ABSTRACT
Pre-mRNA splicing entails the stepwise assembly of an inactive spliceosome, its catalytic activation, splicing catalysis and spliceosome disassembly. Transitions in this reaction cycle are accompanied by compositional and conformational rearrangements of the underlying RNA-protein interaction networks, which are driven and controlled by 8 conserved superfamily 2 RNA helicases. The Ski2-like helicase, Brr2, provides the key remodeling activity during spliceosome activation and is additionally implicated in the catalytic and disassembly phases of splicing, indicating that Brr2 needs to be tightly regulated during splicing. Recent structural and functional analyses have begun to unravel how Brr2 regulation is established via multiple layers of intra- and inter-molecular mechanisms. Brr2 has an unusual structure, including a long N-terminal region and a catalytically inactive C-terminal helicase cassette, which can auto-inhibit and auto-activate the enzyme, respectively. Both elements are essential, also serve as protein-protein interaction devices and the N-terminal region is required for stable Brr2 association with the tri-snRNP, tri-snRNP stability and retention of U5 and U6 snRNAs during spliceosome activation in vivo. Furthermore, a C-terminal region of the Prp8 protein, comprising consecutive RNase H-like and Jab1/MPN-like domains, can both up- and down-regulate Brr2 activity. Biochemical studies revealed an intricate cross-talk among the various cis- and trans-regulatory mechanisms. Comparison of isolated Brr2 to electron cryo-microscopic structures of yeast and human U4/U6•U5 tri-snRNPs and spliceosomes indicates how some of the regulatory elements exert their functions during splicing. The various modulatory mechanisms acting on Brr2 might be exploited to enhance splicing fidelity and to regulate alternative splicing.
Introduction
Eukaryotic protein-coding transcripts are produced as precursors (pre-mRNAs), which undergo several maturation steps to be converted to mature mRNAs that can serve for protein biosynthesis on the ribosome. The vast majority of higher eukaryotic protein-coding genes contain non-coding sequences (introns) interspersed among coding regions (exons), and maturation of their pre-mRNAs thus requires pre-mRNA splicing, during which intronic sequences are removed and exonic sequences are ligated.Citation1 Each splicing reaction consists of 2 consecutive transesterification steps. In the first step, the 2′-hydroxyl group of a conserved adenosine in the branch point sequence (BPS) of an intron attacks the phosphodiester bond at the 5′-splice site (5SS), generating a 5′-exon that bears a free 3′-hydroxyl group and an intron-lariat-3′-exon. In the second step, the 3′-hydroxyl group of the 5′-exon attacks the 3′-splice site (3SS), leading to the ligation of the exons and the excision of the intron lariat. Pre-mRNA splicing is mediated by a highly dynamic, multi-megadalton RNA-protein (RNP) molecular machine, the spliceosome, which encompasses 5 small nuclear (sn) RNPs (U1, U2, U4, U5 and U6 in the case of the major spliceosome) and numerous non-snRNP proteins.Citation2,3 Each snRNP comprises a specific snRNA, a set of 7 common Sm proteins (or LSm proteins in the case of U6) and different numbers of snRNP-specific proteins.Citation4
For each round of splicing, a spliceosome is assembled de novo on a substrate by the stepwise recruitment of snRNPs and non-snRNP proteins ().Citation2,3,5-7 With respect to the snRNPs, the U1 snRNP first recognizes the 5SS (E complex), followed by the binding of the U2 snRNP at the BPS (A complex). Subsequently, the pre-formed U4/U6•U5 tri-snRNP loosely associates with the spliceosome, giving rise to the catalytically inactive pre-B complex.Citation8 Within the tri-snRNP, the U4 and U6 snRNAs are extensively base-paired via 2 regions (stems I and II)Citation9-14 and decorated by several proteins.Citation12-14 Association of the U4/U6 di-snRNP with the U5 snRNP is mainly accomplished by protein-protein interactions.Citation12-15 After pre-B complex formation, the U1 snRNP is released and the tri-snRNP is stably integrated to produce the pre-catalytic B complex, followed by displacement of U4 snRNAs and associated proteins, which leads to the activated spliceosome (Bact).Citation16 Further remodeling of Bact gives rise to the catalytically activated spliceosome (B*).Citation17,18 The B* complex is poised to perform the first step of splicing. The ensuing C complex is subsequently converted to complex C*,Citation19 which carries out the second step of splicing. After the splicing reaction, the product mRNA is released as an mRNP from the post-splicing complex, the resulting intron-lariat spliceosome is disassembled and the remaining subunits are recycled for a new round of splicing.
Figure 1. Top, pre-mRNA splicing by the spliceosome, showing characterized spliceosomal assembly, activation, catalysis and disassembly intermediates (complexes E, A, Pre-B, B, Bact, B*, C, C*, PSC [post-splicing complex] and ILS [intron-lariat spliceosome]). For clarity, the cycle only depicts the compositions with respect to the snRNPs, the many non-snRNP proteins that are also participating have been omitted. Step 1, step 2, first/second transesterification reaction of splicing. Spliceosomal RNA helicases are indicated at the steps during which they are required (blue). Traffic lights refer to the observed or assumed (with question mark) states of Brr2 at the respective stages and during the various transitions (red, fully inhibited; yellow, partially inhibited, green, activated). Bottom, schemes illustrating combinatorial regulation of Brr2 via its NTR (magenta) and via the Prp8 Jab1 domain (gold).
![Figure 1. Top, pre-mRNA splicing by the spliceosome, showing characterized spliceosomal assembly, activation, catalysis and disassembly intermediates (complexes E, A, Pre-B, B, Bact, B*, C, C*, PSC [post-splicing complex] and ILS [intron-lariat spliceosome]). For clarity, the cycle only depicts the compositions with respect to the snRNPs, the many non-snRNP proteins that are also participating have been omitted. Step 1, step 2, first/second transesterification reaction of splicing. Spliceosomal RNA helicases are indicated at the steps during which they are required (blue). Traffic lights refer to the observed or assumed (with question mark) states of Brr2 at the respective stages and during the various transitions (red, fully inhibited; yellow, partially inhibited, green, activated). Bottom, schemes illustrating combinatorial regulation of Brr2 via its NTR (magenta) and via the Prp8 Jab1 domain (gold).](/cms/asset/0e9b16c9-ee92-4db6-b455-586fad920520/kccy_a_1249549_f0001_c.gif)
Each assembly, activation, catalysis and disassembly step involves profound rearrangements of the spliceosomal RNP interaction networks, mediated by at least 8 conserved superfamily 2 (SF2) NTPases/RNA helicases ().Citation20-22 SF2 helicases can be grouped into 5 families, 3 of which are represented among the spliceosomal remodeling enzymes: 3 DEAD box proteins (Prp5, Sup2/UAP56, Prp28) act during initial spliceosome assembly and activation, a single Ski2-like helicase (Brr2) is involved in spliceosome activation and 4 DEAH/RHA enzymes (Prp2, Prp16, Prp22, Prp43) are required during spliceosome activation, catalysis and disassembly.Citation20 The most dramatic rearrangements occur during spliceosome activation, where the Prp28 helicase aids in the displacement of U1 snRNA from the 5SS,Citation8,23,24 followed by Brr2 unwinding the U4 and U6 snRNAsCitation25-27 and leading to displacement of U4 snRNA and U4/U6-bound proteins.Citation28,29 These processes allow U6 to engage in alternative interactions with the 5SS and U2 snRNA, as well as to form a catalytically important internal stem-loop, thereby building up the spliceosome's active site.Citation30-33 Here we review results from structural and functional studies, which have begun to shed light on an intricate network of regulatory principles controlling the Brr2 RNA helicase, and discuss how Brr2 regulation may influence splicing fidelity and alternative splicing.
Brr2 requires tight regulation
Several lines of evidence indicate that Brr2 activity has to be tightly controlled during all stages of snRNP assembly and splicing. Brr2 is a specific subunit of the U5 snRNP. However, initial assembly of yeast U5 snRNP in the cytoplasm leads to a pre-U5 particle that contains the assembly factor Aar2 instead of Brr2.Citation34-39 Thus, Brr2 is transported to the nucleus independent of other U5 snRNP componentsCitation35 and its helicase activity may have to be shut off during this phase to avoid detrimental off-target effects. Once assembled in the nucleus, mature U5 snRNP joins the U4/U6 di-snRNP to form the U4/U6•U5 tri-snRNP, in which Brr2 already encounters its U4/U6 di-snRNA substrate before incorporation into the spliceosome. As U4/U6 di-snRNA disruption in the tri-snRNP or premature unwinding during spliceosome assembly would counteract productive splicing, Brr2 must be held in check by specific mechanisms in the tri-snRNP and during assembly of the pre-catalytic spliceosome. On the other hand, isolated Brr2 is a comparatively weak helicase,Citation25,40,41 and its U4/U6 di-snRNA substrate is stabilized by extensive base pairing and bound proteins,Citation12-14,29,42,43 suggesting that the helicase may also depend on specific activation to efficiently unwind the U4/U6 duplex at the right time.
Most spliceosomal helicases associate only transiently with the spliceosome at the stages during which their activities are needed. Thus, their activities during splicing are regulated in part by the timing of their recruitment and release. Brr2, in contrast, stays associated with the spliceosome after recruitment of the tri-snRNP during the remaining phases of splicing. Moreover, in addition to spliceosome activation, Brr2 has been found to be important for the retention of U5 and U6 snRNAs during spliceosome activationCitation44,45 and to play a role during splicing catalysisCitation46,47 and spliceosome disassembly.Citation48 However, not all of these additional functions apparently rely on the enzyme's ATPase and helicase activities. E.g., based on RNA-seq and cross-linking analyses of wt yeast Brr2 in comparison to a particular variant (G858R)Citation46 and the suppression of a secondary structure-induced step 2 splicing defect by this Brr2 variant,Citation49 Brr2 has been proposed to promote correct positioning of the 3SS for second step catalysis.Citation46 This activity might involve ATP-independent, Brr2-mediated modulation of secondary structures between the BPS and the 3SS and indirect promotion of exon-U5 snRNA interactions by Brr2-mediated recruitment of the Slu7 and Prp18 proteins.Citation46 Furthermore, spliceosome disassembly can proceed in the presence of nucleotide tri-phosphates other than ATP, which Brr2 does not utilize.Citation50 Taken together, molecular mechanisms must be at work, through which Brr2 can be up- or down-regulated repeatedly during snRNP assembly and splicing ().
Brr2 exhibits a unique structure
In line with its unique regulatory requirements, the structure of Brr2 differs decisively from that of other spliceosomal helicases. Brr2 is the only spliceosomal helicase that comprises a tandem array of 2 helicase cassettes, each of which is made up of 2 RecA-like domains (RecA1 and RecA2), a winged helix domain (WH) and a Sec63 homology unit.Citation51 The Sec63 homology units consist of a helical bundle (HB), a helix-loop-helix (HLH) and an immunoglobulin-like (IG) domain ().Citation41,52 Only the N-terminal helicase cassette (NC) is active in nucleotide hydrolysis and RNA unwinding,Citation51 and the enzymatic activity of the NC alone is required for splicing in vivo.Citation27 The C-terminal cassette (CC) is still able to bind ATP,Citation51 but it contains non-canonical residues in the ATP binding and hydrolysis motifs, which abrogate the ATPase activity.Citation27,51 Crystal structures of Brr2 helicase regions confirmed a similar globular architecture of the 2 helicase cassettes and revealed that the cassettes can intimately interact with each other via a large interface.Citation38,51,53
Figure 2. Domain organization and structure of Brr2 in complex with the Prp8 Jab1 domain. (A) Domain organization of the NTR and NC; the CC has the same domain organization as the NC and is not depicted in detail. (B) Diametric view of a full-length yeast Brr2-Jab1 complex (PDB ID 5M52).Citation56 The NTR and the Jab1 domain are highlighted by semi-transparent surfaces. NTR, magenta; RecA1, light gray; RecA2, dark gray; WH, black; HB, blue; HLH, red; IG, green; CC, beige; Jab1, gold. (C) Accommodation of the U4/U6 di-snRNA substrate by Brr2 as seen in a yeast tri-snRNP structure (PDB ID 5GAO).Citation12 Color coding as in (B). The rotation symbol indicates the orientation relative to (B), left panel. (D) Interactions involving the E890 residue of human Brr2 (equivalent to E909 in yeast Brr2) in a crystal structure with ADP bound at the NC (PDB ID 4F93).Citation51 Dashed lines, hydrogen bonds or salt bridges. The rotation symbol indicates the orientation relative to (C).
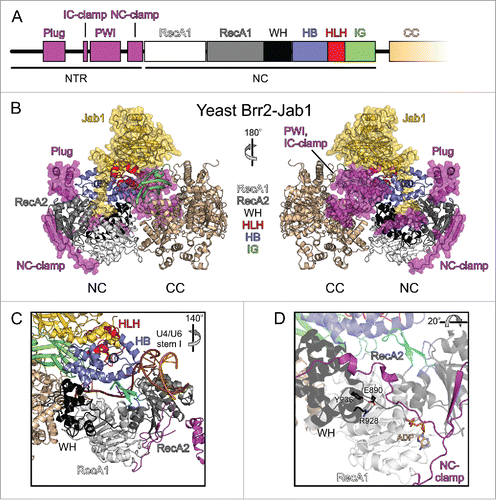
Preceding its 2 helicase cassettes, Brr2 contains a ∼450-residue N-terminal region (NTR; ). Within this NTR, a helical “plug” domain and a PWI-like domain are interspersed among extended, intrinsically disordered regions.Citation54,55 In crystal structures of yeast Brr2 in complex with a C-terminal Jab1/MPN-like (Jab1) domain of the Prp8 protein, the NTR runs along one entire flank of the helicase cassettes and extensively contacts both NC and CC.Citation55,56 Following ∼100 disordered N-terminal residues, the plug domain wedges between the HB and the RecA2 domains of the NC. The plug is followed by an “inter-cassette clamp” (IC-clamp) that bridges NC and CC. C-terminal of the IC-clamp, a non-canonical PWI domain is located next to the CC. Chemical crosslinking and mass spectrometry indicated that in solution the PWI-like domain is further displaced toward and contacts the CC.Citation55 Immediately preceding the NC, an irregularly structured “N-terminal cassette clamp” (NC-clamp) tightly wraps around the NC and connects its RecA and WH domains.
Multiple layers of helicase-associated domains in Brr2
As in other RNA helicases, the dual RecA-like domains of the Brr2 NC harbor conserved sequence motifs, through which the enzyme binds ATP and the RNA substrate, and through which it couples ATP hydrolysis, release and rebinding via conformational changes to translocation on an RNA substrate strand to achieve RNA duplex unwinding. Thus, the 2 N-terminal RecA-like domains comprise the core motor domains of the enzyme. Notably, this central motor region makes up only about 20 % of the entire molecular mass of the protein (). Thus, most molecular parts of Brr2 seem to serve modulatory and/or regulatory functions. Based on structural and functional investigations, the helicase-associated regions of Brr2 can be groups into several layers, which will be described in the following.
Layer I – helicase-associated domains of the NC as modulators of the helicase mechanism and as mediators of intermolecular regulation
In the NC, the active RecA-like domains are connected via a WH domain to a HB domain, forming a circular arrangement with a central tunnel (), in which conserved RNA-binding elements of the RecA domains carpet the floor and a long helix of the HB domain runs along the roof.Citation51 This organization resembles the structure of the related Ski2-like DNA helicase, Hel308, in complex with a DNA substrate,Citation57 which suggested that the long α-helix of the HB domain serves as a ratchet that holds on to the substrate during cycles of RecA detachment from the RNA, repositioning and rebinding. Additionally, a long β hairpin loop in the RecA2 domain of Hel308, the separator loop, is positioned between the 2 strands of a duplex DNA region located upstream of the bound single-stranded region. Upon translocation of the helicase in 3′-to-5′ direction on the entrapped DNA strand, the separator loop is thought to be driven between the 2 strands of the base-paired region, forcing the 2 substrate strands apart. In contrast, in DEAH/RHA helicases an element equivalent to the separator loop (termed “5′HP”) has been suggested to mainly control access to the single-stranded RNA binding site.Citation58,59
Comparative modelingCitation41,51,52 and a recent electron cryo-microscopic (cryo-EM) structure of a yeast U4/U6•U5 tri-snRNPCitation12 showed that Brr2 engages its U4/U6 di-snRNA substrate in a similar manner as Hel308. A single-stranded region of the U4 snRNA is threaded through the central tunnel of the NC between the RNA-binding motifs of the RecA and HB domains, with 3′-portions of the RNA extending toward the CC (). Consistent with the HB ratchet helix being involved in RNA unwinding, some brr2 mutations linked to the RP33 form of autosomal dominant retinitis pigmentosa in humansCitation60-62 lead to exchanges of RNA-contacting residues in this element (S1087L, R1090L). The S1087L RP33 exchange in human Brr2 reduces RNA affinity as well as ATPase and helicase activitiesCitation51 and RP33-like N1104L and R1107L exchanges in yeast Brr2 are associated with reduced ATP-dependent U4/U6 unwinding in tri-snRNP preparations.Citation60 Mutation of a Brr2 element equivalent to the Hel308 separator loop resulted in reduction or loss of cell viability,Citation41,52 in line with a similar duplex disruption mechanism as in Hel308. However, in a yeast U4/U6•U5 tri-snRNP structure,Citation12 Brr2 is loaded on the U4/U6 di-snRNA with the putative separator loop distant from the U4/U6 duplex portion to be unwound and instead with an edge of the RecA2 domain abutting the end of this duplex region (). Thus, the exact mechanism of RNA strand separation by Brr2 and the role of the separator loop remain to be clarified. As revealed by mutationalCitation51 and structuralCitation12 studies, the N-terminal HLH domain acts as an additional RNA-binding element, which engages a 3′-stem-loop of the U4 snRNA, most likely supporting substrate loading and guiding the downstream part of the bound substrate strand through the NC. It may thus provide an additional means to hold on to the substrate while RecA domains are repositioned on the RNA during translocation.
Apart from modulating the enzymatic mechanism of Brr2, the N-terminal Sec63-homology unit serves as a landing pad for a Brr2 regulator, the C-terminal Jab1 domain of the Prp8 protein (further discussed below).Citation38,53 Thus, the first layer of helicase-associated domains in Brr2 directly modulate the enzymatic mechanism of the enzyme and contribute to its regulation by trans-acting factors.
Layer II – the CC as an intra-molecular helicase cofactor
Close contacts between the active NC and inactive CC in crystal structures of Brr2Citation38,51,53 suggest that the NC and CC together form a larger functional unit. Indeed, deletion of the entire CC, exchange of residues in a long, irregularly structured linker that connects the cassettes, or exchange of residues that mediate NC-CC contacts affect the ATPase and helicase activities of the NC.Citation51 Strikingly, although the nucleotide-binding pockets of the 2 cassettes are separated by about 75 Å, a Brr2 variant, in which the nucleotide-binding site of the CC was changed to abrogate ATP binding, also led to reduced RNA duplex unwinding by the NC but left the NC ATPase activity unchanged.Citation51 These findings show that the CC acts as an intra-molecular cofactor of the NC, and that the 2 cassettes can communicate via long-range, intramolecular communication lines. These observations provide an explanation for why the CC is essential in yeastCitation52 although it is inactive as an enzyme. The molecular basis for CC-mediated activation of the NC is presently unclear. Mutational analyses are consistent with the idea that the CC acts as a stable scaffold, possibly further rigidified by bound nucleotide, which is exploited by the NC to efficiently cycle through different conformational states during RNA unwinding.Citation51
Y2H, pulldown and structural studies revealed that the CC also acts as a protein-protein interaction platform.Citation12,63-65 Given the long-range communication between NC and CC, it is conceivable that protein binding at the CC can also modulate the activity of the NC from a distance. An interesting candidate for such regulation is the tri-snRNP protein Snu66. In a cryo-EM structure of a yeast tri-snRNP,Citation12 the ∼200 C-terminal residues of Snu66 adopt an extended, partially helical conformation, and run around the CC. Upon spliceosome activation, Snu66 is released from the spliceosome,Citation28 restricting its putative influence on Brr2 activity to stages before activation.
Layer III – the NTR as an auto-inhibition device
Recent structural and functional studies showed that Brr2 activity is also regulated by its long NTRCitation55 and that this region is required for efficient splicing.Citation44,55 In vitro experiments assessing Brr2 activity upon stepwise NTR truncations revealed that the NTR down-regulates Brr2.Citation55 NTR-based auto-inhibition of Brr2 is implemented by at least 2 mechanisms. First, comparison of structures of full-length Brr2 in complex with the Prp8 Jab1 domainCitation55,56 with the structure of Brr2 bound to U4/U6 di-snRNA in the framework of a yeast tri-snRNPCitation12 indicated that the plug domain of the NTR upon folding back onto the helicase cassettes competes with accommodation of stem I of the substrate, which Brr2 has to unwind during spliceosome activation.Citation29 In full agreement with this structural analysis, RNA binding studies showed that the NTR has no effect on single-stranded RNA binding by Brr2, but inhibits binding of substrates that additionally include a duplex resembling U4/U6 stem I.Citation56 Second, the NC-clamp connects the RecA-like and WH domains of the active NC and locks the RecA1 and the RecA2 domains in a conformation not conducive to ATP hydrolysis.Citation55
The NTR is essential for yeast viability, splicing efficiency, stable association of Brr2 with the tri-snRNP and tri-snRNP stability in the presence of ATP.Citation44,55 Shortening of the NTR led to unconventional, ATP-dependent, Brr2-mediated tri-snRNP disruption into U4/U6 di-snRNP and U5 snRNP,Citation55 and removal of the first 120 residues of the NTR in yeast Brr2 elicited an increased loss of U5 and U6 snRNAs during spliceosome activation.Citation44 Similar to the CC, Y2HCitation54 and pulldown studiesCitation44 suggested that the NTR interacts with a large number of splicing factors implicated in different steps of splicing, consistent with the NTR supporting recruitment or anchoring of Brr2 to U5 snRNP and the tri-snRNP.Citation55 The diverse protein interactions of the NTR might help to reinforce, fine-tune or release Brr2 auto-inhibition via the NTR.
For efficient Brr2-mediated U4/U6 unwinding, auto-inhibition by the NTR has to be relieved. The NTR elements in their auto-inhibitory states interact with the helicase cassettes via a large, combined interface (∼4500 Å2). A continuous interface of this extent may be difficult to disrupt when Brr2 activity is required during splicing. However, distributing this area among several smaller interfaces may allow for a more facile, stepwise “peeling off” of the NTR to relieve Brr2 auto-inhibition. A similar principle has been suggested to facilitate release of bacterial σ factors from the RNA polymerase core enzyme during the transition from transcriptional initiation to elongation.Citation66
Similarities to other RNA helicases
Many SF2 helicases, e.g. all members of the DEAH/RHA and Ski2-like families, contain accessory domains that together with 2 RecA-like domains form active helicase cassettes, reminiscent of the NC of Brr2. Within these cassettes, all DEAH/RHA and Ski2-like helicases contain a Brr2-like ratchet (HB) domain connected via a WH domain to the RecA domains. However, additional domains are restricted to individual proteins or sub-groups of helicases. E.g., while the Ski2-like DNA helicase, Hel308, contains a HLH domain (which improves coupling of ATPase and unwinding activitiesCitation67) following the ratchet domain as also seen in Brr2, the Ski2-like RNA helicase, Mtr4, contains an arch-like domain inserted into its core (which aids in the recognition of specific substratesCitation68,69 and modulates helicase activityCitation70). The DEAH/RHA helicases Prp2, Prp16, Prp22 and Prp43, in contrast, contain a C-terminal OB-fold,Citation58,59,71,72 which in Prp43 stimulates RNA binding and ATPase activity.Citation59
Together with Brr2, only very few other helicases, including yeast Slh1pCitation73 and human ASCC3,Citation74 exhibit 2 helicase cassettes. These enzymes form a small clade within the Ski2-like family. Whether all of these dual-cassette enzymes contain an active and an inactive helicase cassette and whether similar principles of regulation as in Brr2 are at work is presently not known. At least the sequence of active and inactive cassettes does not seem to be universally conserved; in contrast to Brr2, the C-terminal cassette of the human ASCC3 enzyme has been found to be an active DNA helicase.Citation75
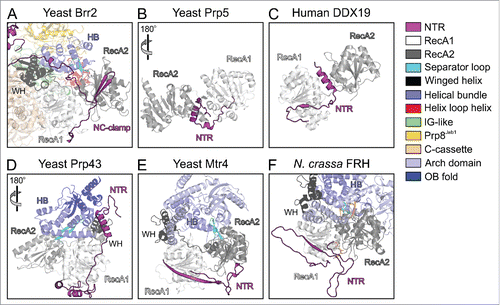
While helicase cassettes are largely composed of structured domains that can be identified based on the protein sequences, and whose functions may be deduced from those of corresponding domains in structurally and functionally characterized enzymes, RNA helicases often also contain N- or C-terminal extensions, which frequently lack recognizable domains. The lengths and sequences of these appendices are highly variable, and their functions can therefore not be predicted based on sequence information alone. Surveying known helicase structures, we found that in many cases N-terminal appendices fold back onto the helicase cassettes and might act as auto-inhibition elements as seen in Brr2 (). The DEAD box helicases Prp5Citation76 and DDX19Citation77 contain N-terminal extensions that locally fold as α-helices to wedge between the 2 RecA-like domains (). In these enzymes, the N-terminal extensions seem to stabilize an inactive relative orientation of the 2 RecA-like domains, with the surfaces that have to come together for ATP and RNA binding remote from each other. In contrast, the N-terminal extensions of Prp43,Citation58,59,71 yeast Mtr4Citation68,69 and Neurospora crassa frequency-interacting RNA helicase (FRH)Citation78 together with accessory domains stabilize a relative orientation of the RecA-domains that resembles the productive conformation (). Similar to Brr2, the N-terminal regions of Prp43, Mtr4 and FRH might thus serve to reduce conformational flexibility in the helicase cores, counteracting intrinsic NTPase, RNA binding and thus helicase activity. Structural bioinformatics analyses have revealed PWI-like domains at the very N-termini of the human Prp2 and Prp22 spliceosomal RNA helicases (but not in the respective yeast proteins),Citation79 but whether the enzymes' NTRs in general or the PWI-like domains in particular play similar regulatory roles as in Brr2 is presently not known.
Figure 3. Comparison of the N-terminal extensions of various SF2 helicases. Structures were spatially aligned with respect to their RecA1 domains. (A) Truncated yeast Brr2-Jab1 (PDB ID 5M5P).Citation56 (B) Yeast Prp5 (PDB ID 4LYJ).Citation76 (C) Human DDX19 (PDB ID 3EWS).Citation77 (D) Yeast Prp43 (PDB ID 2XAU).Citation59 (E) Yeast Mtr4 (PDB ID 4QU4).Citation70 (F) N. crassa FRH bound to ADP (PDB ID 5E02).Citation78 The color code for the various domains and regions is indicated on the right. Rotation symbols indicate views relative to panel (A).
Brr2 regulation via trans-acting factors
In addition to its intricate intra-molecular regulation, Brr2 is influenced by several other spliceosomal proteins, first and foremost Prp8. Prp8 is the largest and most highly conserved spliceosomal proteinCitation80 and forms a scaffold, on which the catalytic core of the spliceosome is assembled.Citation39,81 At its C-terminus, Prp8 comprises an RNase H-like (RH) domainCitation82-84 followed by a Jab1 domain,Citation85,86 both of which can regulate Brr2 function. The RH domain can bind to the single-stranded region of the U4 snRNA neighboring U4/U6 stem I (the U4 central domain), thus hindering Brr2 entry and inhibiting the helicase by substrate competition.Citation87 The Jab1 domain can directly and stably bind on top of the N-terminal Sec63 unit of Brr2 ().Citation38,53 In this position, it can insert an intrinsically disordered C-terminal tail into the Brr2 RNA-binding tunnel, thereby inhibiting accommodation of a substrate strand and consequently RNA duplex unwinding ().Citation53,56 The functional relevance of this mode of Brr2 inhibition is underscored by the observation that mutations in Prp8, which affect residues in the Jab1 C-terminal tail and interfere with its function, lead to a severe form of retinitis pigmentosa in human.Citation53,88 The tail insertion was first observed in the structure of a human Brr2-Jab1 complex,Citation53 in which the Brr2 subunit lacked all elements of the NTR except the NC-clamp, but was not seen in a slightly further truncated yeast Brr2-Jab1 complex.Citation38 These findings raised the question whether the Jab1 tail insertion is conserved among organisms. We addressed this question in the accompanying research paper and found that the tail insertion mechanism is conserved in the yeast, C. thermophilum and human systems.Citation56 Interestingly, however, the interaction network of the Jab1 tail with Brr2 differs between organisms. A structure of full-length yeast Brr2-Jab1Citation56 possibly explains the lack of a stably bound Jab1 tail in N-terminally truncated yeast Brr2-Jab1 complexesCitation38,56: the RecA2 domain can move relative to the remaining parts of the NC and is likely held fixed by the plug of the NTR, which is missing in these latter structures.
Remarkably, removal of the C-terminal 16 residues of the tail (Jab1ΔC) not only abrogates Jab1-mediated Brr2 inhibition but converts Jab1 into a potent activator of Brr2.Citation53,88 Likewise, Jab1 bearing an intact tail can enhance Brr2 activity and efficiency in vitro after displacement of the tail by larger amounts of substrate RNA.Citation53,88 Recent studies suggested that Jab1-mediated stimulation of Brr2 might be required at the stage of spliceosome catalytic activation, during which Brr2 needs to unwind U4/U6 di-snRNA decorated with U4/U6 proteins; addition of the U4/U6 di-snRNP proteins Snu13, Prp31 and Prp3 led to a stepwise decrease in unwinding activity, which could be efficiently restored upon addition of Jab1ΔC.Citation29 On the molecular level, Jab1-mediated Brr2 activation may in part be due to bridging of the N-terminal HB and RecA2 domains by the proximal portion of the Jab1 tail (), via which the U4 snRNA strand might be more efficiently entrapped at the NC. This notion is in line with the finding of an increased processivity of Brr2 in the presence of Jab1.Citation89
In addition to Prp8, the single spliceosomal G protein, Snu114, has been implicated in Brr2 regulation.Citation48,90,91 Like Brr2, Snu114 is a U5 snRNP protein required for spliceosome activationCitation91,92 and is also involved in spliceosome disassembly.Citation48 Furthermore, recent analyses suggested that spliceosome activation is also regulated by reversible ubiquitination of U4/U6-associated proteins.Citation93 We will briefly discuss possible roles of Snu114 and ubiquitin in Brr2 regulation during splicing later in this article.
Functional cross-talk among regulatory levels
Multiple lines of evidence support the view that the various cis- and trans-regulatory mechanisms of Brr2 do not merely act in isolation but are functionally coupled. Systematic RNA binding and unwinding analyses with Brr2 variants containing or lacking the CC revealed that the CC is important for the intra-molecular regulation via the NTR.Citation56 Furthermore, these analyses showed that different regulatory elements within the NTR mutually influence each other.Citation56 These findings can be understood by various regions of the NTR folding back onto neighboring surfaces of the NC and CC to achieve Brr2 auto-inhibitionCitation55,56 Similarly, comparison of the effects of Jab1 with or without its C-terminal tail on Brr2 variants containing or lacking the inactive CC showed that the Jab1 domain likewise requires the presence of the CC to take full effect on the NC.Citation53 As the Jab 1 domain does not foster direct contacts to the CC (), this effect must be mediated indirectly via contacts of the CC to the NC.
In addition to their cooperation with the CC, we recently uncovered an intricate regulatory interplay between the Brr2 NTR and the Prp8 Jab1 domain.Citation56 The NTR has no effect on the binding of single-stranded RNA, but it efficiently inhibits binding of substrates that contain a duplex region resembling stem I of U4/U6 di-snRNA. Conversely, the Jab1 domain in the absence of the NTR is only able to inhibit binding of single-stranded RNA but does not modulate binding of RNAs bearing a stem I-like duplex. The different behavior of the NTR and Jab1 domain with respect to single- and double-stranded substrates is fully in line with the plug domain of the NTR sterically hindering accommodation of the RNA duplex region to be unwound and the Jab1 C-terminal tail competing with accommodation of a single-stranded substrate strand at the RNA-binding tunnel. Moreover, crystal structure analysis of a full-length Brr2-Jab1 complex showed that auto-inhibition by the NTR and inhibition by the Jab1 domain can take effect at the same time.Citation56 Consistently, the NTR and the Jab1 domain mutually reinforce each other in inhibiting RNA binding and unwinding.Citation56 On the other hand, Jab1ΔC enhances Brr2-mediated U4/U6 unwinding irrespective of the presence of the NTR, and the NTR still exerts an inhibitory effect in the presence of Jab1ΔC, showing that NTR- and Jab1-based regulatory mechanisms can also act independently of each other. The two inhibitory mechanisms acting alone or in concert in principle afford opportunities for tuning Brr2 activity to various levels.
Brr2 regulation in the tri-snRNP
In a recent cryo-EM structure of a human U4/U6•U5 tri-snRNP,Citation14 Brr2 was found in an auto-inhibited conformation with the NTR folded back onto the helicase cassettes, very similar to the situation in isolated Brr2-Jab1 complexesCitation55,56 (). Furthermore, in that structure the Brr2 active site is positioned about 80 Å away from the Brr2 entry site on U4 snRNA (). Consistent with Brr2 held in an inhibited state, human tri-snRNP preparations are stable in the presence of ATP,Citation94 indicating that the human tri-snRNP structure represents a resting state of the tri-snRNP before delivery to the spliceosome. Furthermore, in full agreement with the suggested role as a multi-protein interaction device and as an anchor to the tri-snRNP,Citation54,55 the Brr2 NTR interacts with several other proteins in the human tri-snRNP, with the plug contacting Prp6 and the RH domain of Prp8 and the PWI domain interacting with Snu114 and Sad1 (). Sad1 is one of 3 proteins (together with Snu66 and 27K) that are part of the tri-snRNP but absent from the isolated U4/U6 and U5 snRNPs,Citation95,96 suggesting that it is crucial for restraining Brr2 in its inactive position and conformation in the tri-snRNP. In contrast to the NTR-mediated auto-inhibition, additional inhibition of Brr2 via the Jab1 C-terminal tail is not evident from the human tri-snRNP structure, possibly due to the limited resolution, but appears feasible as the globular part of the Prp8 Jab1 domain is bound to Brr2 in the same fashion as seen in isolation.
Figure 4. State of Brr2 in isolation and in tri-snRNPs. Diametric views of (A) an isolated yeast Brr2-Jab1 complex (PDB ID 5DCA),Citation55 (B) human U4/U6•U5 tri-snRNP (PDB ID 3JCR)Citation14 and (C) yeast U4/U6•U5 tri-snRNP (PDB ID 5GAN).Citation12 Brr2, U4 snRNA, U6 snRNA and proteins interacting with the Brr2 NTR or the CC are highlighted by colors. NTR, magenta; NC, dark gray; CC, beige; Prp8 RH domain, light orange; Jab1, gold; Snu114, dark red; Sad1, cyan; Prp6, dark green; Snu66, light green; Prp3, dark blue; U4 snRNA, brown; U6 snRNA, orange. Green asterisks in (B), right panel, location of the Brr2 active site; orange asterisks in (B), right panel, approximate location of the U4 central domain, where Brr2 initially binds the U4/U6 duplex. Views with Brr2 in the same orientation as in (A).
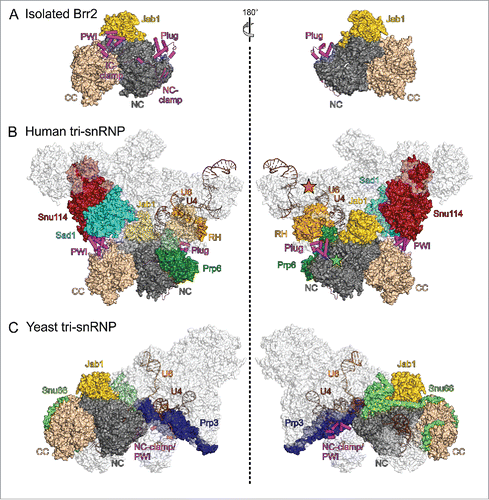
In stark contrast to the human tri-snRNP structure, the Brr2-Jab1 module is detached from Snu114, Prp6 and the Prp8 RH domain in cryo-EM structures of yeast tri-snRNP preparations, and Brr2 is loaded onto the central domain of U4 snRNACitation12,13 (). As required for productive RNA binding, the NTR is completely removed from the helicase core, instead reaching out toward the U4/U6 region of the tri-snRNP, and the single-stranded region of U4 displaces the Jab1 C-terminal tail from the Brr2 RNA-binding tunnel (). As the NTR is completely peeled off the Brr2 helicase region bound at U4 snRNA, U4-loaded Brr2 resembles a Brr2 variant that lacks the entire NTR, which is most active in in vitro unwinding assays.Citation55 Thus, in principle Brr2 in the yeast tri-snRNP structures is ready to unwind U4/U6 stem I. Consistently, the corresponding tri-snRNP preparations disintegrated upon addition of ATP,Citation97 as also observed previously with yeast tri-snRNP in extracts,Citation26,98 suggesting that the state captured in these structures does not correspond to that of a stable tri-snRNP outside the spliceosome in vivo. Notably, the yeast tri-snRNP preparations used for cryo-EM analyses lacked the Sad1 protein,Citation12,13 supporting the notion that Sad1 is crucial for keeping Brr2 inactive in the tri-snRNP and suggesting that the resting state of the yeast tri-snRNP, which includes Sad1, resembles the human tri-snRNP structure. Knockdown of Sad1 in yeast has been shown to lead to ATP-dependent, Brr2-mediated, non-canonical disruption of the tri-snRNP into U4/U6 and U5 particles,Citation99 recapitulating the effects seen with Brr2 NTR truncationsCitation55 and thus corroborating that Sad1 reinforces NTR-mediated Brr2 inhibition and takes advantage of the Brr2 NTR to position the helicase remote from its U4/U6 entry site in the resting state of the yeast tri-snRNP as well.
It is possible that the yeast tri-snRNP structures, in which Brr2 is apparently prematurely primed for U4/U6 disruption, resemble a situation in the pre-catalytic spliceosome directly before Brr2-mediated spliceosome activation. Under this assumption, the position of the Brr2 NTR in one of the yeast tri-snRNP structuresCitation12 would provide an explanation for the observed non-canonical Brr2-mediated tri-snRNP disruption into U4/U6 and U5Citation55 and for the increased loss of U5 and U6 during spliceosome activationCitation44 upon NTR truncation. In this structure, only the PWI domain and NC-clamp regions of the NTR could be traced and are seen to engage in a completely different interaction network than in the ATP-resistant human tri-snRNP structure. Here, part of the NC-clamp adopts a helical structure that together with part of the PWI domain interacts with Prp3 and U4/U6 stem II (). Thus, while in this state the NTR has given up its role of Brr2 auto-inhibition, it may now aid in the proper segregation of subunits during productive tri-snRNP disruption.
Brr2 regulation during splicing
NTR- and Jab1-based regulation during splicing
Very recently, cryo-EM structures of yeast spliceosomal Bact complexes,Citation100,101 of 2 states of the yeast spliceosome after step 1 (C complex)Citation102,103 and of a post-catalytic intron-lariat spliceosomeCitation81 have been determined. These structures together with previous functional analyses support a picture, in which Brr2 aids in recruiting and possibly regulating other spliceosome remodeling enzymes, during which Brr2 itself is apparently inhibited by different combinations of NTR- and Jab1-based mechanisms. Thus, these results illustrate potential roles of Brr2 during the catalytic phase of splicing and during spliceosome disassembly, in which it was previously implicated,Citation46-48 which do not necessarily rely on the enzyme's ATPase/helicase activities.Citation46,50
Brr2 is located on one side of a triangular central body of the spliceosomal Bact complex, close to the DEAH/RHA helicase Prp2 ().Citation100,101 In Bact, the Brr2 NTR adopts its auto-inhibited state, wrapped around the Brr2 helicase cassettes, as in the Brr2-Jab1 complexes and as in the human tri-snRNP structure. Additionally, the globular part of the Prp8 Jab1 domain remains bound to the Brr2 NC and the Jab1 C-terminal tail is occupying the Brr2 RNA-binding tunnel, indicating that the NTR and Jab1 domain together shut Brr2 down after spliceosome activation (). Prp2 is required for the transition from the Bact to the B* complex,Citation104 during which it remodels binding of the multimeric SF3b complex to the branch point region,Citation17,105 reduces association of SF3a and SF3b proteins,Citation106 leads to displacement of the Cwc24 and Cwc27 components of the 19 complex (NTC) as well as of the Bud13 subunit of the RNA retention and splicing (RES) complexCitation106 and proofreads the catalytic core of the spliceosome.Citation107 Consistent with some of these functions, the SF3B1 (Hsh155) protein in the Bact complex bridges between the plug domain of Brr2 and Prp2. Brr2 additionally contacts the splicing factors SF3B3 (Res1), Cwc24, Cwc27 and the endonuclease domain of Prp8. While Brr2 and Prp2 do not directly interact in Bact, Y2H analyses have suggested direct contacts between the proteins involving the C-terminal HB domain of Brr2.Citation64 Thus, Brr2 might aid in recruiting Prp2 to the spliceosome but direct interactions between the proteins may be lost upon accommodation of Prp2 in the Bact complex.
Figure 5. State of Brr2 in different spliceosomal contexts and interaction with other spliceosomal helicases. Views of (A) a yeast spliceosomal Bact complex (PDB ID 5GM6)Citation100 and (B) a yeast spliceosomal C complex (PDB ID 5LJ5).Citation102 Brr2 and proteins interacting with the Brr2 NTR, and the Prp2 and Prp16 helicases are highlighted by colors. NTR, magenta; NC, dark gray; CC, beige; Jab1, gold; SF3B1, dark teal; Prp2, red; Prp16, dark red. Views with Brr2 in the same orientation as in . , left, is turned 30° around the vertical axis compared to .
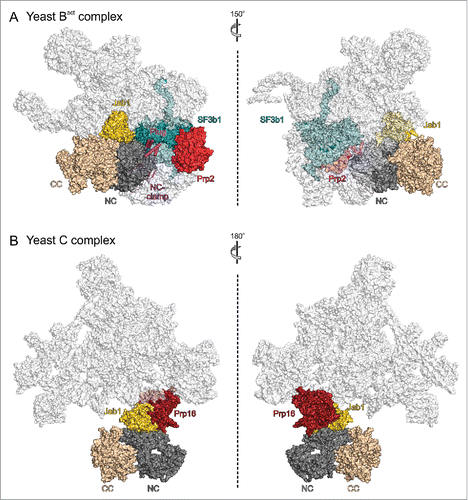
In one structure of a yeast post-step 1 spliceosome,Citation102 Brr2 uses the HLH domain of its NC to directly interact with the DEAH/RHA helicase Prp16 (). Prp16 is required for spliceosome rearrangements that enable second step catalysis,Citation108,109 including the displacement of the NTC-related protein Yju2 and the C complex protein Cwc25.Citation110 Furthermore, it contributes to the fidelity of branchingCitation111-113 and enables alternative 3′SS selection in an ATP-dependent mannerCitation114 and in cooperation with the ISY1 protein of the intron-binding complex.Citation115 Similar to the situation involving Brr2 and Prp2 in the Bact complex, Y2H analyses had indicated interactions of the C-terminal Sec63 homology unitCitation64 and of the NTRCitation54 of Brr2 with Prp16, which are not seen in this structure of the yeast C complex. Thus, Brr2 might again capitalize on different contacts to Prp16 to aid in its recruitment, or it might interact differently with Prp16 during proofreading or in different alternative splicing scenarios to regulate Prp16 activity via protein-protein interactions. Consistent with the first suggestion, Brr2 is not held in a fixed position but is flexibly connected to the spliceosomal body in another structure of a post-step 1 spliceosome,Citation103 which lacks Prp16. This post-step 1 spliceosome might thus represent a stage preceding stable integration of Prp16, where Brr2 is acting as a flexibly tethered recruitment platform for Prp16. Consistent with the notion of Brr2 regulating Prp16 activity via alternative interactions, in vivo and in vitro studies suggested a model in which the C-terminal Sec63 unit of Brr2, in particular the HB domain, inhibits the ATPase activity of Prp16 by interfering with its ability to bind RNA; this inhibition was suggested to involve stabilization of a 5′HP/separator loop-OB domain interaction in Prp16, resulting in an obstruction of its RNA-binding site.Citation64 Upon correct positioning of Prp16 in the C complex this inhibition may be released and Brr2 may remain associated with Prp16 through interactions of its N-terminal cassette HLH domain. Mutations within the HB domain of the Brr2 CC also led to changes in interactions with Prp2,Citation64 indicating that both helicases may engage in similar contacts with and are possibly inhibited by Brr2 during their recruitment, until they are accommodated in the spliceosome.
While it exerts Prp16-related functions, Brr2's own helicase activity might have to be turned off. As there is no evidence in the C complex structure for the NTR auto-inhibiting Brr2, Brr2 inhibition at this stage might depend on the Jab1 C-terminal tail (), which would be in line with the Prp8 Jab1 domain being important for Brr2 function during splicing catalysis.Citation47 This organization would leave the Brr2 NTR available to engage in contacts with and possibly regulate the activity of Prp16.Citation54 However, the limited local resolution in the corresponding region of the Prp16-containing C complex structure does not unequivocally reveal the state of the NTR or the Jab1 domain.
Following mRNP displacement from the post-catalytic spliceosome by the Prp22 helicase, the Prp43 enzyme elicits disassembly of the remaining intron-lariat spliceosome.Citation50,116 Prp43 is recruited in complex with the Ntr1 and Ntr2 proteins during C complex formation, before the second step of splicing.Citation16 As Brr2 can interact with the Ntr2 subunit of this complex,Citation117 it again may support recruitment of the Prp43 machinery for intron-lariat spliceosome disassembly. However, Ntr2 is not essential for association of Prp43 with the spliceosome, yet its presence is required to prevent Prp43-mediated disruption of intact spliceosomal intermediates other than the intron-lariat spliceosome.Citation118 Thus, after accommodation of Prp43, Brr2 might further modulate Prp43 activities based on its intractions with Ntr2. Similar to the structure of a post-step 1 spliceosome lacking Prp16,Citation103 Brr2 is also flexibly tethered in a post-catalytic intron-lariat spliceosome and Prp43 has not been located in that structure,Citation81 presently leaving the structural basis for such putative regulatory interactions unresolved.
Being tethered solely by a long, flexible linker between the Prp8 RH and Jab1 domains in some stages of splicing, Brr2 would be in an ideal configuration for exerting a “fishing” function for other spliceosome remodeling enzymes. A similar role has been attributed to the globular head groups of the bacterial ribosomal L12 proteins, which are flexibly tethered to the large ribosomal subunit and serve as initial docking sites for translation factor GTPases and, after recruitment of these proteins, regulate their GTPase activities.Citation119 As Brr2 in these states obviously lacks contacts to spliceosomal elements other than the Prp8 Jab1 domain, the Brr2-Jab1 complex might well adopt its fully inhibited conformation as seen in an isolated Brr2-Jab1 structureCitation56 ().
Intermittent occupation of the Brr2 RNA-binding tunnel by the Jab1 tail might also be important at stages when Brr2 inhibition has to be maintained, even though the NTR is detached from the helicase core. For example, during priming of a pre-catalytic spliceosome for Brr2-mediated activation, where the NTR has to detach from the helicase cassettes and might interact with other spliceosomal components to guide productive tri-snRNP disruption, the Jab1 tail might inhibit Brr2 until it encounters its cognate U4/U6 substrate to avoid off-target effects (). In this scenario, the Jab1 tail bound at the RNA-binding tunnel of Brr2 may provide a selectivity filter, whose binding strength is appropriately tuned to allow its displacement by the cognate U4/U6 duplex (which comprises multiple regions that associate with Brr2), but not by non-cognate RNAs such as the substrate pre-mRNA (which might predominantly present single-stranded regions that are efficiently competed out by the Jab1 tail). Fine-tuning of the binding strength of the Jab1 C-terminal tail to the Brr2 RNA-binding tunnel likely relies on the intrinsically unstructured nature of this tail.Citation88
Taken together, presently available structures of isolated Brr2-Jab1 complexes, tri-snRNPs and spliceosomes in combination with functional studies support the notion that the NTR is essential to shut off Brr2 activity during tri-snRNP assembly and during several stages of splicing, and that the Jab1 domain remains stably bound to Brr2 during all phases of splicing, in which Brr2 is present (). Jab1-mediated activation of Brr2 is apparently crucial for efficient U4/U6 disruption during spliceosome activation (). NTR- and Jab1-based Brr2 inhibition might take effect individually or in combination, the latter in particular during stages of splicing, in which Brr2 is tethered in a flexible manner to the spliceosomal body ().
Additional mechanisms of Brr2 regulation during splicing
Presently, it is unclear how Brr2 is induced to assume its active conformation and position during priming of the pre-catalytic spliceosome for activation. A candidate protein that might induce Brr2 activation is Snu114. As Snu114 exhibits striking resemblance to the translational translocases, EF-G and eEF2, the protein has been proposed to undergo conformational changes upon GTP hydrolysis that might drive spliceosomal rearrangements.Citation90-92 Another study likened Snu114 to small regulatory G proteins, with the GDP-bound state of Snu114 inhibiting U4/U6 unwinding and spliceosome disassembly and the GTP-bound state activating both processes.Citation48 In the human tri-snRNP structure,Citation14 which might resemble the state of the tri-snRNP immediately following B complex formation, the PWI domain of Brr2 interacts with Snu114 (). Thus, it appears feasible that Snu114 helps to initially lock Brr2 in an inhibited conformation and position in the pre-catalytic spliceosome. Upon GTP hydrolysis, a conformational change could initiate detachment of the Brr2 helicase core from this inhibitory configuration to allow its loading onto U4 snRNA. However, while biochemical analyses provided evidence for the requirement of Snu114-dependent GTP hydrolysis during the activation phase of splicing,Citation90 mutations that affect a putative catalytic residue in Snu114 or a putative Prp8-dependent GTPase inhibition mechanism did not show growth defects in yeast.Citation12 Thus, it presently remains unclear whether and at which stage Snu114-mediated GTP hydrolysis is required and whether and how it might be linked to Brr2 regulation and spliceosome activation.
Likewise, it remains to be elucidated how yet other regulatory mechanisms, established via the Prp8 RH domainCitation87 or via reversible ubiquitination of U4/U6 proteins,Citation93 are implemented on the molecular level. As the RH domain can interact with the NTR of Brr2Citation14 as well as with U4/U6Citation87 and can dramatically change its position during splicing,Citation100-103 it may intermittently contact U4/U6 during priming of a pre-catalytic spliceosome for Brr2-mediated activation, thus guiding the core of Brr2 to its entry site on U4 snRNA. Interestingly, the Prp8 Jab1 domain resembles corresponding domains in deubiquitinating enzymes.Citation85,86 Although it is catalytically inactive as a deubiquitinase, it can still bind ubiquitin.Citation120 Thus, it is conceivable that ubiquitinated spliceosomal proteins contact the Jab1 domain via their ubiquitin units and influence its ability to inhibit or activate Brr2.
Implications for Brr2-dependent proofreading and regulation of alternative splicing
All universally conserved spliceosomal helicases, except Brr2 and UAP56/Sub2, have previously been shown to act as proofreaders by channeling sub-optimal substrates into discard pathways; they thereby enhance splicing fidelity and can influence the outcome of alternative splicing decisions by differentially affecting the kinetics of competing splicing events.Citation107,114,121-124 As Brr2 may be involved in the recruitment of other spliceosomal remodeling enzymes during various stages of splicing and might additionally influence their activities via protein-protein interactions (see above), it could modulate splicing fidelity and alternative splicing indirectly via recruitment and regulation of these other enzymes. This idea is in line with previous proposals of Brr2 functioning as a landing pad for multiple other helicases, which might sequentially interact in similar fashion with the C-terminal cassette.Citation40,41,52,63,64
Based on the observations that a temperature-sensitive yeast Brr2 variant (E909K), encoded by the slt22-1 allele, is synthetically lethal with mutations in U2 or U6 snRNAs that affect the stability or conformation of U2/U6 helix II, and that the ATPase activity of this variant is no longer stimulated by a U2/U6 duplex, it was proposed that Brr2 might proofread U2/U6 interactions.Citation45 Furthermore, different cross-linking profiles of wt Brr2 and a variant exhibiting a residue exchange (G858R) in the NC 5′HP/separator loop with U6 snRNACitation46 together with the latter variant exhibiting a second step defect and showing synthetic lethality with step 2 mutations in Slu7, Prp18 and Prp16Citation46,49 suggest that Brr2 participates in a transient opening of the catalytic core between the 2 steps of splicing, which is characterized by the intermittent disruption of U6-5SS and U2-U6 interactions.Citation125,126 However, whether Brr2 actively disrupts RNA-RNA or RNA-protein interaction in this phase of splicing or whether it simply provides RNA binding sites is presently unclear.Citation46
Curiously, the E909K exchange in Brr2 blocks splicing in extracts at or before the first catalytic step and leads to the appearance of an off-pathway spliceosomal particle following B complex formation, which lacks U4 and U5 snRNAs.Citation45 E909 is located in the linker connecting the RecA2 to the WH domains in the NC of Brr2 and apparently stabilizes the relative orientation of the domains similar to the NC-clamp of the NTR (). As also deletion of the NTR influences the ATPase activity of Brr2 (albeit increasing it)Citation55 and leads to increased loss of U5 and U6 snRNAs during spliceosome activation,Citation44 several Brr2 elements apparently control the Brr2 ATPase (and thus likely helicase) activity by adjusting the relative orientations of NC domains and/or their conformational flexibility. Manipulation of these elements apparently affects the partitioning of spliceosomes between productive activation or a discard pathway that is characterized by the loss of U5 snRNA. Interestingly, several brr2 mutations linked to the RP33 form of autosomal dominant retinitis pigmentosa map to the linker between the RecA domains of the NC (R681C, R681H, V683L) and to the beginning of the RecA2 domain (Y689C), where they mediate inter-RecA contacts or stabilize the fold of the RecA2 domain.Citation51,62 Similar to changes in the elements that affect relative domain arrangements and conformational flexibility of the NC, RP33-linked exchanges in these residues may lead to altered Brr2 ATPase activity and aberrant partitioning of spliceosomes along activation and discard pathways, constituting a novel disease principle.
The above hypothesis is in line with a recent analysis of the mechanism of spliceosome activation using multi-wavelength single-molecule co-localization spectroscopy, which demonstrated that after tri-snRNP binding, the spliceosome can either proceed to activation or release U4 and U5 snRNAs.Citation127 The ATP-dependent loss of U4 and U5 snRNAs was suggested to represent Prp28-mediated displacement of the tri-snRNP.Citation127 However, the observations that Sad1 knockdownCitation99 or truncation of the Brr2 NTRCitation55 lead to non-canonical Brr2-mediated disruption of the tri-snRNP, that NTR truncations lead to increased loss of U5 and U6 snRNAs upon spliceosome activationCitation44 and that a mutation of a Brr2 residue (E909), which supports the relative domain arrangement in the NC, is associated with the loss of U4 and U5 snRNAs from B complex spliceosomesCitation45 indicate that the process observed could alternatively represent a Brr2-mediated discard pathway.
In light of the above considerations, it is conceivable that the NC domain arrangement, the interaction of the NTR with the Brr2 helicase region and/or the reinforcement of this interaction by Sad1 are differentially influenced during tri-snRNP recruitment or during a priming phase for spliceosome activation in different splicing scenarios, leading to a different likelihood of the resulting pre-catalytic complexes to partition along activation or a discard pathway (). This partitioning might be further influenced by a different susceptibility of the pre-catalytic complexes to intermittent Brr2 inhibition by the Jab1 C-terminal tail. Thus, the multiple regulatory mechanisms acting on Brr2 might be used in a combinatorial fashion to affect splicing kinetics and the outcome of alternative splicing decisions ().
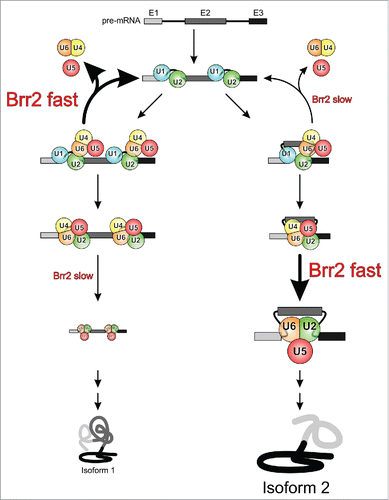
Figure 6. Model for putative Brr2-mediated enhancement of splicing fidelity and regulation of alternative splicing. Depending on the state of the NTR and the Sad1 protein in competing alternative splicing scenarios (left and right branches), Brr2 may be more or less prone to disrupt the tri-snRNP in a non-productive fashion, thus differentially channeling the different substrates along the splicing or discard pathways. Similarly, depending on the level of Brr2 inhibition in competing alternative splicing scenarios, the helicase may elicit spliceosome activation slowly or quickly, kinetically controlling the levels of protein isoforms produced.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Reinhard Lührmann, Berthold Kastner and Holger Stark, Max-Planck-Institute for Biophysical Chemistry, Göttingen, Germany, for sharing data before publication.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (SFB 740 to MCW), the Bundesministerium für Bildung und Forschung (grant 05K10KEC to MCW) and the Einstein Foundation Berlin (grant A-2012-140 to MCW). KFS was supported by a Dahlem International Network PostDoc Fellowship from Freie Universität Berlin.
References
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, Atkins JF, eds. The RNA world First edition. Cold Spring Harbor, NY: Cold Spring Harbor Labratory Press, 1993:303-57
- Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011; 3:1-24; http://dx.doi.org/10.1101/cshperspect.a003707
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009; 136:701-18; PMID:19239890; http://dx.doi.org/10.1016/j.cell.2009.02.009
- Will CL, Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 2001; 13:290-301; PMID:11343899; http://dx.doi.org/10.1016/S0955-0674(00)00211-8
- Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet 2002; 36:333-60; PMID:12429696; http://dx.doi.org/10.1146/annurev.genet.36.043002.091635
- Wahl MC, Lührmann R. SnapShot: Spliceosome Dynamics I. Cell 2015; 161:1474-e1; PMID:26046445; http://dx.doi.org/10.1016/j.cell.2015.05.050
- Wahl MC, Lührmann R. SnapShot: Spliceosome Dynamics II. Cell 2015; 162:456-e1; PMID:26186196
- Boesler C, Rigo N, Anokhina MM, Tauchert MJ, Agafonov DE, Kastner B, Urlaub H, Ficner R, Will CL, Lührmann R. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat Commun 2016; 7:11997; PMID:27377154; http://dx.doi.org/10.1038/ncomms11997
- Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 1988; 334:213-8; PMID:3041282; http://dx.doi.org/10.1038/334213a0
- Hashimoto C, Steitz JA. U4 and U6 RNAs coexist in a single small nuclear ribonucleoprotein particle. Nucleic Acids Res 1984; 12:3283-93; PMID:6201826; http://dx.doi.org/10.1093/nar/12.7.3283
- Bringmann P, Appel B, Rinke J, Reuter R, Theissen H, Luhrmann R. Evidence for the existence of snRNAs U4 and U6 in a single ribonucleoprotein complex and for their association by intermolecular base pairing. EMBO J 1984; 3:1357-63; PMID:6204860
- Nguyen TH, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SH, Nagai K. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. Nature 2016; 530:298-302; PMID:26829225; http://dx.doi.org/10.1038/nature16940
- Wan RX, Yan CY, Bai R, Wang L, Huang M, Wong CCL, Shi Y. The 3.8 angstrom structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science 2016; 351:466-75; PMID:26743623; http://dx.doi.org/10.1126/science.aad6466
- Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Lührmann R, Stark H. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 2016; 351:1416-20; PMID:26912367; http://dx.doi.org/10.1126/science.aad2085
- Liu S, Rauhut R, Vornlocher HP, Lührmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA 2006; 12:1418-30; PMID:16723661; http://dx.doi.org/10.1261/rna.55406
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R, Stark H. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 2009; 36:593-608; PMID:19941820; http://dx.doi.org/10.1016/j.molcel.2009.09.040
- Warkocki Z, Odenwälder P, Schmitzova J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 2009; 16:1237-43; PMID:19935684; http://dx.doi.org/10.1038/nsmb.1729
- Yeh TC, Liu HL, Chung CS, Wu NY, Liu YC, Cheng SC. Splicing factor Cwc22 is required for the function of Prp2 and for the spliceosome to escape from a futile pathway. Mol Cell Biol 2011; 31:43-53; PMID:20956557; http://dx.doi.org/10.1128/MCB.00801-10
- Nguyen THD, Galej WP, Fica SM, Lin PC, Newman AJ, Nagai K. CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast. Curr Opin Struc Biol 2016; 36:48-57; http://dx.doi.org/10.1016/j.sbi.2015.12.005
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 1998; 92:315-26; PMID:9476892; http://dx.doi.org/10.1016/S0092-8674(00)80925-3
- Cordin O, Hahn D, Beggs JD. Structure, function and regulation of spliceosomal RNA helicases. Curr Opin Cell Biol 2012; 24:431-8; PMID:22464735; http://dx.doi.org/10.1016/j.ceb.2012.03.004
- Cordin O, Beggs JD. RNA helicases in splicing. RNA Biol 2013; 10:83-95; PMID:23229095; http://dx.doi.org/10.4161/rna.22547
- Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 1999; 3:55-64; PMID:10024879; http://dx.doi.org/10.1016/S1097-2765(00)80174-4
- Price AM, Görnemann J, Guthrie C, Brow DA. An unanticipated early function of DEAD-box ATPase Prp28 during commitment to splicing is modulated by U5 snRNP protein Prp8. RNA 2014; 20:46-60; PMID:24231520; http://dx.doi.org/10.1261/rna.041970.113
- Laggerbauer B, Achsel T, Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci USA 1998; 95:4188-92; PMID:9539711; http://dx.doi.org/10.1073/pnas.95.8.4188
- Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 1998; 8:847-55; PMID:9705931; http://dx.doi.org/10.1016/S0960-9822(07)00345-4
- Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA 1999; 5:959-71; PMID:10411139; http://dx.doi.org/10.1017/S135583829999012X
- Agafonov DE, Deckert J, Wolf E, Odenwalder P, Bessonov S, Will CL, Urlaub H, Lührmann R. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol Cell Biol 2011; 31:2667-82; PMID:21536652; http://dx.doi.org/10.1128/MCB.05266-11
- Theuser M, Hobartner C, Wahl MC, Santos KF. Substrate-assisted mechanism of RNP disruption by the spliceosomal Brr2 RNA helicase. Proc Natl Acad Sci USA 2016; 113:7798-803; PMID:27354531; http://dx.doi.org/10.1073/pnas.1524616113
- Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol 2004; 11:1237-42; PMID:15543154; http://dx.doi.org/10.1038/nsmb863
- Hang J, Wan R, Yan C, Shi Y. Structural basis of pre-mRNA splicing. Science 2015; 349:1191-8; PMID:26292705; http://dx.doi.org/10.1126/science.aac8159
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature 2013; 503:229-34; PMID:24196718
- Burke JE, Sashital DG, Zuo XB, Wang YX, Butcher SE. Structure of the yeast U2/U6 snRNA complex. RNA 2012; 18:673-83; PMID:22328579; http://dx.doi.org/10.1261/rna.031138.111
- Gottschalk A, Kastner B, Lührmann R, Fabrizio P. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA 2001; 7:1554-65; PMID:11720285
- Boon KL, Grainger RJ, Ehsani P, Barrass JD, Auchynnikava T, Inglehearn CF, Beggs JD. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol 2007; 14:1077-83; PMID:17934474; http://dx.doi.org/10.1038/nsmb1303
- Weber G, Cristao VF, Santos KF, Mozaffari-Jovin S, Heroven AC, Holton N, Lührmann R, Beggs JD, Wahl MC. Structural basis for dual roles of Aar2p in U5 snRNP assembly. Genes Dev 2013; 27:525-40; PMID:23442228; http://dx.doi.org/10.1101/gad.213207.113
- Weber G, Cristao VF, de L. Alves F, Santos KF, Holton N, Rappsilber J, Beggs JD, Wahl MC. Mechanism for Aar2p function as a U5 snRNP assembly factor. Genes Dev 2011; 25:1601-12; PMID:21764848; http://dx.doi.org/10.1101/gad.635911
- Nguyen TH, Li J, Galej WP, Oshikane H, Newman AJ, Nagai K. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure 2013; 21:910-19; PMID:23727230; http://dx.doi.org/10.1016/j.str.2013.04.017
- Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 2013; 493:638-43; PMID:23354046; http://dx.doi.org/10.1038/nature11843
- Maeder C, Kutach AK, Guthrie C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol 2009; 16:42-8; PMID:19098916; http://dx.doi.org/10.1038/nsmb.1535
- Pena V, Mozaffari-Jovin S, Fabrizio P, Orlowski J, Bujnicki JM, Lührmann R, Wahl MC. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell 2009; 35:454-66; PMID:19716790; http://dx.doi.org/10.1016/j.molcel.2009.08.006
- Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Lührmann R, Carlomagno T, Wahl MC. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science 2007; 316:115-20; PMID:17412961; http://dx.doi.org/10.1126/science.1137924
- Liu S, Mozaffari-Jovin S, Wollenhaupt J, Santos KF, Theuser M, Dunin-Horkawicz S, Fabrizio P, Bujnicki JM, Lührmann R, Wahl MC. A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing. Elife 2015; 4:e07320; PMID:26161500
- Zhang L, Li X, Hill RC, Qiu Y, Zhang W, Hansen KC, Zhao R. Brr2 plays a role in spliceosomal activation in addition to U4/U6 unwinding. Nucleic Acids Res 2015; 43:3286-97; PMID:25670679; http://dx.doi.org/10.1093/nar/gkv062
- Xu D, Nouraini S, Field D, Tang SJ, Friesen JD. An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature 1996; 381:709-13; PMID:8649518; http://dx.doi.org/10.1038/381709a0
- Hahn D, Kudla G, Tollervey D, Beggs JD. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev 2012; 26:2408-21; PMID:23124065; http://dx.doi.org/10.1101/gad.199307.112
- Mayerle M, Guthrie C. Prp8 retinitis pigmentosa mutants cause defects in the transition between the catalytic steps of splicing. RNA 2016; 22:793-809; PMID:26968627; http://dx.doi.org/10.1261/rna.055459.115
- Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 2006; 23:389-99; PMID:16885028; http://dx.doi.org/10.1016/j.molcel.2006.05.043
- Lin J, Rossi JJ. Identification and characterization of yeast mutants that overcome an experimentally introduced block to splicing at the 3′ splice site. RNA 1996; 2:835-48; PMID:8752092
- Fourmann JB, Schmitzova J, Christian H, Urlaub H, Ficner R, Boon KL, Fabrizio P, Lührmann R. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev 2013; 27:413-28; PMID:23431055; http://dx.doi.org/10.1101/gad.207779.112
- Santos KF, Mozaffari-Jovin S, Weber G, Pena V, Lührmann R, Wahl MC. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci USA 2012; 109:17418-23; PMID:23045696; http://dx.doi.org/10.1073/pnas.1208098109
- Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol 2009; 16:731-9; PMID:19525970; http://dx.doi.org/10.1038/nsmb.1625
- Mozaffari-Jovin S, Wandersleben T, Santos KF, Will CL, Lührmann R, Wahl MC. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science 2013; 341:80-4; PMID:23704370; http://dx.doi.org/10.1126/science.1237515
- Absmeier E, Rosenberger L, Apelt L, Becke C, Santos KF, Stelzl U, Wahl MC. A noncanonical PWI domain in the N-terminal helicase-associated region of the spliceosomal Brr2 protein. Acta Crystallogr D 2015; 71:762-71; PMID:25849387; http://dx.doi.org/10.1107/S1399004715001005
- Absmeier E, Wollenhaupt J, Mozaffari-Jovin S, Becke C, Lee CT, Preussner M, Heyd F, Urlaub H, Lührmann R, Santos KF. The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation. Genes Dev 2015; 29:2576-87; PMID:26637280
- Absmeier E, Becke C, Wollenhaupt J, Santos KF, Wahl MC. Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2. Cell Cycle 2016; submitted.
- Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol 2007; 14:647-52; PMID:17558417; http://dx.doi.org/10.1038/nsmb1246
- He Y, Andersen GR, Nielsen KH. Structural basis for the function of DEAH helicases. EMBO Rep 2010; 11:180-6; PMID:20168331; http://dx.doi.org/10.1038/embor.2010.11
- Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, Henry Y, Leulliot N. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J 2010; 29:2194-204; PMID:20512115; http://dx.doi.org/10.1038/emboj.2010.102
- Zhao C, Bellur DL, Lu SS, Zhao F, Grassi MA, Bowne SJ, Sullivan LS, Daiger SP, Chen LJ, Pang CP, et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet 2009; 85:617-27; PMID:19878916; http://dx.doi.org/10.1016/j.ajhg.2009.09.020
- Li N, Mei H, MacDonald IM, Jiao X, Hejtmancik JF. Mutations in ASCC3L1 on 2q11.2 are associated with autosomal dominant retinitis pigmentosa in a Chinese family. Invest Ophthalmol Vis Sci 2010; 51:1036-43; PMID:19710410; http://dx.doi.org/10.1167/iovs.09-3725
- Benaglio P, McGee TL, Capelli LP, Harper S, Berson EL, Rivolta C. Next generation sequencing of pooled samples reveals new SNRNP200 mutations associated with retinitis pigmentosa. Hum Mutat 2011; 32:E2246-58; PMID:21618346; http://dx.doi.org/10.1002/humu.21485
- van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 2001; 157:1451-67; PMID:11290703
- Cordin O, Hahn D, Alexander R, Gautam A, Saveanu C, Barrass JD, Beggs JD. Brr2p carboxy-terminal Sec63 domain modulates Prp16 splicing RNA helicase. Nucleic Acids Res 2014; 42:13897-910; PMID:25428373; http://dx.doi.org/10.1093/nar/gku1238
- Chen HC, Tseng CK, Tsai RT, Chung CS, Cheng SC. Link of NTR-mediated spliceosome disassembly with DEAH-box ATPases Prp2, Prp16, and Prp22. Mol Cell Biol 2013; 33:514-25; PMID:23166295; http://dx.doi.org/10.1128/MCB.01093-12
- Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 2003; 13:31-9; PMID:12581657; http://dx.doi.org/10.1016/S0959-440X(02)00005-2
- Woodman IL, Briggs GS, Bolt EL. Archaeal Hel308 domain V couples DNA binding to ATP hydrolysis and positions DNA for unwinding over the helicase ratchet. J Mol Biol 2007; 374:1139-44; PMID:17991488; http://dx.doi.org/10.1016/j.jmb.2007.10.004
- Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J 2010; 29:2205-16; PMID:20512111; http://dx.doi.org/10.1038/emboj.2010.107
- Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci USA 2010; 107:12139-44; PMID:20566885; http://dx.doi.org/10.1073/pnas.1004953107
- Taylor LL, Jackson RN, Rexhepaj M, King AK, Lott LK, van Hoof A, Johnson SJ. The Mtr4 ratchet helix and arch domain both function to promote RNA unwinding. Nucleic Acids Res 2014; 42:13861-72; PMID:25414331; http://dx.doi.org/10.1093/nar/gku1208
- Tauchert MJ, Fourmann JB, Christian H, Luhrmann R, Ficner R. Structural and functional analysis of the RNA helicase Prp43 from the thermophilic eukaryote Chaetomium thermophilum. Acta Crystallogr F 2016; 72:112-20; PMID:NOT_FOUND; http://dx.doi.org/10.1107/S2053230X15024498
- Kudlinzki D, Schmitt A, Christian H, Ficner R. Structural analysis of the C-terminal domain of the spliceosomal helicase Prp22. Biol Chem 2012; 393:1131-40; PMID:23096351; http://dx.doi.org/10.1515/hsz-2012-0158
- Martegani E, Vanoni M, Mauri I, Rudoni S, Saliola M, Alberghina L. Identification of gene encoding a putative RNA-helicase, homologous to SKI2, in chromosome VII of Saccharomyces cerevisiae. Yeast 1997; 13:391-7; PMID:9133744; http://dx.doi.org/10.1002/(SICI)1097-0061(19970330)13:4%3c391::AID-YEA92%3e3.0.CO;2-Q
- Kim HJ, Yi JY, Sung HS, Moore DD, Jhun BH, Lee YC, Lee JW. Activating signal cointegrator 1, a novel transcription coactivator of nuclear receptors, and its cytosolic localization under conditions of serum deprivation. Mol Cell Biol 1999; 19:6323-32; PMID:10454579; http://dx.doi.org/10.1128/MCB.19.9.6323
- Dango S, Mosammaparast N, Sowa ME, Xiong LJ, Wu F, Park K, Rubin M, Gygi S, Harper JW, Shi Y. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol Cell 2011; 44:373-84; PMID:22055184; http://dx.doi.org/10.1016/j.molcel.2011.08.039
- Zhang ZM, Yang F, Zhang J, Tang Q, Li J, Gu J, Zhou J, Xu YZ. Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly. Cell Rep 2013; 5:1269-78; PMID:24290758; http://dx.doi.org/10.1016/j.celrep.2013.10.047
- Collins R, Karlberg T, Lehtio L, Schutz P, van den Berg S, Dahlgren LG, Hammarström M, Weigelt J, Schüler H. The DEXD/H-box RNA Helicase DDX19 Is Regulated by an alpha-Helical Switch. J Biol Chem 2009; 284:10296-300; PMID:19244245; http://dx.doi.org/10.1074/jbc.C900018200
- Conrad KS, Hurley JM, Widom J, Ringelberg CS, Loros JJ, Dunlap JC, Crane BR. Structure of the frequency-interacting RNA helicase: a protein interaction hub for the circadian clock. EMBO J 2016; 35:1707-19; PMID:27340124; http://dx.doi.org/10.15252/embj.201694327
- Korneta I, Magnus M, Bujnicki JM. Structural bioinformatics of the human spliceosomal proteome. Nucleic Acids Res 2012; 40:7046-65; PMID:22573172; http://dx.doi.org/10.1093/nar/gks347
- Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA 2005; 11:533-57; PMID:15840809; http://dx.doi.org/10.1261/rna.2220705
- Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 2015; 349:1182-91; PMID:26292707; http://dx.doi.org/10.1126/science.aac7629
- Yang K, Zhang L, Xu T, Heroux A, Zhao R. Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci USA 2008; 105:13817-22; PMID:18779563; http://dx.doi.org/10.1073/pnas.0805960105
- Ritchie DB, Schellenberg MJ, Gesner EM, Raithatha SA, Stuart DT, Macmillan AM. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat Struct Mol Biol 2008; 15:1199-205; PMID:18836455; http://dx.doi.org/10.1038/nsmb.1505
- Pena V, Rozov A, Fabrizio P, Lührmann R, Wahl MC. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J 2008; 27:2929-40; PMID:18843295; http://dx.doi.org/10.1038/emboj.2008.209
- Zhang L, Shen J, Guarnieri MT, Heroux A, Yang K, Zhao R. Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants. Protein Sci 2007; 16:1024-31; PMID:17473007; http://dx.doi.org/10.1110/ps.072872007
- Pena V, Liu S, Bujnicki JM, Lührmann R, Wahl MC. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell 2007; 25:615-24; PMID:17317632; http://dx.doi.org/10.1016/j.molcel.2007.01.023
- Mozaffari-Jovin S, Santos KF, Hsiao HH, Will CL, Urlaub H, Wahl MC, Lührmann R. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev 2012; 26:2422-34; PMID:23124066; http://dx.doi.org/10.1101/gad.200949.112
- Mozaffari-Jovin S, Wandersleben T, Santos KF, Will CL, Lührmann R, Wahl MC. Novel regulatory principles of the spliceosomal Brr2 RNA helicase and links to retinal disease in humans. RNA Biol 2014; 11:298-312; PMID:24643059; http://dx.doi.org/10.4161/rna.28353
- Ledoux S, Guthrie C. Retinitis Pigmentosa Mutations in Bad Response to Refrigeration 2 (Brr2) Impair ATPase and Helicase Activity. J Biol Chem 2016; 291:11954-65; PMID:27072132; http://dx.doi.org/10.1074/jbc.M115.710848
- Bartels C, Urlaub H, Lührmann R, Fabrizio P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J Biol Chem 2003; 278:28324-34; PMID:12736260; http://dx.doi.org/10.1074/jbc.M303043200
- Bartels C, Klatt C, Lührmann R, Fabrizio P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep 2002; 3:875-80; PMID:12189173; http://dx.doi.org/10.1093/embo-reports/kvf172
- Brenner TJ, Guthrie C. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics 2005; 170:1063-80; PMID:15911574; http://dx.doi.org/10.1534/genetics.105.042044
- Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev 2010; 24:1434-47; PMID:20595234; http://dx.doi.org/10.1101/gad.1925010
- Behrens SE, Lührmann R. Immunoaffinity purification of a [U4/U6.U5] tri-snRNP from human cells. Genes Dev 1991; 5:1439-52; PMID:1831175; http://dx.doi.org/10.1101/gad.5.8.1439
- Fetzer S, Lauber J, Will CL, Luhrmann R. The [U4/U6.U5] tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA 1997; 3:344-55; PMID:9085842
- Makarova OV, Makarov EM, Lührmann R. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J 2001; 20:2553-63; PMID:11350945; http://dx.doi.org/10.1093/emboj/20.10.2553
- Nguyen TH, Galej WP, Bai XC, Savva CG, Newman AJ, Scheres SH, Nagai K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 2015; 523:47-52; PMID:26106855; http://dx.doi.org/10.1038/nature14548
- Cheng SC, Abelson J. Spliceosome assembly in yeast. Genes Dev 1987; 1:1014-27; PMID:2962902; http://dx.doi.org/10.1101/gad.1.9.1014
- Huang YH, Chung CS, Kao DI, Kao TC, Cheng SC. Sad1 Counteracts Brr2-Mediated Dissociation of U4/U6.U5 in Tri-snRNP Homeostasis. Mol Cell Biol 2014; 34:210-20; PMID:24190974; http://dx.doi.org/10.1128/MCB.00837-13
- Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 A resolution. Science 2016; 353:904-11; PMID:27445306; http://dx.doi.org/10.1126/science.aag0291
- Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee CT, Urlaub H, Kastner B, et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 2016; PMID:27562955
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. Cryo-EM structure of the spliceosome immediately after branching. Nature 2016; 537:197-201; PMID:27459055; http://dx.doi.org/10.1038/nature19316
- Wan R, Yan C, Bai R, Huang G, Shi Y. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 2016; 353:895-904; PMID:27445308; http://dx.doi.org/10.1126/science.aag2235
- Kim SH, Lin RJ. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol Cell Biol 1996; 16:6810-9; PMID:8943336; http://dx.doi.org/10.1128/MCB.16.12.6810
- Lardelli RM, Thompson JX, Yates JR, Stevens SW. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 2010; 16:516-28; PMID:20089683; http://dx.doi.org/10.1261/rna.2030510
- Ohrt T, Prior M, Dannenberg J, Odenwalder P, Dybkov O, Rasche N, Schmitzová J, Gregor I, Fabrizio P, Enderlein J, et al. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA 2012; 18:1244-56; PMID:22535589; http://dx.doi.org/10.1261/rna.033316.112
- Wlodaver AM, Staley JP. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome. RNA 2014; 20:282-94; PMID:24442613; http://dx.doi.org/10.1261/rna.042598.113
- Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature 1991; 349:494-9; PMID:1825134; http://dx.doi.org/10.1038/349494a0
- Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J 1992; 11:5033-9; PMID:1464325
- Tseng CK, Liu HL, Cheng SC. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 2011; 17:145-54; PMID:21098140; http://dx.doi.org/10.1261/rna.2459611
- Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell 2004; 14:343-54; PMID:15125837; http://dx.doi.org/10.1016/S1097-2765(04)00217-5
- Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH Box ATPases Prp16 and Prp43 Cooperate to Proofread 5′ Splice Site Cleavage during Pre-mRNA Splicing. Mol Cell 2010; 39:385-95; PMID:20705241; http://dx.doi.org/10.1016/j.molcel.2010.07.014
- Semlow DR, Staley JP. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci 2012; 37:263-73; PMID:22564363; http://dx.doi.org/10.1016/j.tibs.2012.04.001
- Semlow DR, Blanco MR, Walter NG, Staley JP. Spliceosomal DEAH-Box ATPases Remodel Pre-mRNA to Activate Alternative Splice Sites. Cell 2016; 164:985-98; PMID:26919433; http://dx.doi.org/10.1016/j.cell.2016.01.025
- Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev 2005; 19:1894-904; PMID:16103217; http://dx.doi.org/10.1101/gad.1336305
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 2005; 19:2991-3003; PMID:16357217; http://dx.doi.org/10.1101/gad.1377405
- Tsai RT, Tseng CK, Lee PJ, Chen HC, Fu RH, Chang KJ, Yeh FL, Cheng SC. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol Cell Biol 2007; 27:8027-37; PMID:17893323; http://dx.doi.org/10.1128/MCB.01213-07
- Fourmann JB, Dybkov O, Agafonov DE, Tauchert MJ, Urlaub H, Ficner R, Fabrizio P, Lührmann R. The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction. Elife 2016; 5; PMID:27115347; http://dx.doi.org/10.7554/eLife.15564
- Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 2005; 121:991-1004; PMID:15989950; http://dx.doi.org/10.1016/j.cell.2005.04.015
- Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA 2006; 12:292-302; PMID:16428608; http://dx.doi.org/10.1261/rna.2152306
- Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 1993; 73:1377-91; PMID:8324826; http://dx.doi.org/10.1016/0092-8674(93)90363-U
- Mayas RM, Maita H, Semlow DR, Staley JP. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc Natl Acad Sci USA 2010; 107:10020-5; PMID:20463285; http://dx.doi.org/10.1073/pnas.0906022107
- Xu YZ, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell 2007; 28:838-49; PMID:18082608; http://dx.doi.org/10.1016/j.molcel.2007.09.022
- Yang F, Wang XY, Zhang ZM, Pu J, Fan YJ, Zhou JH, Query CC, Xu YZ. Splicing proofreading at 5′ splice sites by ATPase Prp28p. Nucleic Acids Res 2013; 41:4660-70; PMID:23462954; http://dx.doi.org/10.1093/nar/gkt149
- Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell 2006; 21:543-53; PMID:16483935; http://dx.doi.org/10.1016/j.molcel.2006.01.017
- Mefford MA, Staley JP. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA 2009; 15:1386-97; PMID:19458033; http://dx.doi.org/10.1261/rna.1582609
- Hoskins AA, Rodgers ML, Friedman LJ, Gelles J, Moore MJ. Single molecule analysis reveals reversible and irreversible steps during spliceosome activation. Elife 2016; 5:e14166; PMID:27244240; http://dx.doi.org/10.7554/eLife.14166
