314333; http://dx.doi.org/10.1016/j.tcb.2008.01.007
- Aguilo N, Uranga S, Marinova D, Martin C, Pardo J. Bim is a crucial regulator of apoptosis induced by Mycobacterium tuberculosis. Cell Death Dis 2014; 5:e1343; PMID:25032866; http://dx.doi.org/10.1038/cddis.2014.313
- Iyoda T, Takada M, Fukatsu Y, Kumokoshi S, Fujisawa T, Shimada T, Shimokawa N, Matsunaga T, Makino K, Doi N, et al. A novel mechanism underlying the basic defensive response of macrophages against Mycobacterium infection. J Immunol 2014; 192:4254-62; PMID:24663676; http://dx.doi.org/10.4049/jimmunol.1301526
- Oddo M, Renno T, Attinger A, Bakker T, MacDonald HR, Meylan PR. Fas ligand-induced apoptosis of infected human macrophages reduces the viability of intracellular Mycobacterium tuberculosis. J Immunol 1998; 160:5448-54; PMID:9605147
- Balcewicz-Sablinska MK, Keane J, Kornfeld H, Remold HG. Pathogenic Mycobacterium tuberculosis evades apoptosis of host macrophages by release of TNF-R2, resulting in inactivation of TNF-alpha. J Immunol 1998; 161:2636-41; PMID:9725266
- Van Roosbroeck K, Pollet J, Calin GA. miRNAs and long noncoding RNAs as biomarkers in human diseases. Expert Rev Mol Diagn 2013; 13:183-204; PMID:23477558; http://dx.doi.org/10.1586/erm.12.134
- Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle 2007; 6:2127-32; PMID:17786041; http://dx.doi.org/10.4161/cc.6.17.4641
- Xu Z, Zhou A, Ni J, Zhang Q, Wang Y, Lu J, Wu W, Karakousis PC, Lu S, Yao Y. Differential expression of miRNAs and their relation to active tuberculosis. Tuberculosis (Edinb) 2015; 95:395-403; PMID:25936536
- Yi Z, Fu Y, Ji R, Li R, Guan Z. Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PLoS One 2012; 7:e43184; PMID:22900099; http://dx.doi.org/10.1371/journal.pone.0043184
- Singh PP, Li L, Schorey JS. Exosomal RNA from Mycobacterium tuberculosis-Infected Cells Is Functional in Recipient Macrophages. Traffic 2015; 16:555-71; PMID:25753779; http://dx.doi.org/10.1111/tra.12278
- Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol 2011; 49:4246-51; PMID:21998423; http://dx.doi.org/10.1128/JCM.05459-11
- Rajaram MV, Ni B, Dodd CE, Schlesinger LS. Macrophage immunoregulatory pathways in tuberculosis. Semin Immunol 2014; 26:471-85; PMID:25453226; http://dx.doi.org/10.1016/j.smim.2014.09.010
- Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol 2011; 12:861-9; PMID:21785411; http://dx.doi.org/10.1038/ni.2073
- Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol Immunol 2011; 48:1084-90; PMID:21367459; http://dx.doi.org/10.1016/j.molimm.2011.02.001
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell 2008; 133:217-22; PMID:18423194; http://dx.doi.org/10.1016/j.cell.2008.04.001
- Gao X, Zhang R, Qu X, Zhao M, Zhang S, Wu H, Jianyong L, Chen L. MiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myeloma. Leuk Res 2012; 36:1505-9; PMID:22959509; http://dx.doi.org/10.1016/j.leukres.2012.08.021
- Battistella M, Romero M, Castro-Vega LJ, Gapihan G, Bouhidel F, Bagot M, Feugeas JP, Janin A. The high expression of the microRNA 17–92 cluster and its paralogs, and the downregulation of the target gene PTEN, is associated with primary cutaneous B-cell lymphoma progression. J Invest Dermatol 2015; 135:1659-67; PMID:25634356; http://dx.doi.org/10.1038/jid.2015.27
- Xiong Y, Zhang L, Kebebew E. MiR-20a is upregulated in anaplastic thyroid cancer and targets LIMK1. PLoS One 2014; 9:e96103; PMID:24858712
- Wang M, Gu H, Qian H, Zhu W, Zhao C, Zhang X, Tao Y, Zhang L, Xu W. miR-17-5p/20a are important markers for gastric cancer and murine double minute 2 participates in their functional regulation. Eur J Cancer 2013; 49:2010-21; PMID:23333058; http://dx.doi.org/10.1016/j.ejca.2012.12.017
- Philippe L, Alsaleh G, Pichot A, Ostermann E, Zuber G, Frisch B, Sibilia J, Pfeffer S, Bahram S, Wachsmann D, et al. MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann Rheum Dis 2013; 72:1071-9; PMID:23087182; http://dx.doi.org/10.1136/annrheumdis-2012-201654
- Huang G, Nishimoto K, Zhou Z, Hughes D, Kleinerman ES. miR-20a encoded by the miR-17-92 cluster increases the metastatic potential of osteosarcoma cells by regulating Fas expression. Cancer Res 2012; 72:908-16; PMID:22186140; http://dx.doi.org/10.1158/0008-5472.CAN-11-1460
- Zhao S, Yao D, Chen J, Ding N, Ren F. MiR-20a promotes cervical cancer proliferation and metastasis in vitro and in vivo. PLoS One 2015; 10:e120905; PMID:25803820
- Carlsen AL, Schetter AJ, Nielsen CT, Lood C, Knudsen S, Voss A, Harris CC, Hellmark T, Segelmark M, Jacobsen S, et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum 2013; 65:1324-34; PMID:23401079; http://dx.doi.org/10.1002/art.37890
- Geissler A, Haun F, Frank DO, Wieland K, Simon MM, Idzko M, Davis RJ, Maurer U, Borner C. Apoptosis induced by the fungal pathogen gliotoxin requires a triple phosphorylation of Bim by JNK. Cell Death Differ 2013; 20:1317-29; PMID:23832115; http://dx.doi.org/10.1038/cdd.2013.78
- Tan M, Li Z, Ma S, Luo J, Xu S, Lu A, Gan W, Su P, Lin H, Li S, et al. Heroin activates Bim via c-Jun N-terminal kinase/c-Jun pathway to mediate neuronal apoptosis. Neuroscience 2013; 233:1-8; PMID:23262244; http://dx.doi.org/10.1016/j.neuroscience.2012.12.005
- Gargini R, Cerliani JP, Escoll M, Anton IM, Wandosell F. Cancer stem cell-like phenotype and survival are coordinately regulated by Akt/FoxO/Bim pathway. Stem Cells 2015; 33:646-60; PMID:25407338; http://dx.doi.org/10.1002/stem.1904
- Xiong H, Wang J, Guan H, Wu J, Xu R, Wang M, Rong X, Huang K, Huang J, Liao Q, et al. SphK1 confers resistance to apoptosis in gastric cancer cells by downregulating Bim via stimulating Akt/FoxO3a signaling. Oncol Rep 2014; 32:1369-73; PMID:25109605
- Fennelly KP, Jones-Lopez EC, Ayakaka I, Kim S, Menyha H, Kirenga B, Muchwa C, Joloba M, Dryden-Peterson S, Reilly N, et al. Variability of infectious aerosols produced during coughing by patients with pulmonary tuberculosis. Am J Respir Crit Care Med 2012; 186:450-7; PMID:22798319; http://dx.doi.org/10.1164/rccm.201203-0444OC
- Sia JK, Georgieva M, Rengarajan J. Innate Immune Defenses in Human Tuberculosis: An Overview of the Interactions between Mycobacterium tuberculosis and Innate Immune Cells. J Immunol Res 2015; 2015:747543; PMID:26258152; http://dx.doi.org/10.1155/2015/747543
- Moura-Alves P, Fae K, Houthuys E, Dorhoi A, Kreuchwig A, Furkert J, Barison N, Diehl A, Munder A, Constant P, et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature 2014; 512:387-92; PMID:25119038; http://dx.doi.org/10.1038/nature13684
- Dey B, Dey RJ, Cheung LS, Pokkali S, Guo H, Lee JH, Bishai WR. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat Med 2015; 21:401-6; PMID:25730264; http://dx.doi.org/10.1038/nm.3813
- Cappelli G, Volpe P, Sanduzzi A, Sacchi A, Colizzi V, Mariani F. Human macrophage gamma interferon decreases gene expression but not replication of Mycobacterium tuberculosis: analysis of the host-pathogen reciprocal influence on transcription in a comparison of strains H37Rv and CMT97. Infect Immun 2001; 69:7262-70; PMID:11705896; http://dx.doi.org/10.1128/IAI.69.12.7262-7270.2001
- Thuong NT, Dunstan SJ, Chau TT, Thorsson V, Simmons CP, Quyen NT, Thwaites GE, Thi NLN, Hibberd M, Teo YY, et al. Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. Plos Pathog 2008; 4:e1000229; PMID:19057661
- Ciaramella A, Cavone A, Santucci MB, Garg SK, Sanarico N, Bocchino M, Galati D, Martino A, Auricchio G, D'Orazio M, et al. Induction of apoptosis and release of interleukin-1 beta by cell wall-associated 19-kDa lipoprotein during the course of mycobacterial infection. J Infect Dis 2004; 190:1167-76; PMID:15319868; http://dx.doi.org/10.1086/423850
- Arcila ML, Sanchez MD, Ortiz B, Barrera LF, Garcia LF, Rojas M. Activation of apoptosis, but not necrosis, during Mycobacterium tuberculosis infection correlated with decreased bacterial growth: role of TNF-alpha, IL-10, caspases and phospholipase A2. Cell Immunol 2007; 249:80-93; PMID:18160064; http://dx.doi.org/10.1016/j.cellimm.2007.11.006
- Molloy A, Laochumroonvorapong P, Kaplan G. Apoptosis, but not necrosis, of infected monocytes is coupled with killing of intracellular bacillus Calmette-Guerin. J Exp Med 1994; 180:1499-509; PMID:7931080; http://dx.doi.org/10.1084/jem.180.4.1499
- Fratazzi C, Arbeit RD, Carini C, Remold HG. Programmed cell death of Mycobacterium avium serovar 4-infected human macrophages prevents the mycobacteria from spreading and induces mycobacterial growth inhibition by freshly added, uninfected macrophages. J Immunol 1997; 158:4320-7; PMID:9126994
- Schaible UE, Winau F, Sieling PA, Fischer K, Collins HL, Hagens K, Modlin RL, Brinkmann V, Kaufmann SH. Apoptosis facilitates antigen presentation to T lymphocytes through MHC-I and CD1 in tuberculosis. Nat Med 2003; 9:1039-46; PMID:12872166; http://dx.doi.org/10.1038/nm906
- Winau F, Weber S, Sad S, de Diego J, Hoops SL, Breiden B, Sandhoff K, Brinkmann V, Kaufmann SH, Schaible UE. Apoptotic vesicles crossprime CD8 T cells and protect against tuberculosis. Immunity 2006; 24:105-17; PMID:16413927; http://dx.doi.org/10.1016/j.immuni.2005.12.001
- Miller JL, Velmurugan K, Cowan MJ, Briken V. The type I NADH dehydrogenase of Mycobacterium tuberculosis counters phagosomal NOX2 activity to inhibit TNF-alpha-mediated host cell apoptosis. Plos Pathog 2010; 6:e1000864; PMID:20421951; http://dx.doi.org/10.1371/journal.ppat.1000864
- Chen M, Gan H, Remold HG. A mechanism of virulence: virulent Mycobacterium tuberculosis strain H37Rv, but not attenuated H37Ra, causes significant mitochondrial inner membrane disruption in macrophages leading to necrosis. J Immunol 2006; 176:3707-16; PMID:16517739; http://dx.doi.org/10.4049/jimmunol.176.6.3707
- Chen M, Divangahi M, Gan H, Shin DS, Hong S, Lee DM, Serhan CN, Behar SM, Remold HG. Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J Exp Med 2008; 205:2791-801; PMID:18955568; http://dx.doi.org/10.1084/jem.20080767
- Divangahi M, Chen M, Gan H, Desjardins D, Hickman TT, Lee DM, Fortune S, Behar SM, Remold HG. Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat Immunol 2009; 10:899-906; PMID:19561612; http://dx.doi.org/10.1038/ni.1758
- Levy BD, Clish CB, Schmidt B, Gronert K, Serhan CN. Lipid mediator class switching during acute inflammation: signals in resolution. Nat Immunol 2001; 2:612-9; PMID:11429545; http://dx.doi.org/10.1038/89759
- Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M, Nimura Y, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene 2007; 26:6099-105; PMID:17384677; http://dx.doi.org/10.1038/sj.onc.1210425
- Weng H, Huang H, Dong B, Zhao P, Zhou H, Qu L. Inhibition of miR-17 and miR-20a by oridonin triggers apoptosis and reverses chemoresistance by derepressing BIM-S. Cancer Res 2014; 74:4409-19; PMID:24872388; http://dx.doi.org/10.1158/0008-5472.CAN-13-1748
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008; 132:875-86; PMID:18329372; http://dx.doi.org/10.1016/j.cell.2008.02.019
- Bogoyevitch MA, Kobe B. Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases. Microbiol Mol Biol Rev 2006; 70:1061-95; PMID:17158707; http://dx.doi.org/10.1128/MMBR.00025-06
- Haeusgen W, Herdegen T, Waetzig V. Specific regulation of JNK signalling by the novel rat MKK7gamma1 isoform. Cell Signal 2010; 22:1761-72; PMID:20633641; http://dx.doi.org/10.1016/j.cellsig.2010.07.002
- Bubici C, Papa S. JNK signalling in cancer: in need of new, smarter therapeutic targets. Br J Pharmacol 2014; 171:24-37; PMID:24117156; http://dx.doi.org/10.1111/bph.12432
- Sanchez MD, Garcia Y, Montes C, Paris SC, Rojas M, Barrera LF, Arias MA, Garcia LF. Functional and phenotypic changes in monocytes from patients with tuberculosis are reversed with treatment. Microbes Infect 2006; 8:2492-500; PMID:16872859; http://dx.doi.org/10.1016/j.micinf.2006.06.005
- Aguiar LM, Antonangelo L, Vargas FS, Zerbini MC, Sales MM, Uip DE, Saldiva PH. Malignant and tuberculous pleural effusions: immunophenotypic cellular characterization. Clinics (Sao Paulo) 2008; 63:637-44; PMID:18925324; http://dx.doi.org/10.1590/S1807-59322008000500012
- Huang Y, Huang Y, Fang Y, Wang J, Li Y, Wang N, Zhang J, Gao M, Huang L, Yang F, et al. Relatively low level of antigen-specific monocytes detected in blood from untreated tuberculosis patients using CD4+ T-cell receptor tetramers. Plos Pathog 2012; 8:e1003036; PMID:23209409; http://dx.doi.org/10.1371/journal.ppat.1003036
