The International Agency for Research on Cancer classified acetaldehyde related to alcoholic beverage consumption as carcinogenic to humans.Citation1 This classification was based in large part on epidemiological studies of individuals who lack the capacity to metabolize acetaldehyde due to a functional polymorphism in ALDH2. Such individuals have a dramatically elevated risk of esophageal cancer from alcohol consumption than those with fully active ALDH2.Citation2 Acetaldehyde forms DNA adductsCitation3 supporting a genotoxic mechanism of carcinogenicity.
To identify molecular mechanisms that protect against acetaldehyde genotoxicity, Noguchi et al.Citation4 performed a comprehensive mutational analysis of the role of the DNA repair and DNA damage response pathways, using the fission yeast Schizosaccharomyces pombe as a model organism. The experimental design involved exposing wild-type and mutant strains to increasing concentrations of acetaldehyde, then monitoring cell growth as an end point. In some experiments Rad52-YFP was used as a marker of DNA damage. Rad52 is a DNA recombinase that is recruited to sites of DNA damage, forming DNA repair foci.
Homology searches identified 3 possible yeast Aldh genes, denoted Atd1-3. Of these, Atd1 was found to play the major role in protection against acetaldehyde, though cells lacking either Atd1 or Atd2 show elevated levels of DNA damage in response to acetaldehyde. In Atd1Δ cells, deletion of the replication fork protection complex protein Swi1 increased sensitivity to acetaldehyde, and also resulted in a dramatic decrease in chromosome number following acetaldehyde exposure. Swi1 forms a complex with Swi3 that is homologous to the Timeless-Tipin complex in human cells.Citation5 In cells with wild-type Atd1, deletion of both Swi1 and rad26 resulted in a much greater acetaldehyde sensitive than either single mutant. The rad26 gene encodes a protein that interacts with the cell cycle checkpoint kinase Rad3, homologous to ATR in mammals.Citation6
Consistent with work in mammalian cells Citation(3 and references therein), disruption of either nucleotide excision repair (NER), the Fanconi anemia DNA damage response pathway, or homologous recombination repair increased sensitivity to acetaldehyde, as did disruption of multiple translesion synthesis DNA polymerases. Interestingly, acetaldehyde sensitivity was also increased in cells with mutations in in either AP-endonuclease 2 (apn2 Δ) or Endonuclease III (nth1 Δ), consistent with a role for base excision repair (BER) in protecting against acetaldehyde toxicity. This observation is somewhat surprising, since acetaldehyde-DNA lesions are not known to be repaired by BER. However, exposure of living cells to acetaldehyde can result in ethenobase adducts, via an indirect pathway involving lipid peroxidationCitation3; some of these adducts are substrates for BER.Citation6 It is worth noting however that apn2 Δ cells and nth1 Δ cells were only sensitive to acetaldehyde in the context of a cell cycle checkpoint defect (rad3 Δ).
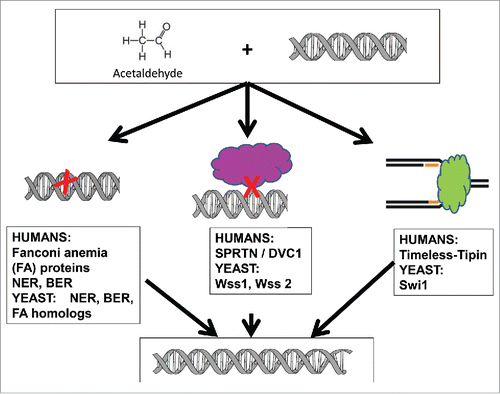
Another notable finding from this workCitation4 is evidence of important roles for Wss-related proteases in protecting against acetaldehyde toxicity. The Wss-related proteases had been implicated in proteolysis related to the repair of DNA – protein crosslinks, which can result from acetaldehyde. These proteases are homologous to the human DVC1-Spartan protease.Citation7 Interestingly, germline mutations in the DVC1-Spartan gene in humans results in premature aging and an increased risk of liver cancer.Citation7 A summary of the main pathways for protecting against acetaldehyde genotoxicity identified by Noguchi et al.Citation4 is given in CitationFig. 1.
One limitation of the current work is that it relied heavily on measures of relative cell growth of different mutant strains under increasing concentrations of acetaldehyde. While this approach is certainly reasonable for rapid screening of many single and multiple mutant strains, future studies could include assays of specific mutagenic events, which might be observed at lower acetaldehyde concentrations, or in strains containing single gene mutations that appeared normal using cell growth assays.
To assess the relevance of these findings for human disease, it will be important to validate these findings in human cells, under acetaldehyde concentrations that are relevant to human exposures. This can be done in a straightforward manner by knocking down the relevant human genes, and assessing effects on the toxicity and mutagenicity of acetaldehyde, either exogenous or generated inside cells by alcohol metabolism. Such experiments could provide a foundation for genetic epidemiology studies to investigate the effects of hypomorphic mutations in the genes identified here on the risk of cancer from alcohol drinking, and possibly in other diseases associated with acetaldehyde exposure.
Figure 1. Schematic depiction of some of the cellular mechanisms for protection against acetaldehyde genotoxicity. Three types of damage are indicated: DNA base lesions, including interstrand crosslinks (left), DNA protein crosslinks (center) and stalled replication forks (right). The yeast genes identified by Noguchi et al. that protect against the different types of damage are indicated, along with the human homologs.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- IARC. A review of human carcinogens. Part E: Personal habits and indoor combustions IARC monographs on the evaluation of carcinogenic risks to humans Vol. 100E. Lyon: France IARC, 2009.
- Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med 2009; 6(3):e50; PMID: 19320537; http://dx.doi.org/10.1371/journal.pmed.1000050
- Brooks PJ, Zakhari S. Acetaldehyde and the genome: beyond nuclear DNA adducts and carcinogenesis. Environ Mol Mutagen 2014; 55(2):77-91; PMID: 24282063; http://dx.doi.org/10.1002/em.21824
- Noguchi C, Grothusen G, Anandarajan V, Martínez-Lage García M, Terlecky D, Corzo K, Tanaka K, Nakagawa H, Noguchi E. Genetic controls of DNA damage avoidance in response to acetaldehyde in fission yeast. Cell Cycle 2017; 16(1):45-58; PMID: 27687866; http://dx.doi.org/10.1080/15384101.2016.1237326
- Leman AR, Noguchi E. Local and global functions of Timeless and Tipin in replication fork protection. Cell Cycle 2012; 11(21):3945-55; PMID: 22987152; http://dx.doi.org/10.4161/cc.21989.
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz R, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: American Society for Microbiology, 2006
- Stingele J, Jentsch S. DNA-protein crosslink repair. Nat Rev Mol Cell Biol 2015; 16(8):455-60; PMID: 26130008; http://dx.doi.org/10.1038/nrm4015
