ABSTRACT
In response to replication-blocking lesions, proliferating cell nuclear antigen (PCNA) can be sequentially ubiquitinated at the K164 residue leading to 2 modes of DNA-damage tolerance, namely translesion DNA synthesis (TLS) and error-free lesion bypass. Ectopic expression of PCNA fused with ubiquitin (Ub) lacking the 2 C-terminal Gly residues resembles PCNA monoubiquitination-mediated TLS. However, if the fused Ub contains C-terminal Gly residues, it is further polyubiquitinated and inhibits cell proliferation. Unexpectedly, the polyubiquitination chain does not require any surface Lys residues and is likely to be head-to-tail linked. Such PCNA polyubiquitination interferes with replication, arrests cells at the S-phase and activates the p53 checkpoint pathway. The above cell-cycle arrest is reversible in an ATR-dependent manner, as simultaneous inhibition of ATR, but not ATM, induces apoptosis. Since ectopic expression of PCNA-Ub also induces double-strand breaks that colocalize with single-stranded DNA, we infer that this non-canonical PCNA poly-Ub chain serves as a signal to activate ATR checkpoint and recruit double-strand-break repair apparatus.
Introduction
Proliferating cell nuclear antigen (PCNA) is a DNA polymerase processivity factor serving as a loading scaffold for the replication machinery through association with various replication-related factors.Citation1 During replication, PCNA is loaded to DNA after chaperon-like replication factor C (RFC) binds to the RNA primer-DNA template junction.Citation2,3 Polα is released and Polε is loaded to mediate leading-strand elongation.Citation4 For the discontinuous lagging strand, short Okazaki fragments are firstly produced by Polα and Polδ.Citation5 The initiator RNA sequences are then removed from the newly synthesized Okazaki fragments by Flap structure-specific endonuclease-1 (FEN-1).Citation6 The processed Okazaki fragments are then ligated via DNA ligase I. During this process, PCNA can directly interact with Polδ, Polε, FEN-1 and DNA ligase I to stabilize their association with DNA and stimulate their enzymatic activity while it encircles DNA.Citation7-9
PCNA plays important roles not only in DNA replication but also in several DNA damage-responsive pathways.Citation1 Living organisms are constantly challenged by various sources of DNA damage including endogenous metabolites and environmental agents such as UV irradiation and chemicals that often cause DNA replication blocks. Living organisms have evolved sophisticated tolerance mechanisms, including translesion synthesis (TLS) and error-free DNA damage tolerance (DDT) to deal with these replication-blocking lesions. Interestingly, all 3 lesion-bypass pathways require PCNA modification by ubiquitination.Citation10-12 In budding yeast, PCNA can be monoubiquitinated by the E2-E3 Rad6-Rad18 complex at the K164 residue and further modified with a K63-linked Ub chain by another E2-E3 complex, Mms2-Ubc13-Rad5.Citation13 The non-canonical K63-linked Ub chain plays crucial roles in regulating various cell-signaling pathways by altering the target protein activity, which is different from conventional K48-linked Ub chains that target proteins for degradation by the 26S proteasome.Citation14,15
Recent studies in mammalian cells suggest a model by which monoubiquitinated PCNA recruits TLS polymerases through an enhanced physical interaction and facilitates TLS.Citation16 Polyubiquitinated PCNA was also detected in cultured mammalian cells upon UV irradiation,Citation17 and 2 Rad5 homologs, SHPRHCitation18,19 and HLTF,Citation20,21 appear to be required for this polyUb chain assembly. Polyubiquitination of PCNA is thought to mediate error-free lesion bypass via template switching,Citation22 although it is unclear how this is achieved. Homologous recombination (HR) is one of the important mechanisms used by cells to resolve replication stress of stalled replication forks,Citation23 which is considered a high-fidelity process since it utilizes homologous sequence as the template. DNA double strand breaks (DSBs) can be sensed by an Mre11-Rad50-NBS1 (MRN) complex, which initiates recruitment and activation of the ATM kinase.Citation24 Meanwhile, large tracts of single strand DNA (ssDNA) emerge due to uncoupling of DNA synthetic enzymes at stalled DNA replication forks.Citation25,26 Binding of replication protein-A (RPA) to ssDNA specifically activates the ATR kinase, which is dependent on the association of Ataxia-Telangiectasia and Rad3-related–ATR-Interacting Protein (ATR–ATRIP), TopBP1 and the Rad9-Rad1-Hus1 (9-1-1) complex.Citation25,27 ATR or ATM then rapidly phosphorylate hundreds of downstream targets including transducers Chk1 or Chk2, respectively, as well as tumor suppressors like p53 and BRCA1, which regulate cell cycle checkpoints and DNA repair.Citation28 Moreover, several pieces of indirect evidence suggest that ATR promotes HR as ATR phosphorylates a number of substrates known to directly affect HR.Citation29-33
In this study, we surprisingly found that monoubiquitinated PCNA can be readily modified to form a Lys-independent polyUb chain, which induces the replication-stress checkpoint and recruits double-strand-repair machinery for error-free lesion bypass. This study sheds light on the role(s) of PCNA polyUb chain in signaling replication-block.
Results
PCNA-Ub fusion inhibits cell proliferation
Our previous studies of PCNA-Ub fusionsCitation34,35 removed 2 C-terminal Gly residues from Ub (UbΔG) to avoid potential polyubiquitin chain assembly. In this study, we made mammalian PCNA-Ub expression vectors so that the only difference between PCNA-Ub and PCNA-UbΔG fusions is whether the Ub contains the 2 C-terminal Gly residues (). We surprisingly found that T-Rex 293 cells carrying PCNA-Ub behave very differently from those carrying PCNA-UbΔG. While PCNA-UbΔG cells were as previously reported,Citation34 PCNA-Ub was further modified in host cells (). Furthermore, PCNA-Ub transfected cells were unable to proliferate once the fusion protein was induced by Dox (), while un-induced PCNA-Ub (Fig, 1C) and Dox-induced PCNA-UbΔG () cells grew equally well (Supplementary Fig. S1).
Figure 1. Expression of PCNA-Ub fusion causes further modification and the arrest of cell proliferation. (A) A schematic diagram of PCNA-UbΔG and PCNA-Ub fusion constructs. The PCNA ORF contains a K164R mutation to prevent in vivo PCNA ubiquitination. In addition, the Ub coding region is fused to PCNA at the C-terminus. The fusion ORFs are cloned into plasmid pcDNA5.0FRT/TO under TetO2 regulation. (B) Expression of PCNA-UbΔG and PCNA-Ub in stably-transfected T-Rex 293 cells. 2 µg/ml Dox was added to induce ectopic fusion gene expression for 2 d followed by protein gel blot (WB) using an anti-PCNA antibody. (C) Cell growth of PCNA-Ub stable transfectants with or without 2 µg/ml Dox treatment. (D) Cell growth of PCNA-UbΔG stable transfectants with or without 2 µg/ml Dox treatment. (E) Cellular distribution of GFP-UbΔG and GFP-Ub in transfected 293 cells. Cells were treated with 2 µg/ml Dox for 2 d followed by ICC with an anti-GFP antibody (green). The nuclei are stained by DAPI. (F) Expression of GFP-UbΔG and GFP-Ub fusion proteins. Experimental conditions were as in . (G,H) Cell growth of GFP-Ub (G) or GFP-UbΔG (H) stable transfectants with or without 2 µg/ml Dox treatment.
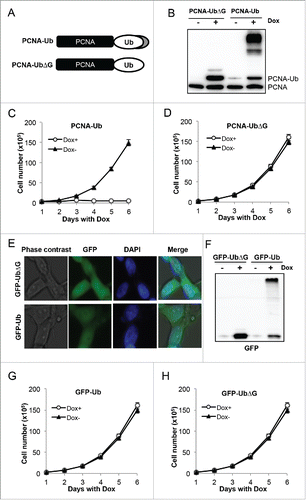
To distinguish whether the inhibitory effect is specific to the PCNA-Ub fusion or to Ub further modification in general, we made GFP-Ub and GFP-UbΔG constructs in a manner similar to PCNA fusions and created stable T-Rex293 cell lines. An immunofluorescence assay () revealed that GFP-Ub and GFP-Ub△G were expressed indistinguishably in both nucleus and cytoplasm, while WB () confirmed that GFP-Ub but not GFP-UbΔG was further modified. In contrast to the PCNA-Ub fusion, Dox-induced expression of GFP-Ub did not affect cell proliferation (). To further explore mechanistic differences between PCNA-Ub and GFP-Ub expressing cells, we separated total cellular proteins into soluble and chromatin-bound fractions. GFP-containing proteins were only found in the soluble fraction while in contrast PCNA-Ub fusion and its further modified proteins were found in both soluble and chromatin-bound fractions (Supplementary Fig. S2). The above observations are consistent with a notion that it is the further modification of chromatin-bound PCNA-Ub that causes the arrest of cell proliferation. Since PCNA as a DNA polymerase processivity factor plays pivotal roles in replication,Citation36-38 it is conceivable that the cell proliferation arrest was due to replication block by modified PCNA.
PCNA-Ub interferes with DNA replication and activates cell cycle checkpoints
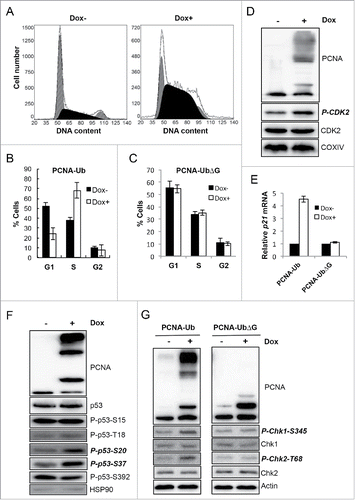
Since cell growth is inhibited strongly by PCNA-Ub, we wish to address its underlying mechanism(s). Flow cytometry analysis () indicates that upon PCNA-Ub expression, cells were arrested mainly at S-phase. In contrast, cell cycle progression was not affected by PCNA-UbΔG expression (). The activity of S-phase-related cyclin-dependent kinase 2 (CDK2) is considered a faithful indicator of S-phase cells.Citation39 While total CDK2 levels between non-induced and Dox-induced cells were comparable, the phosphorylated CDK2 level increased upon PCNA-Ub expression (). We also measured the expression level of other important tumor suppressor genes and found that the p21 transcript level was 4.5-fold higher in Dox-induced PCNA-Ub cells than uninduced cells (). Although the total p53 protein level remains unchanged (), Dox-induced PCNA-Ub expression apparently increased its phosphorylation at Ser20 and Ser37 (), indicative of ATM and ATR activation, respectively,Citation40 but not at Ser15, Thr18 or Ser392. Consistently, PCNA-Ub expression did not alter total cellular levels of Chk1 and Chk2, while enhancing their phosphorylation levels (), further implying that ATM and/or ATR checkpoint pathways were also activated by PCNA-Ub.
Figure 2. PCNA-Ub expression activates cell-cycle checkpoints. (A) PCNA-Ub expression arrested cells at S-phase. Dox-induced expression of ectopic PCNA-Ub was subject to flow cytometry. (B,C) Statistical analysis of flow cytometric data for PCNA-Ub (B) and PCNA-UbΔG (C). The results are the average of at least 3 independent experiments with standard deviations. (D) PCNA-Ub activates CDK2. After PCNA-Ub expression total CDK2 and the phosphorylated CDK2 (P-CDK2) levels were determined by WB while COXIV serves as an internal control. (E) Comparison of p21 expression by real-time RT-PCR upon PCNA-Ub or PCNA-UbΔG expression. (F) Examination of p53 phosphorylation patterns upon PCNA-Ub expression. Cellular HSP90 serves as an internal control. Note that the total p53 protein level was not altered with PCNA-Ub expression. (G) Expression of PCNA-Ub activates Chk1 and Chk2. Upon induction of PCNA-Ub, the Chk1 and Chk2 phosphorylation levels were elevated while total Chk1 and Chk2 levels remained. The expression of PCNA-UbΔG serves as an experimental control and the cellular actin level as an internal control.
PCNA-Ub modification causes DNA damage
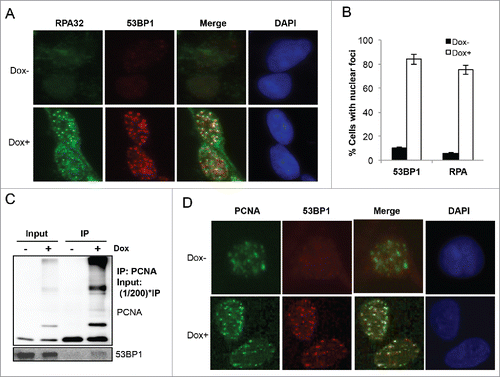
To test a hypothesis that PCNA-Ub expression signals for replication blocks, we examined the RPA nuclear focus formation. RPA is a trimeric protein complex that specifically coats ssDNA and protects ssDNA gaps at stalled replication forks.Citation41,42 In undamaged cells, RPA does not form nuclear foci even in S-phase cells.Citation34 Under our experimental conditions, RPA nuclear foci were barely detectable in uninduced cells; after Dox-induced PCNA-Ub expression discrete RPA nuclear foci were observed in the majority of cells (). Interestingly, these RPA foci are colocalized with PCNA-Ub-induced 53BP1 foci (), indicative of the occurrence of DNA damage responseCitation43 at or near ssDNA gaps. The PCNA-Ub-induced colocalization of 53BP1 and RPA foci is also consistent with the simultaneous activation of ATM and ATR. Indeed, an anti-PCNA antibody can immunoprecipitate native 53BP1 in Dox-induced PCNA-Ub cells but not in uninduced cells (), indicating the role of PCNA polyubiquitination in the recruitment of DNA damage repair apparatus. In an immunocytochemistry assay, PCNA nuclear foci were observed in both Dox-treated and untreated cells; however, 53BP1 nuclear foci were found only in Dox-induced cells and colocalize with PCNA foci (), further confirming that PCNA-Ub expression causes the local accumulation of DNA damage.
Figure 3. PCNA-Ub expression induces ssDNA and DSBs and recruits 53BP1. (A) PCNA-Ub induced 53BP1 and RPA32 nuclear foci and their colocalization. PCNA-Ub cells were treated with Dox for 2 d followed by ICC with anti-53BP1 (red) and anti-RPA32 (green) antibodies. (B) Percentage of cells with 53BP1 and RPA32 nuclear foci, with or without PCNA-Ub expression. Statistical analysis was based on at least 1000 cells for each treatment from different experiments. (C) Co-IP experiment showing the physical interaction between PCNA-Ub and 53BP1. PCNA-Ub-transfected T-Rex 293 cells were treated with Dox for 2 d and the lysates were immunoprecipitated using an anti-PCNA antibody immobilized to Protein G agarose. The immunoprecipitate was then examined by WB using either the anti-PCNA antibody (upper panel) or an anti-53BP1 antibody (lower panel). (D) PCNA-Ub colocalizes with 53BP1 in nuclear foci. PCNA-Ub cells with or without Dox treatment for 48 hours were subject to ICC with anti-53BP1 (green) and anti-PCNA (red) antibodies.
PCNA-Ub induced cell cycle arrest does not cause cell death
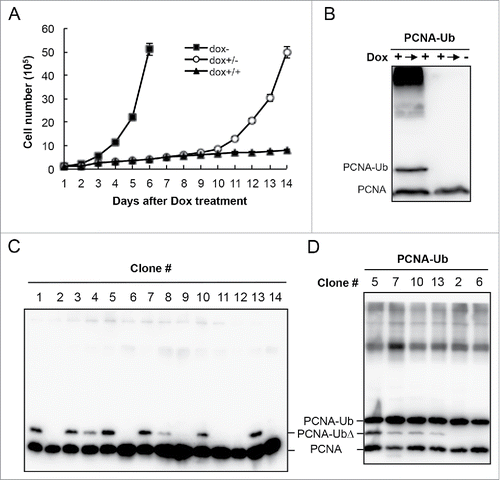
Interestingly, despite the extensive DNA damage and stalled cell proliferation, the arrested cells by PCNA-Ub expression remain viable for at least a week; once Dox is withdrawn from the culture medium, cells began to grow again () and the further modified PCNA-Ub bands disappear (), indicating that the growth-inhibitory effect is reversible. Since PCNA-Ub induced cells are viable but do not grow, we attempted to isolate and characterize PCNA-Ub resistant mutants. After two weeks of incubation in the presence of Dox, a few colonies appeared from plates containing PCNA-Ub transfected cells and WB analyses () reveal that the surviving colonies either expressed truncated PCNA-Ub (PCNA-UbΔ, e.g., Clones 1,3,4,5,7,8,10,13) or did not express PCNA-Ub at all (e.g., Clones 2,6,9,11,12,14); in both cases, heavily modified PCNA-Ub bands disappeared, leading us to believe that the cell cycle arrest is due to PCNA-Ub further modification. The above selected cell lines were transiently transfected with the plasmid expressing PCNA-Ub and monitored for the PCNA expression. All transfectants expressed the full-length PCNA-Ub and regained further PCNA-Ub modifications (), confirming that the survived cell lines had managed not to harbor or express the full-length PCNA-Ub. We conclude from the above observations that the spontaneous PCNA-Ub modification is responsible for the inhibition of mammalian cell proliferation.
Figure 4. PCNA-Ub induced cell-cycle arrest is reversible. (A,B) Effects of Dox withdrawal on cell proliferation (A) and PCNA-Ub modification by WB analysis using an anti-PCNA antibody. The stable PCNA-Ub cells were initially treated with 2 µg/ml Dox and incubated for 7 d. Half of the cells were then washed and incubated in fresh medium without Dox (Dox+/−) and another half remained for incubation (Dox+/+) for an additional 7 d. Cells were collected at the end of the experiment (14th day) for WB. (C) Characterization of cell lines tolerant to PCNA-Ub expression. Stably-transfected PCNA-Ub cells were cultured in the presence of 2 µg/ml Dox for 3 weeks. A few colonies appeared and were selected to inoculate fresh Dox medium. Cells were collected 4 d after incubation and analyzed by WB using an anti-PCNA antibody (left panel). Note that these lines either do not express PCNA-Ub, or express a truncated PCNA-Ub (PCNA-UbΔ). (D) The selected cell lines were re-transfected with PCNA-Ub and cultured in Dox medium for 2 d before WB analysis.
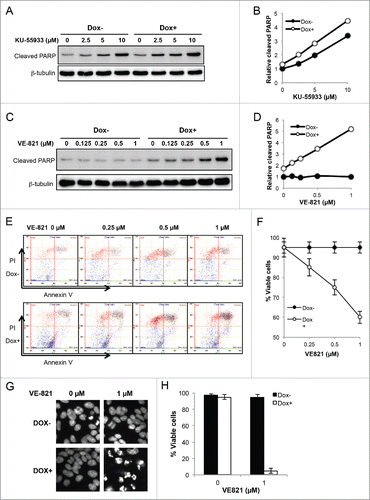
The ATR pathway is required for the maintenance of PCNA-Ub cell viability
Since Chk1 and Chk2 activation () is linked to ATM and ATR pathways, respectively, and ATM mainly senses DNA DSBs while ATR mainly senses stalled replication forks,Citation44,45 to ask which of the above checkpoint pathways is primarily responsible for PCNA-Ub-induced genome instability, we utilized 2 inhibitors, KU-55933 and VE-821, that specifically inhibit ATM and ATR functions, respectively,Citation46-48 and monitored PARP cleavage as an indicator of apoptosis.Citation49 The reported concentration of ATM/ATR inhibitors used to inhibit the ATM or ATR kinase activity is around 10 µM.Citation50-52 Treatment of PCNA-Ub-transfected cells with up to 10 µM KU-55933 induced PARP cleavage; however, there was barely any difference in the PARP cleavage efficiency between uninduced and Dox-induced cell populations (). In contrast, under concentrations of up to 1 µM VE-821 that did not induce PARP cleavage alone in uninduced cells, the inhibitor in combination with PCNA-Ub expression displayed a synergistic effect in inducing PARP cleavage (). Similar results were also obtained by flow cytometry that directly measures apoptosis (). In the above WB and flow cytometry experiments, cells were treated with Dox and inhibitors by only 2 d to collect enough cells or proteins. When cells were treated with 1 µM VE-821 for 4 days, more than 90% PCNA-Ub expression cells were dead and become floated (). We conclude from the above observations that although both ATM and ATR pathways are likely activated in PCNA-Ub expression cells, only ATR plays a critical role in protecting PCNA-Ub cells from undergoing apoptosis. Since the ATR checkpoint pathway functions to stabilize stalled replication forks,Citation53 the above observations indicates that PCNA-Ub expression signals for blocked replication forks.
Figure 5. ATR but not ATM protects PCNA-Ub cells from undergoing apoptosis. (A,B) Inhibition of ATM by KU-55933 for up to 2 d did not affect apoptosis in Dox-induced and uninduced PCNA-Ub cells. KU-55933 was added to the indicated final concentration one day after Dox induction, and apoptosis was measured by PARP cleavage. (C,D) Inhibition of ATR by VE-821 for up to 2 d sensitized PCNA-Ub expression cells to undergoing apoptosis, while it did not affect uninduced PCNA-Ub cells. VE-821 was added to the indicated final concentration one day after Dox induction and apoptosis was measured by PARP cleavage. Note that the concentration of VE-821 was 1/10 that of KU-55933. (E,F) Flow cytometric analysis of PCNA-Ub-transfected cells after 2-day treatment with VE-821 to measure apoptotic (Annexin V) and inviable (PI) cells. (E) Representative flow cytometric data. (F) Statistic analysis of the flow cytometric data with standard deviation. (G,H) Effects of 1 µM VE-821 on PCNA-Ub transfected cells after 3-day treatment. Cells were stained by Hoechst (G) to determine % viability (H).
PCNA-Ub facilitates Lys-independent polyUb chain assembly
To ask in which form(s) PCNA-Ub is modified, we immunoprecipitated PCNA-containing fractions by an anti-PCNA antibody and probed them against both PCNA and Ub antibodies. As shown in , all heavily modified bands can be recognized by an anti-Ub antibody, indicating that they are most likely PCNA-Ub polyubiquitin chains. In mammalian cells, the non-canonical K63-linked Ub chain and canonical K48-linked Ub chain play crucial roles in regulating various cell-signaling pathways by altering the target protein activity and in degradation for target protein by the 26S proteasome, respectively. In yeast cells, the Mms2-Ubc13-Rad5 complex is absolutely required for K63-linked PCNA-polyUb chain formation. Mammalian cells contain 2 Mms2 homologsCitation54,55 and at least 2 Rad5 homologs,Citation18-21 but only one Ubc13. Surprisingly, experimental depletion of Ubc13 did not affect PCNA-Ub further modifications (), nor did Ub-K48R or Ub-K63R mutations ().
Figure 6. PCNA-Ub is polyubiquitinated through an N-to-C linkage. (A) PCNA-Ub can form a polyUb chain in vivo. PCNA-Ub transfected T-Rex 293 cells were treated with 2 µg/ml Dox for 2 d. Immunoprecipitation was performed by an anti-PCNA antibody immobilized to Protein G agarose. The immunoprecipitate was then examined by WB using either an anti-PCNA antibody (left panel) or anti-Ub antibody (right panel). (B) PCNA-Ub further modification is independent of Ubc13. After the stably-transfected PCNA-Ub cells were incubated with Dox for 4 hours, they were transfected with different concentrations of siRNA against UBC13 (siUBC13) and the incubation continued for 3 d followed by WB using an anti-PCNA antibody. The Ubc13 depletion efficiency was also monitored. (C) The PCNA-polyUb chain formed by the PCNA-Ub fusion protein is not mediated by K48 or K63. Stable PCNA-Ub cells were transiently transfected with HA-UbK48R or HA-UbK63R and incubated for 6 hrs followed by Dox treatment for 2 d. Immunoprecipitation was performed by anti-HA antibody immobilized to Protein G agarose, followed by WB using an anti-PCNA antibody. (D) PCNA-Ub polyUb chains formed is not mediated by any surface Lys residues. Experimental conditions are as described in (C). (E) The PCNA-Ub polyubiquitination chain is dependent on the C-terminal Gly of Ub. Experimental conditions are the same as in (D), using HA-UbΔG instead of HA-Ub (All K-R). (F) Incorporation of HA-UbΔG interferes with polyubiquitination chain formation. Stable PCNA-Ub cells were transiently transfected with HA-UbΔG and incubated for 6 hrs followed by Dox treatment for 2 d. Immunoprecipitation was performed by anti-PCNA antibody immobilized to Protein G agarose, followed by WB using an anti-HA and anti-PCNA antibody.
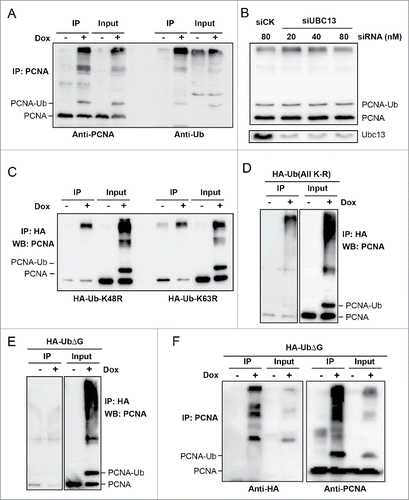
To further address the possible involvement of other surface Lys residues in the polyUb chain formation, we made an HA-tagged Ub-All(K-R) mutation and found that these Ub molecules could still be incorporated into the PCNA-polyUb chains (). In contrast, if a polyUb chain requires the C-terminus but not surface Lys of Ub, it may form an N-to-C linked linear chain. To test this hypothesis, we transiently transfected Dox-induced PCNA-Ub cells with HA-UbΔG followed by immunoprecipitation. When the C-terminal 2 Gly residues were removed from the HA-tagged Ub, the PCNA-polyUb chain could not be detected from the HA IP products (). Compared with the same cells transfected with HA-Ub or HA-Ub containing surface K-to-R mutations (), more intermediate length of molecules containing PCNA and HA-Ub was observed (). We interpret this result as indicating that UbΔG can still be linked to PCNA-Ub or PCNA-Ubn molecules; however, the incorporation of HA-UbΔG prevents further modifications and results in increased intermediate products.
Discussion
In this report, we designed carefully controlled experiments to express the PCNA-Ub fusion protein and examined its biochemical and biological effects. Unexpectedly, the expression of PCNA-Ub has very different biological consequences than those of PCNA-UbΔG expression. While expression of PCNA-UbΔG does not affect cell growth and even enhances cellular tolerance to DNA damage,Citation34 the expression of PCNA-Ub arrests cell proliferation. Since this effect is directly correlated to the further modification of PCNA-Ub and absolutely requires the C-terminal Gly residues, the cell cycle arrest by PCNA-Ub expression is attributed to the covalent modification of Ub in PCNA-Ub. Perhaps the most surprising finding from this study is that PCNA-Ub is rapidly polyubiquitinated to form most likely an N-to-C linked linear Ub chain, in contrast to the K63-linked polyUb chain as observed in budding yeastCitation13 or the canonical K48-linked polyUb chain in all eukaryotic cells. This conclusion is based on several observations. Firstly, K48R, K63R or even all K-to-R substitutions in Ub does not affect the polyUb chain formation, indicating that the polyUb chain does not require Ub surface Lys residues. Secondly, efficient depletion of cellular Ubc13 does not affect the PCNA-polyUb formation, which effectively rules out the involvement of the Ubc13-Uev in this modification. Thirdly, PCNA-Ub lacking the C-terminal Gly residues completely prevents the polyUb chain formation, indicating that the C-terminal Gly is absolutely required to accept the next Ub. Finally, if HA-tagged Ub lacks the C-terminal Gly residues, it can be incorporated into PCNA-Ub or its polyUb chain but serves as a chain terminator.
The linear ubiquitin chain has recently emerged as an important means of protein modification controlling immune responses, cell survival, proliferation and development by regulating the NF-κB pathway.Citation56-59 To date the linear polyubiquitination has only been found in NEMO and RIP1, and is mediated by LUBAC (linear ubiquitin chain assembly complex), composed of SHARPIN, HOIL-IL and HOIP.Citation60,61 Can native PCNA be another linear Ub target upon DNA damage? Although we do not have direct evidence, several observations support this notion. Firstly, we have previously shown that our PCNA-UbΔG fusion protein functionally behaves like native PCNA monoubiquitination,Citation34 making its polyubiquitination a possible physiological event. Secondly, despite a well-demonstrated sequential PCNA ubiquitination model in budding yeast and intensively studied PCNA monoubiquitination in mammalian cells, it remains inconclusive whether PCNA is polyubiquitinated in mammals upon DNA damage,Citation13,62 although a mammalian ZARANB3/AH2 has been identified to bind PCNA and K63-linked Ub chain through its 2 PCNA-binding domains and a specialized NPL4 zinc finger-binging domain, respectively.Citation63 Thirdly, based on the Ub structure, the K63 residue is adjacent to the Met1 residue and hence the linear Ub chain topologically resembles K63-linked polyUb chain and differs from K48-linked polyUb chain, suggesting that PCNA linear polyubiquitination could functionally resemble its K63-linked polyubiquitination. However, one has to be aware that for PCNA-K164 ubiquitination, the C-terminal Gly of Ub is linked to K164, making its N-terminus and surface Lys available, whereas in the PCNA-Ub fusion, its C-terminal Gly is absolutely required while its N-terminus is unavailable. Hence, the incoming Ub must use its N-terminus to form linear polyUb chains. Furthermore, we have not been able to prevent PCNA-Ub from been polyubiquitinated by suppressing the LUBAC complex (data not shown), which is either due to inefficient depletion, or this linear polyubiquitination is promoted by an unknown protein or protein complex.
What are the biological consequences of PCNA polyubiquitination? In budding yeast, PCNA polyubiquitination is known to promote error-free DDT, and several pieces of evidence support a notion that it mediates recruitment of HR factors via a template switch model.Citation22,64,Citation65 It has been suspected that PCNA polyubiquitination may serve as a signal to recruit HR factors; however, no such polyUb-binding protein has been identified in budding yeast that mediates such a process, although ZARANB3/AH2 is proposed to be involved in template switch.Citation66 Since this study demonstrates that PCNA-Ub can be readily polyubiquitinated by a linear Ub chain, we argue that perhaps PCNA-polyUb (including both linear and K63-linked) in both budding yeast and mammals serves as a replication-blocking signal. Indeed, several observations support this notion. Firstly, neither PCNA-UbΔG nor GFP-polyUb arrests cell proliferation, indicating that PCNA-polyUb is a rather specific signal. Secondly, PCNA-polyUb can be found in the chromatin-bound fraction and within nuclear foci, indicating that the signal most likely indicats an arrested replication fork. Thirdly, it has recently been reported that chemically-synthesized PCNA-polyUb protein inhibits both TLS by Polη across an abasic site and DNA synthesis by Polδ in vitro.Citation67 If the same effect occurs in vivo, it could satisfactorily explain our observations.
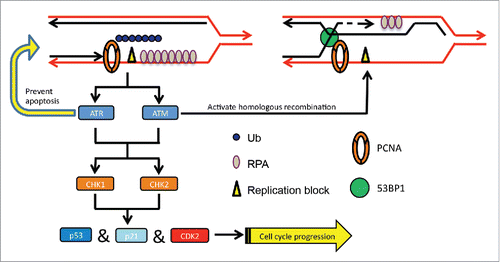
In this study we also investigated possible mechanisms of downstream events after PCNA polyubiquitination. Although cell cycle progression is blocked at the S-phase, cells are kept viable mainly by functional ATR. DSBs are then induced adjacent to ssDNA gaps, which in turn recruit 53BP1 and possibly other HR factors required for error-free lesion bypass via HR (). We do not know the exact nature of the DSB, but suspect that it may be formed between the 2 sister chromatids at the stalled replication fork. This process is reminiscent of the template switch in budding yeast that is required to complete error-free DDT.Citation22,64,Citation65 We infer that under physiological conditions, a very small fraction of PCNA molecules are polyubiquitinated at stalled replication forks upon DNA damage, which may activate the ATR and replication checkpoint, but does not severely inhibit cell proliferation. Meanwhile, activated ATM initiates error-free lesion bypass by recruiting the HR machinery to the damage site (). Exactly how this is achieved awaits future investigations.
Figure 7. A working model for PCNA-polyUb-induced error-free DDT signaling. PCNA-polyUb serves as a replication-blocking signal to activate the ATR checkpoint and stabilize the replication fork to prevent collapse. A DSB signal may be formed due to annealing of the 2 sister chromatids adjacent to the ssDNA gap, which in turn activates the ATM checkpoint and recruits 53BP1 and other HR proteins for error-free lesion bypass possibly via template switching.
Materials and methods
Plasmids and plasmid construction
The PCNA-K164R mutation was created by a mega-primer PCR methodCitation68 and the open reading frame (ORF) was cloned into plasmid vector pcDNA5.0FRT/TO (Invitrogen) as a HindIII-BamHI fragment. The full-length UB gene and UB lacking the C-terminal Gly-Gly codons (UBΔG) was cloned into the above resulting plasmid as a BamHI-XhoI fragment to make a fusion protein. HA-UB with K48 or K63 mutation was created by a PCR-based method, while HA-UB with all K-to-R mutations were generous gifts from Dr. H. Zhou (Penn State University, USA). Plasmid YCp-LEU2-POL30-UB was made as previously describedCitation35 except that the UB gene contains the 2 C-terminal Gly codons.
Cell culture and cell growth assay
The Flp-In™ T-Rex™ 293 cell line (T-Rex 293, Catalog no. R780-07, Invitrogen) were cultured in DMEM (SH30243.01B, Hyclone) plus 100 µg/ml Zeocin, 15 µg/ml blasticidin and 10% FBS (SH30256.01B, Hyclone) in a 37 °C, 5% CO2 humidified incubator. T-Rex 293 cells were transfected with a desired plasmid, incubated in the presence of 100 µg/ml hygromycin B, and stably-transfected cell lines were selected after a 3-day incubation. To induce ectopic PCNA expression, 2 µg/ml doxycycline (Dox) was added to the culture medium and the incubation continued for 2 d or as indicated. It was found that under the above conditions, the ectopic PCNA was noticeably induced after as little as 2 hours and lasted for several days. To assess cell growth, transfected T-Rex 293 cells were seeded in 6D cell dishes and with appropriate treatments. The cells were collected at the given time intervals and counted by haemocytometry or the Countess™ automated cell counter from Invitrogen.
Yeast cells were grown and transfected, and the experimental conditions were as previously described.Citation35
Western blotting
Samples were collected and placed on ice in a cell lysis solution (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 10% glycerol) containing 0.5% SDS and 2 µM PMSF with a protease inhibitor cocktail (Sigma P-8340, 1:100) and 10 mM N-ethylmalimide (NEM). Cellular proteins were resolved on a 12% SDS-PAGE gel run at 4°C. Gels were blotted in 5% Carnation instant skim milk in PBST, and the membrane was probed with anti-PCNA (Calbiochem: NA03), anti-PARP(Asp214) (CST: 9541), anti-53BP1 (CST: 4937), anti-P53 (sc-6248), anti-p-P53(Ser15) (CST: 9284), anti-p-P53(Thr18) (CST: 2529), anti-p-P53(Ser20) (CST: 9287), anti-p-P53(Ser37) (CST: 9289), anti-p-P53(Ser392) (CST: 9281), anti-CDK2(sc-6248), anti-p-CDK2(Thr14/Tyr15) (sc-28435), anti-Chk1(CST: 9917), anti-Chk2 (CST: 2662), anti-p-Chk1(Ser345) (Cell signal: 133D3), anti-p-Chk2 (Thr68) (Cell Signaling: C13C1). To prepare soluble and the chromatin-bound fractions, cultures were first rinsed with ice-cold PBS and exposed to PBS containing 0.4% NP-40, 10 mM NEM and protease inhibitors for 5 min. After centrifugation at 13,000 rpm for 10 min, the supernatant was collected as the soluble fraction. To obtain the chromatin-bound fraction, the plate was rinsed 3 times with ice-cold PBST and the pellet was resuspended in a 2x loading buffer (250 mM Tris-HCl, pH6.8, 10% SDS, 0.5% BPB, 50% glycerol, 5% β-mercaptoethanol), boiled for 10 min followed by SDS-PAGE and WB against anti-PCNA, anti-MDC1 (1:10,000 overnight) and anti-tubulin antibodies. Wherever necessary, the WB images were scanned and band intensity measured using a Luminescent Image Analyzer (LAS-4000).
For co-immunoprecipitation of 53BP1 or AH2 with PCNA-Ub, T-Rex 293 cells stably transfected with PCNA-Ub were induced by Dox for 2 d. The cell lysates were incubated with protein G beads from GE (17-0618-01) and the anti-PCNA or anti-Flag (Sigma F1804) antibody for 4 hrs or overnight. Beads were boiled with a 5x loading buffer for 5 min followed by 3 washes with the lysis buffer to remove unbound proteins. The input and the co-immunoprecipitated proteins were detected by western blotting.
Nuclear focus formation assay
Cultured cells were routinely seeded onto poly-lysine-coated cover slips, rinsed with ice-cold PBS and treated with 0.4% NP-40 in PBS for 20 min on ice prior to fixation in 4% formaldehyde for 30 min. Fixed cells were rinsed 4 times over 30 min with PBST (PBS with 0.1% Tween-20) prior to incubation with a mouse anti-RPA (Abcam, Ab2175), anti-53BP1 and anti-PCNA antibodies. Following four washes with PBST, the Alexa 488 goat anti-mouse secondary antibody (A11001), the Alexa 546 goat anti-rabbit secondary antibody (A11035) and 1.5 µg/ml 4,6-diamidino-2-phenylindole (DAPI) were added for one hour and then washed again 4 times. Microscopy was performed with an inverted Olympus 10×22 microscope equipped with a 40x immersion lens and images were acquired using the CCD RoHs (Q26053). Samples for comparison in each panel were always included in the same experiment and treated identically. Within each experiment, images containing at least 1000 cells for each treatment were captured and analyzed.
For ICC, cultured cells were routinely seeded onto poly-lysine-coated coverslips, rinsed with ice-cold PBS, fixed with 4% formaldehyde for 30 min and then treated with 0.5% Triton X-100 prior to immunocytochemistry.
Flow cytometry
For cell cycle analysis, cells were first washed with PBS and then fixed in 70% ethanol. Cells were stored at 4 °C overnight, pelleted at 800 rpm for 5 min, resuspended in 1ml PBS and pelleted again. The pellet was resuspended in PBS with RNase A and then washed twice with PBS. Before test by flow cytometry cells were stained by DAPI. For cell apoptosis analysis, cells were collected in PBS and kept alive at a low temperature. Before staining with annexin V and propidium iodide (PI), cells were washed with PBS and filtered with a 300-mesh membrane filter. All the work needs to be finished in 1 hour after annexin V and PtdIns staining.
Real-time RT-PCR (qRT-PCR)
Total RNA was extracted using Trizol reagent (Invitrogen). cDNA synthesis was performed with 500 ng of total RNA using RevertAid™ First Strand cDNA Synthesis kit (Fermentas) according to the manufacturer's instructions. Endogenous mRNA levels were measured by qRT-PCR analysis based on SYBR Green detection (Fermentas) with the BioRad real-time PCR machine. Results were normalized with GAPDH. p21 primers used in the study gave rise to a single product of expected size in an agarose gel analysis. p21-1F, TCTCCAAGAGGAAGCCCTAA;p21-1R, AGGAACCTCTCATTCAACCG; p21-2F, GCAGACCAGCATGACAGATT; p21-2R, TTAGGGCTTCCTCTTGGAGAp21-3F, CTTCGACTTTGTCACCGAGA; p21-3R, ATGGTCTTCCTCTGCTGTCC.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1245247_Supplemental_Material.pdf
Download PDF (592.6 KB)Acknowledgments
The authors wish to thank members of the Xiao laboratory for helpful discussion and Michelle Hanna for proofreading the manuscript.
Funding
This work was supported by Chinese National 973 Project 2013CB911003, Capital Normal University 211 Special Fund 043145303300 and Canadian Institutes of Health Research operating grant MOP-93612 to W.X.
References
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell 2007; 129:665-79; PMID:17512402; http://dx.doi.org/10.1016/j.cell.2007.05.003
- Bowman GD, O'Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 2004; 429:724-30; PMID:15201901; http://dx.doi.org/10.1038/nature02585
- Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol 2004; 78:227-60; PMID:15210332; http://dx.doi.org/10.1016/S0079-6603(04)78006-X
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem 2000; 69:497-529; PMID:10966467; http://dx.doi.org/10.1146/annurev.biochem.69.1.497
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell 2008; 30:137-44; PMID:18439893; http://dx.doi.org/10.1016/j.molcel.2008.02.022
- Chai Q, Zheng L, Zhou M, Turchi JJ, Shen B. Interaction and stimulation of human FEN-1 nuclease activities by heterogeneous nuclear ribonucleoprotein A1 in alpha-segment processing during Okazaki fragment maturation. Biochemistry 2003; 42:15045-52; PMID:14690413; http://dx.doi.org/10.1021/bi035364t
- Prelich G, Tan CK, Kostura M, Mathews MB, So AG, Downey KM, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature 1987; 326:517-20; PMID:2882424; http://dx.doi.org/10.1038/326517a0
- Dua R, Levy DL, Li CM, Snow PM, Campbell JL. In vivo reconstitution of Saccharomyces cerevisiae DNA polymerase epsilon in insect cells. Purification and characterization. J Biol Chem 2002; 277:7889-96; PMID:11756442; http://dx.doi.org/10.1074/jbc.M108546200
- Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J 1998; 17:3786-95; PMID:9649448; http://dx.doi.org/10.1093/emboj/17.13.3786
- Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res 2008; 18:162-73; PMID:18157158; http://dx.doi.org/10.1038/cr.2007.114
- Zhang W, Qin Z, Zhang X, Xiao W. Roles of sequential ubiquitination of PCNA in DNA-damage tolerance. FEBS Letters 2011; 585:2786-94; PMID:21536034; http://dx.doi.org/10.1016/j.febslet.2011.04.044
- Gazy I, Kupiec M. The importance of being modified: PCNA modification and DNA damage response. Cell Cycle 2012; 11:2620-23; PMID:22732495; http://dx.doi.org/10.4161/cc.20626
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002; 419:135-41; PMID:12226657; http://dx.doi.org/10.1038/nature00991
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 2004; 8:610-6; PMID:15556404; http://dx.doi.org/10.1016/j.cbpa.2004.09.009
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet 1996; 30:405-39; PMID:8982460; http://dx.doi.org/10.1146/annurev.genet.30.1.405
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 2005; 310:1821-24; PMID:16357261; http://dx.doi.org/10.1126/science.1120615
- Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet 2006; 2:e116; PMID:16789823; http://dx.doi.org/10.1371/journal.pgen.0020116
- Unk I, Hajdú I, Fátyol K, Szakál B, Blastyák A, Bermudez V, Hurwitz J, Prakash L, Prakash S, Haracska L. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci U S A 2006; 103:18107-12; PMID:17108083; http://dx.doi.org/10.1073/pnas.0608595103
- Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, Myung K. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol 2006; 175:703-8; PMID:17130289; http://dx.doi.org/10.1083/jcb.200606145
- Motegi A, Liaw HJ, Lee KY, Roest HP, Maas A, Wu X, Moinova H, Markowitz SD, Ding H, Hoeijmakers JH, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci U S A 2008; 105:12411-6; PMID:18719106; http://dx.doi.org/10.1073/pnas.0805685105
- Unk I, Hajdú I, Fátyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci U S A 2008; 105:3768-73; PMID:18316726; http://dx.doi.org/10.1073/pnas.0800563105
- Xu X, Blackwell S, Lin A, Li F, Qin Z, Xiao W. Error-free DNA-damage tolerance in Saccharomyces cerevisiae. Mutation research. Rev Mutation Res 2015; 764:43-50; PMID:26041265; http://dx.doi.org/10.1016/j.mrrev.2015.02.001
- Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007; 6:923-35; PMID:17363343; http://dx.doi.org/10.1016/j.dnarep.2007.02.006
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 2005; 308:551-4; PMID:15790808; http://dx.doi.org/10.1126/science.1108297
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300:1542-8; PMID:12791985; http://dx.doi.org/10.1126/science.1083430
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 2005; 19:1040-52; PMID:15833913; http://dx.doi.org/10.1101/gad.1301205
- Yang XH, Zou L. Checkpoint and coordinated cellular responses to DNA damage. Results Probl Cell Differ 2006; 42:65-92; PMID:16903208; http://dx.doi.org/10.1007/b136684
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316:1160-6; PMID:17525332; http://dx.doi.org/10.1126/science.1140321
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol 2004; 24:1279-91; PMID:14729972; http://dx.doi.org/10.1128/MCB.24.3.1279-1291.2004
- Pichierri P, Rosselli F, Franchitto A. Werner's syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene 2003; 22:1491-1500; PMID:12629512; http://dx.doi.org/10.1038/sj.onc.1206169
- Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev 2000; 14:2989-3002; PMID:11114888; http://dx.doi.org/10.1101/gad.851000
- Li W, Kim SM, Lee J, Dunphy WG. Absence of BLM leads to accumulation of chromosomal DNA breaks during both unperturbed and disrupted S phases. J Cell Biol 2004; 165:801-12; PMID:15197177; http://dx.doi.org/10.1083/jcb.200402095
- Hu B, Wang H, Wang X, Lu HR, Huang C, Powell SN, Huebner K, Wang Y. Fhit and CHK1 have opposing effects on homologous recombination repair. Cancer Res 2005; 65:8613-6; PMID:16204026; http://dx.doi.org/10.1158/0008-5472.CAN-05-1966
- Qin Z, Lu M, Xu X, Hanna M, Shiomi N, Xiao W. DNA-damage tolerance mediated by PCNA*Ub fusions in human cells is dependent on Rev1 but not Poleta. Nucleic Acids Res 2013; 41:7356-69; PMID:23761444; http://dx.doi.org/10.1093/nar/gkt542
- Pastushok L, Hanna M, Xiao W. Constitutive fusion of ubiquitin to PCNA provides DNA damage tolerance independent of translesion polymerase activities. Nucleic Acids Res 2010; 38:5047-58; PMID:20385585; http://dx.doi.org/10.1093/nar/gkq239
- Bravo R, Celis JE. A search for differential polypeptide synthesis throughout the cell cycle of HeLa cells. J Cell Biol 1980; 84:795-802; PMID:6892640; http://dx.doi.org/10.1083/jcb.84.3.795
- Mathews MB, Bernstein RM, Franza BR Jr, Garrels JI. Identity of the proliferating cell nuclear antigen and cyclin. Nature 1984; 309:374-6; PMID:6145097; http://dx.doi.org/10.1038/309374a0
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 1994; 79:1233-43; PMID:8001157; http://dx.doi.org/10.1016/0092-8674(94)90014-0
- Woo RA, Poon RY. Cyclin-dependent kinases and S phase control in mammalian cells. Cell Cycle 2003; 2:316-24; PMID:12851482; http://dx.doi.org/10.4161/cc.2.4.468
- Chen Y, Poon RY. The multiple checkpoint functions of CHK1 and CHK2 in maintenance of genome stability. Front Biosci 2008; 13:5016-29; PMID:18508566
- Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst) 2006; 5:1495-8; PMID:16956796; http://dx.doi.org/10.1016/j.dnarep.2006.07.002
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM-and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 2006; 8:37-45; PMID:16327781; http://dx.doi.org/10.1038/ncb1337
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 2000; 151:1381-90; PMID:11134068; http://dx.doi.org/10.1083/jcb.151.7.1381
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 2001; 276:42462-7; PMID:11571274; http://dx.doi.org/10.1074/jbc.C100466200
- Shechter D, Costanzo V, Gautier J. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair (Amst) 2004; 3:901-8; PMID:15279775; http://dx.doi.org/10.1016/j.dnarep.2004.03.020
- Chatterjee P, Plesca D, Mazumder S, Boutros J, Yannone SM, Almasan A. Defective chromatin recruitment and retention of NHEJ core components in human tumor cells expressing a Cyclin E fragment. Nucleic Acids Res 2013; 41:10157-69; PMID:24021630; http://dx.doi.org/10.1093/nar/gkt812
- Leung WH, Vong QP, Lin W, Janke L, Chen T, Leung W. Modulation of NKG2D ligand expression and metastasis in tumors by spironolactone via RXRgamma activation. J Exp Med 2013; 210:2675-92; PMID:24190430; http://dx.doi.org/10.1084/jem.20122292
- Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, Golec JM, Pollard JR. Selective killing of ATM-or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 2011; 7:428-30; PMID:21490603; http://dx.doi.org/10.1038/nchembio.573
- He J, Whitacre CM, Xue LY, Berger NA, Oleinick NL. Protease activation and cleavage of poly(ADP-ribose) polymerase: an integral part of apoptosis in response to photodynamic treatment. Cancer Res 1998; 58:940-6; PMID:9500454
- Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 2015; 518:254-7; PMID:25642960; http://dx.doi.org/10.1038/nature14157
- Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 2015; 528:422-6; PMID:26649820; http://dx.doi.org/10.1038/nature16142
- Xu G, Chapman JR, Brandsma I, Yuan J, Mistrik M, Bouwman P, Bartkova J, Gogola E, Warmerdam D, Barazas M, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 2015; 521:541-4; PMID:25799992; http://dx.doi.org/10.1038/nature14328
- Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell 2002; 111:779-89; PMID:12526805; http://dx.doi.org/10.1016/S0092-8674(02)01113-3
- Xiao W, Lin SL, Broomfield S, Chow BL, Wei YF. The products of the yeast MMS2 and two human homologs (hMMS2 and CROC-1) define a structurally and functionally conserved Ubc-like protein family. Nucleic Acids Res 1998; 26:3908-14; PMID:9705497; http://dx.doi.org/10.1093/nar/26.17.3908
- Andersen PL, Zhou H, Pastushok L, Moraes T, McKenna S, Ziola B, Ellison MJ, Dixit VM, Xiao W. Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J Cell Biol 2005; 170:745-55; PMID:16129784; http://dx.doi.org/10.1083/jcb.200502113
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell 2008; 132:344-62; PMID:18267068; http://dx.doi.org/10.1016/j.cell.2008.01.020
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep 2008; 9:536-42; PMID:18516089; http://dx.doi.org/10.1038/embor.2008.93
- Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep 2009; 10:706-13; PMID:19543231; http://dx.doi.org/10.1038/embor.2009.144
- Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat Rev Immunol 2009; 9:778-88; PMID:19855404; http://dx.doi.org/10.1038/nri2655
- Tokunaga F, Iwai K. Linear ubiquitination: a novel NF-kappaB regulatory mechanism for inflammatory and immune responses by the LUBAC ubiquitin ligase complex. Endocr J 2012; 59:641-52; PMID:22673407; http://dx.doi.org/10.1507/endocrj.EJ12-0148
- Tokunaga F, Iwai K. LUBAC, a novel ubiquitin ligase for linear ubiquitination, is crucial for inflammation and immune responses. Microbes Infect 2012; 14:563-72; PMID:22309894; http://dx.doi.org/10.1016/j.micinf.2012.01.011
- Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair 2004; 3:1503-14; PMID:15380106; http://dx.doi.org/10.1016/j.dnarep.2004.06.015
- Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell 2012; 47:396-409; PMID:22704558; http://dx.doi.org/10.1016/j.molcel.2012.05.024
- Branzei D, Foiani M. Template switching: from replication fork repair to genome rearrangements. Cell 2007; 131:1228-30; PMID:18160033; http://dx.doi.org/10.1016/j.cell.2007.12.007
- Ball LG, Zhang K, Cobb JA, Boone C, Xiao W. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol Microbiol 2009; 73:89-102; PMID:19496932; http://dx.doi.org/10.1111/j.1365-2958.2009.06748.x
- Zeman MK, Cimprich KA. Finally, polyubiquitinated PCNA gets recognized. Mol Cell 2012; 47:333-4; PMID:22883622; http://dx.doi.org/10.1016/j.molcel.2012.07.024
- Yang K, Gong P, Gokhale P, Zhuang Z. Chemical protein polyubiquitination reveals the role of a noncanonical polyubiquitin chain in DNA damage tolerance. ACS Chem Biol 2014; 9:1685-91; PMID:24918305; http://dx.doi.org/10.1021/cb500133k
- Picard V, Ersdal-Badju E, Lu A, Bock SC. A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res 1994; 22:2587-91; PMID:8041621; http://dx.doi.org/10.1093/nar/22.13.2587
