Cell division is essential to the development and proliferation of all organisms. Each and every step of cell division has to occur and resume at a certain time to ensure the correct propagation of the genetic material to the daughter cells. More than a century of research in the field has identified and described a fascinating network of factors that finely regulate each step of the process. This tight regulation includes, for instance, establishment and resolution of sister chromatid cohesion during cell cycle.Citation1 Most of the factors involved in cell division, either proteins or RNAs, are themselves regulated in terms of abundance across the cell cycle.Citation2 Moreover, several of them, which physiologically control cell cycle, become dysfunctional in cancer, where they no longer drive accurate cell proliferation.Citation2
Over the last decade, the rapid growth of genomics has revealed that the human genome is more widely transcribed than previously thought, and that protein-coding genes represent only a small portion of the total. In fact, while less than 2% of the human genome encodes for proteins, at least 75% of its total is actively transcribed into noncoding RNAs. A subset of these transcripts are long (>200 nt), capped and polyadenylated RNAs transcribed by RNA polymerase II, and are collectively called long noncoding RNAs (lncRNAs).Citation3 Although some lncRNAs are known to be functional, thousands of them are still uncharacterized, and their roles remain an open question.
Our work describes a novel human lncRNA, named CONCR (cohesion regulator noncoding RNA) involved in DNA replication and sister chromatid cohesion.Citation4 We show that CONCR expression is periodic in the cell cycle, suggesting that the lncRNA has a tightly-regulated temporal function in the cell. Furthermore, as previously shown for protein coding genes, such as cyclins or growth factors, may represent an example of noncoding gene fundamental to cycling cells. Indeed, we observed that depletion of CONCR by siRNA or genome editing affects DNA replication and cell cycle progression, leading to decreased cell proliferation and increased cell death.
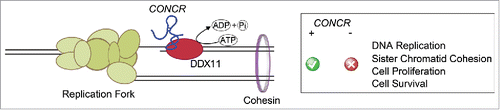
Cells depleted of CONCR show severe defects in sister chromatid cohesion. Both the aspect and the percentages of the phenotypic classifications of the chromatids aspect, i.e. “loosely paired” or “completely separated," appeared comparable across different cell lines, suggesting a general role for CONCR in cohesion establishment during cell division. CONCR physically interacts with DDX11 (DEAD/H box protein 11), a DNA helicase known to be involved in DNA replication and sister chromatid cohesion.Citation5 Mutations in DDX11 have been in fact associated to a rare pathological condition known as Warsaw breakage syndrome; a syndrome characterized at cellular level by sister chromatid cohesion defects.Citation5 We found that CONCR and DDX11 co-localize on the chromatin and that silencing of the lncRNA reduces the binding of DDX11 to DNA at replicating regions. An interesting question that remains to be explored in more detail is why DDX11 needs an accessory lncRNA to function. In our attempt to understand this point, we performed DDX11 ATPase assays in vitro and observed that the binding of CONCR to DDX11 enhances the ATPase activity of the helicase. Although further evidences are needed, our findings are in line with the known common features of the DEAD-box proteins. In fact, a common property of this family of enzymes is that their ATPase activity is stimulated by RNA.Citation6 Indeed, many DEAD-box proteins have been shown to bind cooperatively RNA and adenosine nucleotides, which ultimately results in enhanced ATPase kinetics.Citation6
CONCR may act as an oncogene. It is transcriptionally controlled by c-MYC and presents higher levels in tumor cells with inactive p53. CONCR expression is significantly increased in a number of tumor types, suggesting a contribution of the lncRNA to the development and maintenance of the tumor.
In summary, our results illustrate how, through the modulation of the activity of a protein, a lncRNA is able to regulate DNA replication and chromosome dynamics (). It is plausible that other lncRNAs, either by tuning-up the function of proteins involved in the replication machinery, or by regulating the structure of the chromatin, impact the capacity of cells to accurately replicate their DNA and propagate their genomes faithfully to the next generation. These RNAs, which represent a so far unknown component of the cell networks, may influence the likelihood of a cell to become malignantly transformed.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
FPM and MH are supported by the European Research Council Starting Grant 281877 and the Spanish Ministry of Science SRYC1100I008347XV0.
References
- Peters JM, Nishiyama T. Sister chromatid cohesion. Cold Spring Harb Perspect Biol 2012; 4; PMID:23043155; http://dx.doi.org/10.1101/cshperspect.a011130
- Whitfield ML et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 2002; 13:1977-2000; PMID:12058064; http://dx.doi.org/10.1091/mbc.02-02-0030
- Djebali S et al. Landscape of transcription in human cells. Nature 2012; 489:101-8; PMID:22955620; http://dx.doi.org/10.1038/nature11233
- Marchese FP et al. A long noncoding RNA regulates sister chromatid cohesion. Mol Cell 2016; 63:397-407; PMID:27477908; http://dx.doi.org/10.1016/j.molcel.2016.06.031
- Bharti SK et al. Molecular functions and cellular roles of the ChlR1 (DDX11) helicase defective in the rare cohesinopathy Warsaw breakage syndrome. Cell Mol Life Sci 2014; 71:2625-39; PMID:24487782; http://dx.doi.org/10.1007/s00018-014-1569-4
- Russell R, Jarmoskaite I, Lambowitz AM. Toward a molecular understanding of RNA remodeling by DEAD-box proteins. RNA Biol 2013; 10:44-55; PMID:22995827; http://dx.doi.org/10.4161/rna.22210