ABSTRACT
We have recently shown that HMGA2 is overexpressed in esophageal squamous cell carcinoma (ESCC) and its detection allows to discriminate between cancer and normal surrounding tissue proposing HMGA2 as a novel diagnostic marker. Interestingly, esophageal adenocarcinoma shows an opposite behavior with the overexpression of HMGA1 but not HMGA2. Moreover, we show that the suppression of HMGA2 in 2 ESCC cell lines reduces the malignant phenotype. Then, this paper highlights a differential induction of the HMGA proteins, depending on the cancer histological type, and reinforces the perspective of an innovative esophageal cancer therapy based on the suppression of the HMGA protein function and/or expression.
We have recently published a study aiming to analyze the expression of the HMGA proteins in human esophageal carcinomas.Citation1 Esophageal Cancer (EC) is the tumor originating from the esophagus.Citation2 Since 2012, EC responds for the eighth position in incidence and the sixth in mortality among all tumor types, being characterized as a highly lethal tumor.Citation2,3 The high lethality rates exihibited by this tumor are mostly due to the lack of symptoms until late stages of the disease progression which leads to EC late diagnosis and poor prognosis.Citation2,3 In addition to this scenario, there is also a lack of deep knowledge on the mechanisms involved in the genesis and progession of the disease. There are 2 main EC histotypes: EAC (esophageal adenocarcinoma) and ESCC (esophageal squamous cell carcinoma) which widely differ in associated etiological factors, geographical incidence and molecular alterations involved in disease genesis and progression. Nevertheless, ESCC is the most prevalent histotype, responding for about 80% of all EC cases.Citation4
EAC arises from the glandular cells present in the lower third of the esophagus, more precisely in the gastroesophageal junction. Epidemiologic studies indicate that EAC results from a complex etiology, figuring among the main risk factors associated with its genesis and progression: gastroesophageal reflux disease (GERD), Barrett's Esophagus (BE), obesity and tobacco consumption. Citation5 Continuos GERD has already been described as the main risk factor leading to a premalignant condition characterized by the replacement of the normal stratified squamous epithelium lining of the esophagus by simple columnar epithelium with goblet cells, a phenomenon known as BE, and is highly related with EAC development. Citation5 BE patients posses an increased risk for EAC of around 30 times.Citation6,7 EAC incidence is particularly higher in developed countries, such as United States, United Kingdom, Ireland and The Netherlands,Citation5 where obesity represents an endemic condition.
Differently from EAC, ESCC originates from the epithelial cells that coat the esophagus. This cancer also possesses a highly complex etiology and presents a particular characteristic which is the remarkable great geographical variation of its incidence worldwide, with low incidence areas surrounded by areas in which the incidence is up to 10 times higher. The high ESCC incidence areas are represented by the “Central Asian Esophageal Cancer Belt,” region ranging from Caspian Sea to Central China, south and east of Africa and parts of South America. Exposure to different etiological agents may be involved in development of ESCC in the high incidence areas. For instance, the typical risk factors associated with ESCC genesis and development are alcohol, tobacco and opium consumption, very hot beverages drinking habits, dietary exposure to N-nitrose compounds and mycotoxins. Other related factors are nutritional deficiencies, low socioeconomic status and poor oral hygiene. ESCC is more incident among men than women, what could be explained by the greater exposition of men to the risk factors associated with this disease (reviewed in Citation8 ).
The High Mobility Group A protein family includes HMGA1a, HMGA1b (coded for by the HMGA1 gene through an alternative splicing) and HMGA2, product of the homonimous gene.Citation11These proteins have not only been associated to neoplastic malignancies, but they have a causal role in cell transformation and cancer progression. Indeed, they are overexpressed in several human tumors, whereas they are not expressed, or just at low levels, in normal adult tissues. Conversely, their expression is very abundant during embryogenesis.Citation9 Moreover, several studies have shown that the silencing of HMGA expression reverts the neoplastic phenotypeCitation10,11 and transgenic mice carrying the HMGA1 or HMGA2 genes develop many types of neoplasias. Citation9
Even though HMGA1 and HMGA2 have a similar structure including 3 AT hook domains and a similar expression pattern, they surely have different functions as demonstrated by the different phenotype of the HMGA1 and HMGA2 null mice.Citation9 Indeed, Hmga1-null and heterozygous mice develop cardiac hypertrophy and type 2 diabetes showing that a suitable quantity of HMGA1 protein is needed for cardiomyocyte cell growth and modulation of the insulin pathway.Citation12,13 On the other hand, Hmga2-null and heterozygous mice exhibited a pygmy phenotype with a reduction in body size of 60%, in comparison with the wild-type mice, and a strong decrease of fat tissue,Citation14,15 supporting a central role of HMGA2 in the regulation of body size and adipocyte differentiation and proliferation. Interestingly, a correlation between overexpression of HMGA family members and a poor prognosis has been widely reported.Citation9,16 The analysis of HMGA protein in our study revealed a very high HMGA2 expression levels in ESCC samples, whereas it was almost absent in EAC tumors. Conversely, we detected a low HMGA1 protein expression in ESCC tumors whereas it was high in EAC samples. This result sounds very interesting, since the differential expression pattern of HMGA1 and HMGA2 in EAC and ESCC could reflect the complex and widely divergent etiology and genesis of these tumors.Citation17,8 Moreover, the correlation of a type of HMGA with a specific histotype further suppports the diversity between HMGA1 and HMGA2 as already suggested from their respective KO mice. Interestingly, preliminary results show an increased expression of HMGA2, but not HMGA1, in squamous skin (Chiappetta et al., manuscript in preparation) and larynx (Palumbo Júnior et al., manuscript in preparation) carcinomas, suggesting that the HMGA2 specific induction is associated somehow with the squamous histotype.
Intriguingly, it has been previously reported that crescent HMGA1 expression along BE progression may be associated with the malignant development of EAC.Citation18 Nonetheless, despite the clues correlating HMGA1 overexpression and the development of the esophageal glandular tumor, the functional consequences of this altered expression along EAC progression has not been fully addressed yet. Therefore, due to the notorious absence of HMGA1 and HMGA2 expression in adult tissues,Citation9,16 tracking their overexpression along tumorigenesis could represent a great opportunity to reveal the distinct “malignant roots” that differentially guide EAC and ESCC appearance.
In our study we demonstrate that HMGA2 mRNA expression was capable of clearly distinguishing ESCC from healthy esophageal tissue and tumor surrounding mucosa that, even though histologically normal, already harbors molecular alterations not found in esophageal healthy tissues.Citation8 In this way, the identification of a molecular marker differentially expressed in ESCC samples, tumor surrounding mucosa and healthy esophageal tissue represents a potential improvement for ESCC early detection in the near future. Nonetheless, despite the high levels of HMGA2 transcript and protein detected in the ESCC samples evaluated in our study, its overexpression did not significantly impact on ESCC patients overall survival. This could be due to the fact that most of the ESCC samples used in our study were derived from advanced tumor stages, due to the lack of clinical symptoms and screening approaches, which consists in a technical limitation to assess early ESCC stage samples and efficiently study the development of ESCC by translational studies. Moreover, while the development of EAC is commonly preceded by pathological alterations, such as Barrett's metaplasia and dysplastic lesions, which represent the initial steps of this tumor evolution, Citation5 ESCC does not present such a clear evolution represented by well defined pathological conditions. Furthermore, the advanced stage of the cancer patients analyzed in our study has not allowed us to detect early molecular alterations involved in tumorigenesis. It has already been reported that driver molecular alterations represent the early molecular events in malignant transformation that generate a genetic imbalance which leads to additional secondary alterations and a consequent highly undifferentiated and aggressive tumor phenotype.Citation19 In line with this, it has been reported that the alterations in HMGA family members represent an early event in tumorigenesis, Citation9,16,20,21 thus suggesting that HMGA overexpression may take place as an initial step in ESCC genesis and lead to a more aggressive tumor phenotype along its progression by regulating a cascade of downstream molecular events.
Therefore, despite the lack of prognostic value, in our study, HMGA2 mRNA expression levels presented a high diagnostic potential, being capable of clearly distinguishing ESCC from healthy and surrounding esophageal tissues, as revealed by the receiver operation characteristics (ROC) analysis, which allows to discriminate between 2 patient states (“diseased” and “nondiseased”).Citation22 This finding is of great importance for ESCC course, specially if we consider that a new methodology for detecting HMGA2 mRNA in the plasma of patients affected by ovarian carcinomas has been recently reported.Citation23 Hence, it suggests the possibility to test the discriminatory potential of HMGA2 mRNA expression in individuals from ESCC risk populations, by using a minimally invasive approach. Moreover, taking into account that HMGA1 seems to play an important role for EAC genesis and development, similar approaches should be employed in order to test the HMGA1 prognostic and diagnostic potential for this tumor. In line with this, an efficient method to measure HMGA1 protein in the plasma of patients with diverse neoplasms is currently being developed by our group (manuscript in preparation).
In our study we have also analyzed possible epigenetic mechanisms that could account for the HMGA2 overexpression in ESCC. The HMGA1-pseudogenes HMGA1P6 and HMGA1P7, that have been demonstrated to regulate both the HMGA proteins,Citation24,25 are not induced in ESCC (unpublished results). Moreover, some ananalyzed miRNAs already reported to target HMGA2 gene do not show changes in their expression (unpublished results). These results suggest that at least these epigenetic mechanisms are not involved in the regulation of HMGA2 protein levels in ESCC.
Finally, we have also reported that the inhibition of HMGA2 expression reduces the malignant phenotype of esophageal squamous carcinoma cells. Therefore, these results support an approach of ESCC therapy based on the suppression of HMGA protein synthesis. This is corroborated by other recent studies reporting that HMGA1 silencing restores normal stem cell characteristics in colon cancer stem cells,Citation26 besides rescuing the sensitivity of colon and thyroid cancer cells to antineoplastic drugs.Citation27 Moreover, this study highlights the need of investigating the possible malignant phenotype reversion of EAC upon HMGA1 silencing, in order to understand and propose this pathway as a potential future approach for EAC therapy.
In conclusion, this study supports the critical role of HMGA proteins in esophageal cancer and suggests new perspectives for the development of future strategies for the management of this disease, thus improving esophageal cancer diagnosis, prognosis and treatment ().
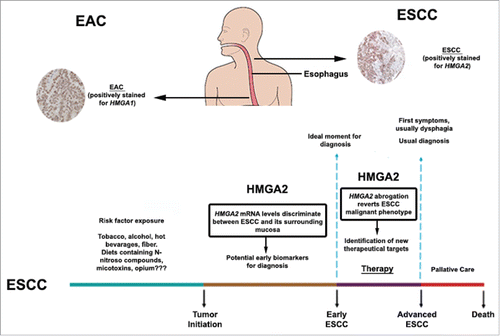
Figure 1. HMGA involvement in esophageal carcinogenesis. As shown by Palumbo Júnior et al., 2016, HMGA proteins are differentially induced in esophageal cancer, depending on the histolopathological tumor type. HMGA2, but not HMGA1, is overexpressed in esophageal squamous cell carcinoma (ESCC) and its mRNA levels discriminate ESCC from tumor surrounding mucosa, pointing out its potential role as an ESCC early diagnostic marker. In addition, HMGA2 abrogation is capable of reverting ESCC malignant phenotype, revealing HMGA2 as an innovative target for ESCC cancer therapy, as shown in the figure ilustrating ESCC progression accross time (Adpated from Meireles Da Costa et al., 2013 – reference Citation8). Conversely, HMGA1, but not HMGA2, is overexpressed in esophageal adenocarcinoma (EAC).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Palumbo Jr A, Meireles Da Costa N, Esposito F, De Martino M, D'Angelo D, de Sousa VP, Martins I, Nasciutti LE, Fusco A, Ribeiro Pinto LF. HMGA2 overexpression plays a critical role in the progression of esophageal squamous carcinoma. Oncotarget 2016; 7:25872-84.
- Montgomery EA, Bosman FT, Brennan P, Malekzadeh R. Oesophageal Cancer. In World Cancer Report 2014. World Health Organization; 2014: 528-43.
- Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut 2015; 64(3):381-7; PMID:25320104; http://dx.doi.org/10.1136/gutjnl-2014-308124
- Vizcaino AP, Moreno V, Lambert R, Parkin D M. Time trends incidence of both major histologic types of esophageal carcinomas in selected countries. Int J Cancer 2002; 99(6):860-8
- Rubenstein JH, Shaheen NJ. Epidemiology, Diagnosis, and Management of Esophageal Adenocarcinoma. Gastroenterology (Reviews in Basic and Clinical Gastroenterology and Hepatology) 2015; 149:302-17
- Yousef F, Cardwell C, Cantwell MM, Galway K, Johnston BT, Murray L. The incidence of esophageal cancer and high-grade dysplasia in Barrett's esophagus: a systematic review and meta-analysis. Am J Epidemiol 2008; 168(3):237-49; PMID:18550563; http://dx.doi.org/10.1093/aje/kwn121
- Sharma P, Marcon N, Wani S, Bansal A, Mathur S, Sampliner R, Lightdale C. Non-biopsy detection of intestinal metaplasia and dysplasia in Barrett's esophagus: a prospective multicenter study. Endoscopy 2006; 38(12):1206-12; http://dx.doi.org/10.1055/s-2006-944974
- Meireles Da Costa NM, Lima SCS, de Almeida Simão T, Ribeiro Pinto LF. The potential of molecular markers to improve interventions through the natural history of oesophageal squamous cell carcinoma. Biosci Rep 2013; 33(4):e00057; PMID:23837802
- Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer 2007; 7:899-910; PMID:18004397; http://dx.doi.org/10.1038/nrc2271
- Berlingieri MT, Manfioletti G, Santoro M, Bandiera A, Visconti R, Giancotti V, Fusco A. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol Cell Biol 1995; 15:1545-53; PMID:7862147; http://dx.doi.org/10.1128/MCB.15.3.1545
- Berlingieri MT, Pierantoni GM, Giancotti V, Santoro M, Fusco A. Thyroid cell transformation requires the expression of the HMG1 proteins. Oncogene 2002; 21:2971-80; PMID:12082527; http://dx.doi.org/10.1038/sj.onc.1205368
- Foti D, Chiefari E, Fedele M, Iuliano R, Brunetti L, Paonessa F, Manfioletti G, Barbetti F, Brunetti A, Croce CM, et al. Lack of the architectural factor HMGA1 causes insulin resistance and diabetes in humans and mice. Nat Med 2005; 11:765-73; PMID:15924147; http://dx.doi.org/10.1038/nm1254
- Fedele M, Fidanza V, Battista S, Pentimalli F, Klein-Szanto AJ, Visone R, De Martino I, Curcio A, Morisco C, Del Vecchio L, et al. Haploinsufficiency of the Hmga1 gene causes cardiac hypertrophy and myelo-lymphoproliferative disorders in mice. Cancer Res 2006; 66:2536-43; PMID:16510570; http://dx.doi.org/10.1158/0008-5472.CAN-05-1889
- Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995; 376:771-4; PMID:7651535; http://dx.doi.org/10.1038/376771a0
- Anand A, Chada K. In vivo modulation of Hmgic reduces obesity. Nat Genet 2000; 24:377-80; PMID:10742101; http://dx.doi.org/10.1038/74207
- Pallante P, Sepe R, Puca F, Fusco A. High mobility group a proteins as tumor markers. Front Med 2015; 2:15; PMID:25859543; http://dx.doi.org/10.3389/fmed.2015.00015
- Gevaert O, Daemen A, De Moor B, Libbrecht L. A taxonomy of epithelial human cancer and their metastases. BMC Med Genomics 2009; 2(1):1; PMID:19128478; http://dx.doi.org/10.1186/1755-8794-2-69
- Chen X, Lechago J, Ertan A, Ergun G, Verm R, Bridges M, Younes M. Expression of the high mobility group proteins HMGI (Y) correlates with malignant progression in Barrett's metaplasia. Cancer Epidemiol Biomarkers Prev 2004; 13(1):30-33; http://dx.doi.org/10.1158/1055-9965.EPI-03-0151
- Yap TA, Gerlinger M, Futreal P A, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Translati Med 2012; 4(127):10; http://dx.doi.org/10.1126/scitranslmed.3003854
- Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review). Int J Oncol 2008; 32(2):289-305; PMID:18202751
- Fedele M, Fusco A. HMGA and cancer. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 2010; 1799(1):48-54; http://dx.doi.org/10.1016/j.bbagrm.2009.11.007
- Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Int Med 2013; 4(2):627; PMID:24009950
- Galdiero F, Romano A, Pasquinelli R, Pignata S, Greggi S, Vuttariello E, Losito NS. Detection of high mobility group A2 specific mRNA in the plasma of patients affected by epithelial ovarian cancer. Oncotarget 2015; 6(22):19328; PMID:25749380; http://dx.doi.org/10.18632/oncotarget.2896
- Esposito F, De Martino M, Petti MG, Forzati F, Tornincasa M, Federico A, Arra C, Pierantoni GM, Fusco A. HMGA1 pseudogenes as candidate proto-oncogenic competitive endogenous RNAs. Oncotarget 2014; 5:8341-54; PMID:25268743; http://dx.doi.org/10.18632/oncotarget.2202
- Esposito F, De Martino M, D'Angelo D, Mussnich P, Raverot G, Jaffrain-Rea ML, Fraggetta F, Trouillas J, Fusco A. HMGA1-pseudogene expression is induced in human pituitary tumors. Cell Cycle 2015; 14:1471-75; PMID:25894544; http://dx.doi.org/10.1080/15384101.2015.1021520
- Puca F, Colamaio M, Federico A, Gemei M, Tosti N, Bastos AU, Fusco A. HMGA1 silencing restores normal stem cell characteristics in colon cancer stem cells by increasing p53 levels. Oncotarget 2014; 5(10):3234-45; PMID:24833610; http://dx.doi.org/10.18632/oncotarget.1914
- D'Angelo D, Borbone E, Palmieri D, Uboldi S, Esposito F, Frapolli R, Fusco A. The impairment of the High Mobility Group A (HMGA) protein function contributes to the anticancer activity of trabectedin. Eur J Cancer 2013; 49(5):1142-51; http://dx.doi.org/10.1016/j.ejca.2012.10.014
