Mitochondria are organelles of symbiotic origin that have retained a gene expression machinery during evolution. However, the large majority of the genes encoding mitochondrial proteins have been transferred to the nuclear genome, requiring cytoplasmic translation and mitochondrial import of about 1000 different mitochondrial proteins. In human cells, 13 proteins are encoded by the mitochondrial genome and their translation occurs on mitochondrial ribosomes in the matrix. While the mitochondrial ribosomal proteins and translation factors need to be imported from the cytoplasm, the 2 ribosomal (r)RNAs and the minimalistic set of 22 transfer (t)RNAs are encoded in the organelle and are transcribed by the mitochondrial RNA polymerase. In contrast to both the bacterial and the cytoplasmic translation systems, where separate tRNAs exist that mediate incorporation of methionine either during translation initiation or elongation, mitochondria contain only a single tRNA that facilitates incorporation of methionine during translation (). In addition, this single mitochondrial tRNAMet (mt-tRNAMet) is employed to not only read the conventional AUG codon, but is also responsible for integration of methionine at AUA and AUU codons during translation initiation and at AUA codons during elongation, thereby playing a key role in implementing the non-conventional genetic code of mitochondria. It was previously suggested that RNA modifications in the anticodon of the mt-tRNAMet could expand its codon recognition, however, how the modifications are installed and which enzymes are involved had remained unknown.
Figure 1. Wobble position modifications in the mitochondrial tRNAMet expand codon recognition during translation. While cytoplasmic translation employs 2 different tRNAMet for translation initiation (tRNAiMet) and elongation (tRNAeMet), mitochondria contain only one (mt-)tRNAMet. Cytosine 34 (C34) of the mt-tRNAMet can be methylated by the RNA methyltransferase NSUN3 to generate m5C34, which can be further oxidised by the dioxygenase ABH1/ALKBH1 to 5-formylcytosine (f5C34).
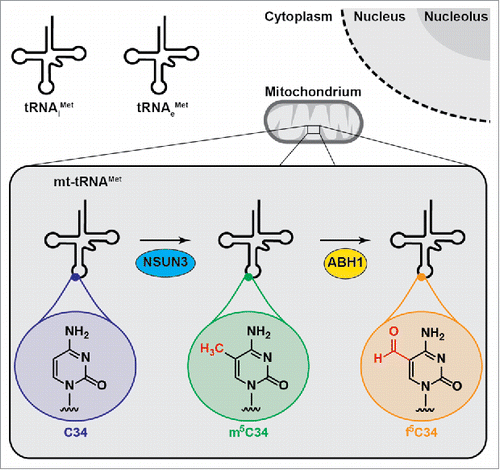
In parallel with 2 independent studies, we found that cytosine 34 (C34) in the "wobble position" of mt-tRNAMet is methylated at position 5 of the pyrimidine ring by the RNA methyltransferase NSUN3 ().Citation1,2,3 This enzyme is a member of the Nol1/Nop2/SUN domain (NSUN) family, which also contains the RNA methyltransferases NSUN2 and NSUN6 that modify cytoplasmic tRNAs.Citation4,5 In contrast, NSUN3 localizes to the mitochondrial matrix where it specifically recognizes the anticodon stem loop (ASL) of mt-tRNAMet. Interestingly, mutations that compromise basepairing in the ASL, including a pathogenic mutation, reduce C34 methylation by NSUN3, implying that lack of this modification in mt-tRNAMet can lead to disease.Citation1,2 This is further supported by Van Haute and colleagues, who describe a patient lacking functional NSUN3 and suffering from mitochondrial dysfunction. Interestingly, previous reports have suggested the presence of 5-formylcytosine (f5C) at position 34 of mt-tRNAMet, implying that the methyl group of m5C installed by NSUN3 can be oxidised to generate the formyl group of f5C. We have identified the Fe(II)/α-ketoglutarate-dependent dioxygenase, ALKBH1/ABH1 as the enzyme responsible for this oxidation ().Citation3 The related TET proteins, which oxidise m5C in DNA, have been shown to form f5C and 5-carboxycytosine (ca5C) via a distributive mechanism that leads to accumulation of 5-hydroxymethylcytosine (hm5C), as the first oxidation intermediate. In contrast in vitro and in vivo data imply that ABH1 primarily produces f5C in mt-tRNAMet. Cytosine 34 of mt-tRNAMet is almost fully modified in vivo,Citation1 however, the relative abundance of tRNAs carrying the different modifications at this position requires further clarification; while both mass spectrometry analysis of isolated mt-tRNAMet and bisulfite sequencing predominantly identified f5C,Citation1,2,3 the presence of m5C could also be detected,Citation1,3 suggesting that although the majority of mt-tRNAMet is oxidised by ABH1, a portion may remain in the methylated state. It has been discussed that the localization of ABH1 may differ between cell types, raising the possibility that the extent of oxidation of m5C34 of mt-tRNAMet may also vary. Ribosome binding studies using differently modified forms of mt-tRNAMet (or the ASL) indicate that these modifications serve to expand codon recognition by mt-tRNAMet, enabling this single methionine tRNA to fulfil its diverse functions in mitochondrial translation.Citation1,6 The importance of the increased decoding capacity of mt-tRNAMet generated by modification of C34 is highlighted by the requirement for NSUN3 and ABH1 for efficient mitochondrial translation in vivo.Citation1,2,3
The newly identified 2-step modification pathway involving the m5C RNA methyltransferase NSUN3 and the dioxygenase ABH1 explains how codon recognition by mt-tRNAMet is extended by RNA modifications at the “wobble position” of its anticodon. This enables the single mt-tRNAMet to mediate the incorporation of methionine on different codons and to act in both translation initiation and elongation in human mitochondria. Interestingly, such complex, multi-step modifications are also observed at the “wobble position” of other mitochondrial tRNAs and similarly function to alter codon recognition during mitochondrial translation (reviewed in ref.Citation7 and references therein). For example, the 5-taurinomethyluridine (τm5U) modification at position 34 of mt-tRNATrp, mediated by GTPBP3 and MTO1, allows incorporation of tryptophan at the UGA codon, which is normally read as a stop codon by the cytoplasmic translation machinery. Analogous to NSUN3, mutations in both these enzymes have been shown to cause mitochondrial dysfunction. Therefore, RNA modifications at key positions in the anticodon emerge as important features that expand codon recognition by specific tRNAs and thereby enable use of the minimalistic mitochondrial translation system. Furthermore, these findings add to the growing body of evidence for genetic diseases that are caused by a lack of tRNA modifications or compromised mitochondrial function.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, Hübner B, Seikowski J, Dennerlein S, Rehling P, et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J 2016; 35:2104–2119; PMID: 27497299; https://dx.doi.org/10.15252/embj.201694885
- Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNAMet. Nat Chem Biol 2016; 12(7):546–51; PMID: 27214402; https://dx.doi.org/10.1038/nchembio.2099
- Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S, et al. Deficient methylation and formylation of mt-tRNAMet wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun 2016; 7:12039. PMID: 27356879; https://dx.doi.org/10.1038/ncomms12039
- Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res 2006; 34(20):6034–43. PMID:17071714; https://dx.doi.org/10.1093/nar/gkl765
- Haag S, Warda AS, Kretschmer J, Günnigmann MA, Höbartner C, Bohnsack MT. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 2015; 21(9):1532–43; PMID: 26160102; https://dx.doi.org/10.1261/rna.051524.115
- Bilbille Y, Gustilo EM, Harris KA, Jones CN, Lusic H, Kaiser RJ, Delaney MO, Spremulli LL, Deiters A, Agris PF. The human mitochondrial tRNAMet: structure/function relationship of a unique modification in the decoding of unconventional codons. J Mol Biol; 406(2):257–74; PMID: 21168417; https://dx.doi.org/10.1016/j.jmb.2010.11.042
- Powell CA, Nicholls TJ, Minczuk M. Nuclear-encoded factors involved in post-transcriptional processing and modification of mitochondrial tRNAs in human disease. Front Genet 2015; 6:79; PMID: 25806043; https://dx.doi.org/10.3389/fgene.2015.00079
