ABSTRACT
Damaged DNA is repaired by specialized repair factors that are recruited in a well-orchestrated manner to the damage site. The DNA damage response at UV inflicted DNA lesions is accompanied by posttranslational modifications of DNA repair factors and the chromatin environment sourrounding the lesion. In particular, mono- and poly-ubiquitylation events are an integral part of the DNA damage signaling. Whereas ubiquitin signaling at DNA doublestrand breaks has been subject to intensive studies comparatively little is known about the intricacies of ubiquitylation events occurring during nucleotide excision repair (NER), the major pathway to remove bulky helix lesions. Both, the global genomic (GG-NER) and the transcription-coupled (TC-NER) branches of NER are subject to ubiquitylation and deubiquitylation processes.Here we summarize our current knowledge of the ubiquitylation network that drives DNA repair in the NER pathway and we discuss the crosstalk of ubiquitin signaling with other prominent post-translational modfications that might be essential to time the DNA damage recognition step.
Ubiquitin: An essential cellular signal transmitter
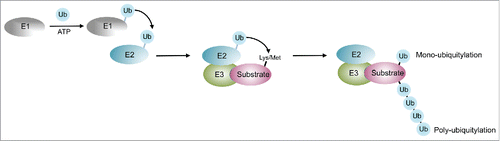
Ubiquitin is a small regulatory protein that is expressed in almost all tissues of eukaryotic organisms. Ubiquitylation, the attachment of ubiquitin residues to a substrate protein, can affect the fate and the function of the substrate in various ways. It triggers their degradation via the proteasome, it may alter their cellular localization, affect their enzymatic activity, and facilitate or prevent protein interactions.Citation1-3 Ubiquitylation is catalyzed by a set of specific enzymes, ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin-ligase enzymes (E3) (). In conjunction these enzymes generate an isopeptide bond between the c-terminus of ubiquitin and either a lysine or the n-terminal Methionine of the substrate, leading to its mono-ubiquitylation. Mono-ubiquitylation for instance occurs at a specific lysine residue, such as Lys164 in PCNA, a key factor in DNA replication.Citation4 Considerable mono-ubiquitylation exists at histone H2A throughout the cell cycle. About 5 to 10 percent of histone H2A proteins are modifiedCitation5 and deposition of the mono-ubiquitin mark was demonstrated to play a role in pluripotency of embryonic stem cells and during cell fate decisions,Citation6 during the DNA damage response at DSBsCitation7 and in NER.Citation8,9 Substrate-attached ubiquitin harbors 7 lysine residues, which may also be modified with polymeric chains. In particular homogenous ubiquitin chains where the same lysine residue is modified during chain extension induce distinct outcomes in the cell.Citation10 Both, homogenous Lysine48- (K48) and Lysine63- (K63) linked poly-ubiquitin chains, and mono-ubiquitylation of histone H2A provide important signals in the DNA damage response in NER and other DNA repair pathways. Although many E3 ubiquitin ligases have been identified it is less clear how they functionally interact and crosstalk to each other and how the respective E3-catalyzed ubiquitin signals regulate the DNA damage response (DDR). In this review we discuss recent discoveries in ubiquitin signaling during NER and how ubiquitylation events time the transition from recognition to verification of UV-light inflicted DNA damage.
Figure 1. The cellular ubiquitylation system. Ubiquitin is activated by an ATP consuming Ubiquitin acitvating enzyme (E1), and transferred from E1 to the active site cysteine of the Ubiquitin-conjugating enzyme (E2). E3 enzymes are capable of interacting with both E2 and the substrate and catalyze the transfer of the ubiquitin moeity to a lysine or the n-terminal Methionine of the substrate. Repeated cycles of the ubiquitylation cascade lead to poly-ubiquitylation, where any of the lysines of ubiquitin may be utilized as attachment site.
The enzymatic machinery of nucleotide excision repair
Exposure of DNA to UV light results in the generation of pyrimidine crosslinks namely cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts (6–4PPs). Such bulky, helix distorting DNA lesions are removed by nucleotide excision repair (NER), one of the major cellular DNA repair pathways.Citation11,12 Mammalian NER comprises of two sub-branches that differ in the way of DNA lesion recognition. Transcription-coupled NER (TC-NER) is limited to lesions located in the transcribed strand during the expression of genes. Stalled RNA Polymerase II elicits the DNA damage response by recruiting several specific DNA repair factors including the Cockayne syndrome proteins A and B (CSA and CSB).Citation13,14 In contrast, in global genome NER (GG-NER) DNA lesions are recognized by two lesion recognition factors, XPC and DDB2 (XPE). XPC, the main damage recognition factor, scans the DNA for helix distortionsCitation15,16 and operates as a trimeric complex with the Rad23 homologues RAD23A or RAD23B, respectively, and centrin2.Citation17,18 This trimeric complex recognizes a variety of lesions and induces NER activity.Citation19-21 Notably, RAD23B, which constitutes an adaptor protein that binds ubiquitylated proteins and the proteasome,Citation22,23 enhances the binding of XPC to the damage site but dissociates from XPC upon binding to damaged DNA.Citation21 The stable maintenance of XPC at damaged chromatin is probably mediated by its poly-ubiquitylation as discussed in detail below. Despite its general function as a reader of helix distortions XPC lacks specificity for CPDs, which is compensated by DDB2, an essential factor for the detection of UV light inflicted DNA lesions.Citation24-26 The current model for damage recognition of UV-light inflicted lesions considers DDB2 as the first reader of the damage, which hands the damage over to XPC to trigger the repair process. After damage recognition both sub-pathways converge into a common damage verification step. This step is performed by the repair factor XPA and by the generation of the pre-excision complex, which involves the DNA unwinding function of TFIIH via its helicase subunits XPB and XPD.Citation12 Finally, the DNA lesion is cut out by a dual DNA incision, which is generated by the two endonucleases XPF and XPG. Subsequently the gap is filled by DNA polymerases.Citation13,27
The damage recognition steps of these above outlined repair mechanism are accompanied by a ubiquitylation cascade that is presumably initiated by mono-ubiquitylation of histone H2A
Ubiquitylation events in GG-NER: Mono-ubiquitylation for starters
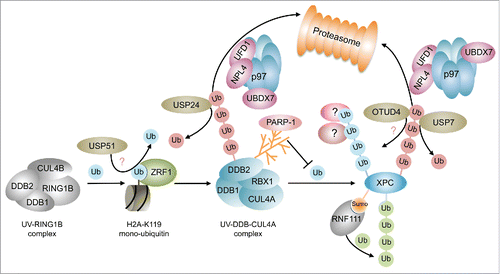
A distinctive histone modification at DNA damage sites is mono-ubiquitylation of histones H2A, H2AX and H1.Citation7,28,29 The role of mono-ubiquitylation of histone H2A has been studied intensively in DSB repair. Ubiquitin-mediated signaling at DSBs commences with MDC1-dependent recruitment of the E3 ligase RNF8, which subsequently brings about the mono-ubiquitylation of histone H1.Citation30 Mono-ubiquitylated histone H1 in turn facilitates the recruitment of RNF168, which in conjunction with RNF8, causes the mono- and poly-ubiquitylation of histones H2A and H2AX at lysines 13–15.Citation28,29,31-33 These poly-ubiquitylated histones provoke the recruitment of effector proteins that promote DSB repair. Parallel to this pathway the E3 ligase RING1B causes mono-ubiquitylation of histone H2A at lysine 119,Citation33-36 which presumably condenses chromatin regions sourrounding the damage site.Citation37 During GG-NER various E3 ligases catalyze ubiquitylation of histone H2A. In contrast to repair at DSBs their sequence of action is less well understood. H2A-ubiquitylation during GG-NER is catalyzed by the E3 ligase RNF8, the UV-DDB-CUL4A/B complexes and the UV-RING1B complex.Citation8,38-41 DDB2 forms a multiprotein complex consisting of DDB1, the E3 ligase RBX1 and either of the scaffold proteins CUL4A or CUL4B (UV-DDB-CUL4A/B), that catalyzes the mono-ubiquitylation of histones H3, H4 and H2A.Citation40,42-45 The UV-DDB2-CUL4A E3 ligase acts downstream of the UV-RING1B complex, which consists of the subunits DDB1, DDB2, CUL4B and the E3 ligase RING1B. The UV-RING1B complex specifically modifies lysine 119 of histone H2A, which is bound by the H2AK119-ubiquitin binding protein ZRF1 (). ZRF1 was originally identified as a factor that promotes cellular differentiationCitation46-48 and only recently it was demonstrated that it facilitates the exchange of the cullin and E3 ligase proteins from E3 multiprotein complexes.Citation41,49 At the damage site it removes the CUL4B-RBX1 module from the DDB2-DDB1 dimer and facilitates the incorporation of CUL4A-RBX1 thus converting the UV-RING1B complex into the UV-DDB CUL4A complex (). In contrast, it is not known which lysine in H2A is targeted by the UV-DDB-CUL4A complex. It seems reasonable that it ubiquitylates either a specific lysine other than K119 or that it operates less specific targeting different lysine residues. In agreement with the latter, the UV-DDB-CUL4B complex, which differs from the aforementioned complex only in its cullin subunit, seems to have a relatively broad substrate specificity potentially promoting ubiquity-lation at multiple lysine residues.Citation50 Mono-ubiquitylation at histone H2A is additionally catalyzed by the RING-containing E3 ligase RNF8 at later stages of NER causing a continuous H2A-ubiquitylation.Citation51 However, it seems that RNF8 does not directly affect the NER process but rather links it with the DSB-induced DNA damage response. It is currently not known which lysine is targeted by RNF8, but given its importance in recruiting DSB repair factors like 53BP1 and BRCA151 one might speculate that RNF8-mediated H2A-ubiquitylation might occur at lysines 13–15. Which information does mono-ubiquitylation of histone H2A at different lysines encode? Although speculative, it seems possible that the ubiquitin mark at the C-terminus of histone H2A (Lys119) might confer a different topology than ubiquitylation at the flexible N-terminal histone tails (Lys13–15) and hence lead to the recruitment of different downstream acting factors. Ubiquitylation at K119 and subsequent recruitment of ZRF1 causes a remodelling of multiprotein complexesCitation41,49 whereas it has been suggested that mono-ubiquitylation of histones by the UV-DDB-CUL4A complex facilitates the decondensation of chromatin thereby promoting access of the NER repair machinery to damaged DNA.Citation52 Hence, ubiquitylation at different lysine residues might have different outcomes either promoting remodelling of chromatin or chromatin-associated protein complexes. Taken together these two remodelling events might contribute to the DDR by orchestrating the well-timed recruitment of DNA repair factors in a decondensed chromatin environment. Interestingly, the UV-DDB-CUL4A complex not only carries out mono-ubiquitylation events but it is well established that it catalyzes the poly-ubiquitylation of DNA repair factors. Hence, by remodelling the E3 ligase complexes at the damage site ZRF1 occupies a central role in the GG-NER pathway switching the signal transmission from mono- to poly-ubiquitylation.
Figure 2. Ubiquitin signaling during lesion recognition in the GG-NER pathway. The ubiquitylation cascade during GG-NER is initiated the UV-RING1B E3 ligase complex that sets the H2A-ubiquitin mark specifically at lysine 119. At DSBs H2A-ubiquitin at lysines 13–15 is removed by USP51. ZRF1 reads the mono-ubiquitin mark and causes the assembly of the UV-DDB-CUL4A complex. This complex or its subunit DDB2 undergoes K48 linked self-ubiquitylation, which leads to its segregation by the p97 complex and its subsequent proteasomal degradation. Ubiquitylation of XPC by the UV-DDB-CUL4A ligase causes a poly-ubiquitylation of XPC, which is inhibited by parylation of DDB2. The linkage type and potential interactors of this ubiquitin chain are not known. XPC may be regarded a control center of damage recognition where many ubiquitin signals concur. XPC, like DDB2, is extracted from chromatin upon poly-ubiquitylation and also decorated by a K63-linked ubiquitin chain catalyzed by the SUMO-dependent E3 ligase RNF111. Poly-ubiquitin chains at XPC are edited by USP7 and potentially by OTUD4. Green spheres depict K63-linked ubiquitylation, red spheres depict K48-linked ubiquitylation. Blue spehres indicate mono-ubiquitylation or poly-ubiquitylation of unknown linkage.
Signal transmission through poly-ubiquitylation of DNA repair proteins
In analogy to DSB repair poly-ubiquitylation events are an integral part of the ubiquitin signaling cascade during NER.Citation7 The UV-DDB-CUL4A complex poly-ubiquitylates XPC thereby increasing its binding affinity for DNA in vitro and in vivo and it thus confers a stable binding of XPC to photolesions.Citation41,53,54 Hence, the linkage of this ubiquitin chain should be distinct from the K48-linkage, which is essential for proteasomal degradation (). Still, the UV-DDB-CUL4A complex is competent in assembling K48-linked ubiquitin chains as it generates a self-ubiquitylation of its subunit DDB2 promoting its proteasomal degradationCitation53,55 (). Interestingly, this E3 ligase complex catalyzes mono-ubiquitylation of histone H2A and poly-ubiquitylation of different linkage types. This variation in linkage-specificity of RING-domain E3 ligases is presumably defined by employing different E2 enzymes during the ubiquitylation reactions.Citation10 Additionally, XPC is modified by K63-linked poly-ubiquitylation via the SUMO-targeted E3 ligase (STUbL) RNF111/Arkadia56,57 (). XPC sumoylation at lysine 8 via RNF111 is a prerequisite for K63-linked ubiquitylation of XPC and the timely removal of DDB2 and XPC from damaged DNA.Citation56-58 In disagreement with these findings, an earlier study reported that upon UV-irradiation XPC was modified by SUMO-1 in a DDB2 and XPA dependent manner and that this sumoylation led to its stabilization.Citation59 Hence, it might be possible that sumoylation of XPC occurs at different lysines or, depending on the respective chromatin context, leads to the recruitment of different STUbLs, which either promote retention or removal of XPC. Besides its sumoylation and K63-linked ubiquitylation XPC is subject to proteasomal degradation. Poly-ubiquitylated XPC and DDB2 are extracted from damaged chromatin by p97 in complex with its co-factors UFD1-NPL4 and UBDX760 () indicating that XPC is also decorated by a K48 ubiquitin chain. Depletion of p97 was shown to cause retention of XPC and DDB2, which eventually leads to genotoxicityCitation60 underlining the importance of the timely removal of damage recognition factors to proceed with the verification and incision reactions. It is still not known which E3 ligase or which pair of E2 and E3 enzymes catalyzes K48-linked ubiquitylation to facilitate p97-mediated segregation of XPC and which lysine residues in XPC are ubiquitylated. Further, it is not resolved whether p97-mediated XPC extraction is linked to the RNF111 mediated removal of XPCCitation56 or whether both processes are part of independent signaling pathways.
The ubiquitin chains that are covalently linked to XPC and other DNA repair factors are additionally edited by deubiquitylases which further increases the complexity of the ubiquitin dependent DNA repair regulation network.
Regulation of poly-ubiquitylation by parylation and deubiquitylation
It has been demonstrated that DDB2 and XPC are targeted by poly(ADP-ribose) polymerase-1 (PARP-1). The activity of PARP-1 is triggered by DNA damage utilizing NAD+ to generate ADP-ribose polymers to modify various proteins involved in recombination and DNA repair events.Citation61,62 PARP-1 modifies DDB2 and it stimulates recruitment of XPC by DDB2 thus ensuring the efficiency of NERCitation62 (). Parylation of DDB2 inhibits ubiquitylation and subsequent proteasomal degradation of DDB2 and hence provides a means of stabilizing DDB2 at the damage site allowing it more time to facilitate chromatin remodelingCitation63 as for example by recruitment of ALC1.Citation64 It was further demonstrated that both XPC and RAD23B are parylated by PARP1 in response to UV irradiation.Citation65 Whether parylation of these factors is linked to ubiquitylation events as observed for DDB2 is currently unclear but an attractive speculation. Further, XPA shows high affinity for long PAR chainsCitation66 suggesting that parylation might also play a role during damage verification. This non-covalent interaction with PAR chains lowers the DNA binding affinity of XPA in vitro emphasizing that parylation in general attenuates damage signaling. Taken together, parylation is an important means to regulate the timing of DNA repair factors at chromatin and at least in some cases it seems to be linked to ubiquitylation events. Ubiquitylation of DNA repair factors is reversed by deubiquitylating enzymes (DUBs), which are responsible for removing covalently attached ubiquitin molecules from substrates or poly-ubiquitylated chains.Citation67 The human genome encodes for about a hundred deubiquitylases which belong to the families of Otubain domain-containing proteases (OTUs),Citation68,69 ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), JAMM (JAB1/MPN/Mov34) proteases and Machado-Joseph domain-containing proteins (MJDs).Citation10 Without any doubt, the function of the USP family in maintaining genome integrity is understood best when compared with the other classes of DUBs. USPs act in many genome surveillance and repair pathways probably owing to their broad substrate spectrum. Whereas many other DUBs specifically cleave K48 and K63 chains, USPs are fairly promiscous with respect to their substrates.Citation10 One prominent example is USP7, which acts during DNA replication, during DSB repair and during NER. DNA replication is regulated by USP7 via deubiquitylation of sumoylated proteins at the replication fork.Citation70 SUMOylation was proposed to target a protein group rather than individual proteins.Citation71 Hence, SUMO-dependent deubiquitylation might be an effective means of generating and maintaining a Ubiquitin-poor environment at sites of DNA replication as recently suggested.Citation70,72 Further, USP7 disassembles Rad18-dependent poly-ubiquitin chains and compromises UV-induced PCNA mono-ubiquitylation in the DNA damage tolerance pathway.Citation73-75 As detailed below, USP7 operates with UVSSA during TC-NER.Citation76,77 In the GG-NER sub-branch USP7 was shown to physically interact with XPC and to counteract p97-mediated segregation of XPC presumably by removing the K48-linked ubiquitin chainCitation78 (). Thus, USP7 contributes to stabilization of XPC at the DNA damage site safeguarding it from proteasomal degradation. As previously mentioned, XPC undergoes K63-linked poly-ubiquitylation via RNF111 and poly-ubiquitylation via the UV-DDB-CUL4A complex. It is currently not clear which DUBs apart from USP7 might additionally contribute to editing XPC-ubiquitylation or whether USP7 has any specifity toward a certain chain linkage. One potential candiate for editing XPC-ubiquitylation is OTUD479 (). OTUD4 was identified as an interaction partner of XPC and it was demonstrated that it deubiquitylates preferentially K48-linked ubiquitin chains.Citation68 It seems therefore likely that it removes K48-linked ubiquitin chains from XPC thereby counteracting p97-mediated extraction and proteasomal degradation. Also, the DUB USP24 is involved in the deubiquitylation events during the UV triggered DNA damage responseCitation80,81 (). USP24 deubiquitylates DDB2 thereby protecting it from proteasomal degradation and stabilizing the UV-DDB-CUL4 E3 ligase at the damage site. This might in turn cause an increased poly-ubiquitylation of XPC, which likewise would stabilize XPC at damaged chromatin. USP45 is another DUB that operates further downstream in the NER pathway regulating the ubiquitylation of the XPF interacting protein ERCC1.Citation82 Although its impact on the repair of CPDs was demonstrated it not clear how it is recruited to chromatin and how it is linked to the ubiquitylation events occurring during lesion recognition.
The ubiquitylation cascade at UV damage sites is initiated by mono-ubiquitylation of histone H2A. Thus, deubiquitylation of histone H2A will also impact on the DNA damage response. Specific DUBs may exist that remove the ubiquitin moiety from the lysines of the histone. During transcriptional activation for instance many DUBs have been demonstrated to deubiquitylate H2A at lysine 119.Citation83 Whether these DUBs operate during DNA repair is still obscure. Very recently, USP51 was shown to deubiquitylate histone H2A at lysines 13–15 regulating the DNA damage resonse at DSBs.Citation84 However, how H2A-ubiquitylation signals are erased during NER stays enigmatic. Hence, in the future it will be essential to identify DUBs that specifically operate at CPDs or 6–4 photoproducts.
Ubiquitylation events in the TC-NER pathway
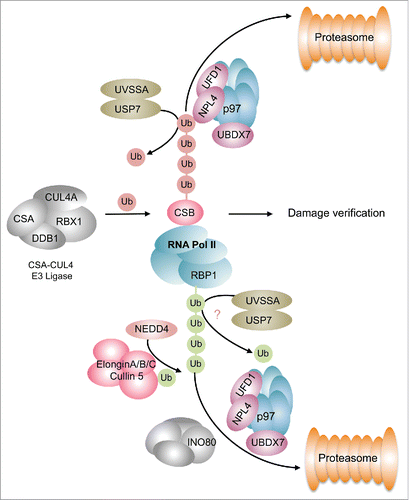
Whereas ubiquitylation events in GG-NER seem quite prevalent our insights into ubiquitin-dependent processes in TC-NER are relatively sparse. During lesion recognition in TC-NER stalled RNA Pol II recruits CSB. Notably, CSB is part of a multiprotein complex consisting of DDB1, the scaffold proteins CUL4A or CUL4B, respectively, and the E3 ubiquitin ligase RBX1 (). This E3 ligase complex is involved in fine-tuning the progression of TC-NER presumably via ubiquitylation and subsequent proteasomal degradation of CSB. Ubiquitylated CSB is extracted from chromatin by the p97/VCP segregase and its cofactors UFD1 and UBXN7,Citation85 which commit CSB to proteasomal degradation (). The ubiquitin chains attached to CSB may be removed by USP7, which is recruited to the damage site via its interaction partner UV-stimulated scaffold protein A (UVSSA).Citation76,86,87 UVSSA in turn is recruited to chromatin by either of its interaction partners RNA-Pol II or CSA.Citation76,86,88 Thus, UVSSA may counteract proteasomal degradation and stabilize CSB at chromatin through USP7-mediated deubiquitylation (). Recently it was demonstrated that the deubiquitylation activity of USP7 is suppressed by its interaction with UVSSA,Citation89 which suggests that the regulation of CSB ubiquitylation might be more complex that anticipated earlier. Additionally, UVSSA facilitates ubiquitylation of stalled Pol II but not its proteasomal degradation.Citation90,91 This implies that UVSSA does not promote the generation of a K48-linked ubiquitin chain on Pol II but it probably facilitates K63-linked ubiquitylation as suggested by the authors of the underlying study.Citation90 In contrast, ubiquitylation and degradation of the Pol II subunit RBP1 is a two-step process that involves at least two E3 ligases.Citation92 The HECT domain E3 ligase NEDD4 catalyzes the mono-ubiquitylation of RBP1 or a K63-linked poly-ubiquitylation, which is subsequently processed by the deubiquitylase UBP2Citation92-95 (). The mono-ubiquitin moiety on RBP1 is then extended via the Elongin A ubiquitin ligase.Citation92,96 Removal of this poly-ubiquitylated Pol II from sites of DNA damage is facilitated by the remodeler INO80, and the p97 segregase to enable further transcription of genesCitation97 (). Taken together, poly-ubiquitylation events signifcantly participate in the regulation of TC-NER. It still needs to be found out how different ubiquitin linkage types contribute to TC-NER and which lysine residues in the substrates are targeted. Very recently it was reported that site specific ubiquitylation of CSB at Lys991 also participates in the repair of oxidative damageCitation98 foreshadowing that modulation of DNA repair and presumably the timing of DNA damage recognition are subject to a complex network of ubiquitylation events.
Figure 3. Ubiquitylation events during TC-NER. CSB is ubiquitylated by the CSA-CUL4 E3 ligase causing its p97-mediated segregation and proteasomal degradation. This process is counteracted by the UVSSA-USP7 complex that catalyzes the deubiquitylation of CSB. Removal of CSB is an important step handing over the damage site for damage verification. The RBP1 subunit of RNA Pol II undergoes K63-linked ubiquitylation, which is brought about by a two-step reaction involving NEDD4 and the Elongin E3 ligase complex. Ubiquitylated Polymerase is extracted from chromatin by the INO80 and p97 segregase complexes comitting it to proteasomal degradation. This is presumably counteracted by the UVSSA-USP7 complex. Green spheres depict K63-linked ubiquitylation, red spheres indicate K48-linked ubiquitylation.
Timing of ubiquitylation events during DNA damage recognition in NER: An outlook
Ubiquitylation and deubiquitylation of DNA repair factors have an important impact on timing the lesion recognition. The most important player timing the damage recognition step during GG-NER is probably XPC, which seems to function as a control center that integrates different ubiquitin signals (). Besides K48-linked poly-ubiquitylation that is causing its proteasomal degradation, XPC is poly-ubiquitylated by the UV-DDB-CUL4A complex, which evokes its stabilization at chromatin. It is still not understood by which proteins these stabilizing ubiquitin chains are read and how this signal is linked to other XPC-ubiquitylation events. The XPC ubiquitylation network is even more complex as RNF111 catalyzes K63-linked ubiquitylation of XPC, which seems to be important for the removal of XPC and the proper recruitment of XPG.Citation56 Whether these diverse ubiquitylation events describe sequentiell steps occurring during lesion recognition or if they describe perhaps independent signaling pathways that occur in different chromatin environments is still a matter of debate. Further, all ubiquitylation events occurring at XPC are edited by DUBs. Deubiquitylation in general impacts on the dwell time of factors at the damage site and fine-tunes the timing of damage recognition and its handover to damage verification and incision. As XPC modifications presumably constitute a critical control point of damage recognition it will be important to better understand the role of the ubiquitin linkage types decorating XPC. Whereas the fate of K48-ubiquitylated XPC is evident, the role of K63-ubiquitylated XPC needs to be further investigated. Identification of K63-reading proteins or protein complexes harboring a K63-ubiquitin binding protein might give valuable new insights into the ubiquitin-mediated regulation of NER. Further, identification of the lysine residues that serve as ubiquitin attachment sites in XPC would facilitate to dissect the functions of the different ubiquitylation reactions. Directed mutagenesis of these lysine residues or similar mutations in XPC patients might help to further elucidate ubiquitin-related functions during the lesion recognition step. As to TC-NER, more ubiquitin-related processes and the respective E3 ligases and deubiquitylases need to be unveiled to better understand the timing of lesion recognition in this sub-branch. One major task for the future will be to identify more players of the ubiquitin signaling pathways, and more importantly, to comprehend how they crosstalk during the DDR. In general, it would be advantagous to gain more insight into ubiquitin signaling in the context of the chromatin conformation at the damage site. A condensed chromatin configuration will certainly rely on more elaborate remodelling activities to enable DNA repair. Could the different ubiquitin chain linkages and the use of different E3 ligases perhaps reflect DNA repair at different chromatin configurations? It's about time we looked more intensively at chromatin and its localization in the nucleus to better understand DNA repair in the NER pathway.
Abbreviations
| DDR | = | DNA damage response |
| DSB | = | double strand breaks |
| NER | = | nucleotide excision repair |
| GG-NER | = | global genomic nucleotide excision repair |
| TC-NER | = | transcription coupled nucleotide excision repair |
| UV | = | ultraviolet |
| CDP | = | cyclopyrimidine dimer |
| Pol II | = | RNA Polymerase II |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Richly laboratory for critical reading of the manuscript. We apologize to colleagues for being unable to cite their work due to space limitations.
Funding
H.R is supported by grants from the Boehringer Ingelheim Foundation.
References
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002; 82:373-428; PMID:11917093
- Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem 2003; 278:35857-60; PMID:12860974
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 2007; 315:201-5; PMID:17218518
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002; 419:135-41; PMID:12226657
- Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J Biol Chem 1975; 250:7182-7; PMID:1165239
- Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 2010; 35:323-32; PMID:20346678
- Jackson SP, Durocher D. Regulation of DNA Damage Responses by Ubiquitin and SUMO. Mol Cell 2013; 49:795-807; PMID:23416108
- Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Gen Dev 2006; 20:1343-52; PMID:16702407
- Nouspikel T. Multiple roles of ubiquitination in the control of nucleotide excision repair. Mech Ageing Dev 2011; 132:355-65; PMID:21466822
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem 2012; 81:203-29; PMID:22524316
- de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Gen Dev 1999; 13:768-85; PMID:10197977
- Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 2013; 5:a012609; PMID:24086042
- Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res 2008; 18:73-84; PMID:18166977
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JHJ. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Bio 2014; 15:465-81
- Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J 2003; 22:5293-303; PMID:14517266
- Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell 1998; 2:223-32; PMID:9734359
- Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem 2001; 276:18665-72; PMID:11279143
- Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek PJ, Bootsma D, et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J 1994; 13:1831-43; PMID:8168482
- Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Gen Dev 2001; 15:507-21; PMID:11238373
- Hoogstraten D, Bergink S, Ng JM, Verbiest VH, Luijsterburg MS, Geverts B, Raams A, Dinant C, Hoeijmakers JH, Vermeulen W, et al. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J Cell Sci 2008; 121:2850-9; PMID:18682493
- Bergink S, Toussaint W, Luijsterburg MS, Dinant C, Alekseev S, Hoeijmakers JH, Dantuma NP, Houtsmuller AB, Vermeulen W. Recognition of DNA damage by XPC coincides with disruption of the XPC-RAD23 complex. J Cell Biol 2012; 196:681-8; PMID:22431748
- Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem 2002; 277:11691-5; PMID:11805121
- Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell 2004; 15:3357-65; PMID:15121879
- Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell 2000; 5:737-44; PMID:10882109
- Kulaksiz G, Reardon JT, Sancar A. Xeroderma pigmentosum complementation group E protein (XPE/DDB2): purification of various complexes of XPE and analyses of their damaged DNA binding and putative DNA repair properties. Mol Cell Biol 2005; 25:9784-92; PMID:16260596
- Nichols AF, Itoh T, Graham JA, Liu W, Yamaizumi M, Linn S. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J Biol Chem 2000; 275:21422-8
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 2014; 15:465-81; PMID:24954209
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007; 131:887-900; PMID:18001824
- Pan MR, Peng G, Hung WC, Lin SY. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J Biol Chem 2011; 286:28599-607; PMID:21676867
- Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S, et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 2015; 527:389-93; PMID:26503038
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 2012; 150:1182-95; PMID:22980979
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 2009; 136:435-46; PMID:19203579
- Pan MR, Peng G, Hung WC, Lin SY. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J Biol Chem 2011; 286:28599-607; PMID:21676867
- Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol 2010; 191:45-60; PMID:20921134
- Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, Citterio E, van Lohuizen M, Ganesan S BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol 2011; 31:1972-82; PMID:21383063
- Chagraoui J, Hebert J, Girard S, Sauvageau G. An anticlastogenic function for the Polycomb Group gene Bmi1. Proc Natl Acad Sci U S A 2011; 108:5284-9; PMID:21402923
- Ui A, Nagaura Y, Yasui A. Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Mol Cell 2015; 58:468-82; PMID:25921070
- Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapić-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci U S A 2006; 103:2588-93; PMID:16473935
- Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J Cell Biol 2009; 186:835-47; PMID:19797077
- Guerrero-Santoro J, Kapetanaki MG, Hsieh CL, Gorbachinsky I, Levine AS, Rapić-Otrin V. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res 2008; 68:5014-22; PMID:18593899
- Gracheva E, Chitale S, Wilhelm T, Rapp A, Byrne J, Stadler J, Medina R, Cardoso MC, Richly H. ZRF1 mediates remodeling of E3 ligases at DNA lesion sites during nucleotide excision repair. J Cell Biol 2016; 213:185-200; PMID:27091446
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006; 443:590-3; PMID:16964240
- Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 2006; 22:383-94; PMID:16678110
- Shiyanov P, Nag A, Raychaudhuri P. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J Biol Chem 1999; 274:35309-12; PMID:10585395
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 2003; 113:357-67; PMID:12732143
- Richly H, Rocha-Viegas L, Ribeiro JD, Demajo S, Gundem G, Lopez-Bigas N, Nakagawa T, Rospert S, Ito T, Di Croce L. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 2010; 468:1124-8; PMID:21179169
- Papadopoulou T, Kaymak A, Sayols S, Richly H. Dual Role of Med12 in PRC1-dependent Gene Repression and ncRNA-mediated Transcriptional Activation. Cell Cycle 2016; 15:1479-1493; PMID:27096886
- Kaymak A, Richly H. Zrf1 controls mesoderm lineage genes and cardiomyocyte differentiation. Cell Cycle 2016; 15(23):3306–3317; PMID:27754813
- Papadopoulou T, Richly H. On-site remodeling at chromatin: How multiprotein complexes are rebuilt during DNA repair and transcriptional activation. BioEssays 2016; 38(11):1130–1140; PMID:27599465
- Lan L, Nakajima S, Kapetanaki MG, Hsieh CL, Fagerburg M, Thickman K, Rodriguez-Collazo P, Leuba SH, Levine AS, Rapić-Otrin V. Monoubiquitinated histone H2A destabilizes photolesion-containing nucleosomes with concomitant release of UV-damaged DNA-binding protein E3 ligase. J Biol Chem 2012; 287:12036-49; PMID:22334663
- Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J Cell Biol 2009; 186:835-47; PMID:19797077
- Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 2006; 22:383-94; PMID:16678110
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 2005; 121:387-400; PMID:15882621
- Matsumoto S, Fischer ES, Yasuda T, Dohmae N, Iwai S, Mori T, Nishi R, Yoshino K, Sakai W, Hanaoka F, et al. Functional regulation of the DNA damage-recognition factor DDB2 by ubiquitination and interaction with xeroderma pigmentosum group C protein. Nucleic Acids Res 2015; 43:1700-13; PMID:25628365
- El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, Wani AA. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem 2006; 281:13404-11; PMID:16527807
- van Cuijk L, van Belle GJ, Turkyilmaz Y, Poulsen SL, Janssens RC, Theil AF, Sabatella M, Lans H, Mailand N, Houtsmuller AB, et al. SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair. Nature communications 2015; 6:7499; PMID:26151477
- Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, ,van Belle GJ, Streicher W, Wikström M, Choudhary C, Houtsmuller AB, Marteijn JA, et al. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 2013; 201:797-807; PMID:23751493
- Akita M, Tak YS, Shimura T, Matsumoto S, Okuda-Shimizu Y, Shimizu Y, Nishi R, Saitoh H, Iwai S, Mori T, et al. SUMOylation of xeroderma pigmentosum group C protein regulates DNA damage recognition during nucleotide excision repair. Sci Rep 2015; 5:10984; PMID:26042670
- Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res 2005; 33:4023-34; PMID:16030353
- Puumalainen MR, Lessel D, Ruthemann P, Kaczmarek N, Bachmann K, Ramadan K, Naegeli H Chromatin retention of DNA damage sensors DDB2 and XPC through loss of p97 segregase causes genotoxicity. Nat Commun 2014; 5:3695; PMID:24770583
- De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol 2012; 84:137-46; PMID:22469522
- Robu M, Shah RG, Petitclerc N, Brind'Amour J, Kandan-Kulangara F, Shah GM Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci U S A 2013; 110:1658-63; PMID:23319653
- Luijsterburg MS, Lindh M, Acs K, Vrouwe MG, Pines A, van Attikum H, Mullenders LH, Dantuma NP DDB2 promotes chromatin decondensation at UV-induced DNA damage. J Cell Biol 2012; 197:267-81; PMID:22492724
- Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, Hensbergen P, Deelder A, de Groot A, Matsumoto S, et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol 2012; 199:235-49; PMID:23045548
- Maltseva EA, Rechkunova NI, Sukhanova MV, Lavrik OI. Poly(ADP-ribose) Polymerase 1 Modulates Interaction of the Nucleotide Excision Repair Factor XPC-RAD23B with DNA via Poly(ADP-ribosyl)ation. J Biol Chem 2015; 290:21811-20; PMID:26170451
- Fischer JM, Popp O, Gebhard D, Veith S, Fischbach A, Beneke S, Leitenstorfer A, Bergemann J, Scheffner M, Ferrando-May E, et al. Poly(ADP-ribose)-mediated interplay of XPA and PARP1 leads to reciprocal regulation of protein function. FEBS J 2014; 281:3625-41; PMID:24953096
- Komander D. Mechanism, specificity and structure of the deubiquitinases. Subcell Biochem 2010; 54:69-87; PMID:21222274
- Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 2013; 154:169-84; PMID:23827681
- Kulathu Y, Garcia FJ, Mevissen TE, Busch M, Arnaudo N, Carroll KS, Barford D, Komander D. Regulation of A20 and other OTU deubiquitinases by reversible oxidation. Nat Commun 2013; 4:1569; PMID:23463012
- Lecona E, Rodriguez-Acebes S, Specks J, Lopez-Contreras AJ, Ruppen I, Murga M, Muñoz J, Mendez J, Fernandez-Capetillo O. USP7 is a SUMO deubiquitinase essential for DNA replication. Nat Struct Mol Biol 2016; 23:270-7; PMID:26950370
- Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 2012; 151:807-20; PMID:23122649
- Lecona E, Fernandez-Capetillo O. A SUMO and ubiquitin code coordinates protein traffic at replication factories. BioEssays 2016; 38(12):1209-1217; PMID:27667742
- Zlatanou A, Stewart GS. Damaged replication forks tolerate USP7 to maintain genome stability. Mol Cell Oncol 2016; 3:e1063571; PMID:27308573
- Zlatanou A, Sabbioneda S, Miller ES, Greenwalt A, Aggathanggelou A, Maurice MM, Lehmann AR, Stankovic T, Reverdy C, Colland F, et al. USP7 is essential for maintaining Rad18 stability and DNA damage tolerance. Oncogene 2016; 35:965-76; PMID:25961918
- Qian J, Pentz K, Zhu Q, Wang Q, He J, Srivastava AK, Wani AA. USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene 2015; 34:4791-6; PMID:25435364
- Schwertman P, Lagarou A, Dekkers DH, Raams A, van der Hoek AC, Laffeber C, Hoeijmakers JH, Demmers JA, Fousteri M, Vermeulen W, et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet 2012; 44:598-602; PMID:22466611; http://dx.doi.org/10.1038/ng.2230
- Schwertman P, Vermeulen W, Marteijn JA. UVSSA and USP7, a new couple in transcription-coupled DNA repair. Chromosoma 2013; 122:275-84; PMID:23760561; http://dx.doi.org/10.1007/s00412-013-0420-2
- He J, Zhu Q, Wani G, Sharma N, Han C, Qian J, Pentz K, Wang QE, Wani AA. Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. J Biol Chem 2014; 289:27278-89; PMID:25118285; http://dx.doi.org/10.1074/jbc.M114.589812
- Lubin A, Zhang L, Chen H, White VM, Gong F. A human XPC protein interactome–a resource. Int J Mol Sci 2014; 15:141-58; ; http://dx.doi.org/10.3390/ijms15010141
- Zhang L, Gong F. Involvement of USP24 in the DNA damage response. Mol Cell Oncol 2016; 3:e1011888; PMID:27308530; http://dx.doi.org/10.1080/23723556.2015.1011888
- Zhang L, Nemzow L, Chen H, Lubin A, Rong X, Sun Z, Harris TK, Gong F. The deubiquitinating enzyme USP24 is a regulator of the UV damage response. Cell reports 2015; 10:140-7; PMID:25578727; http://dx.doi.org/10.1016/j.celrep.2014.12.024
- Perez-Oliva AB, Lachaud C, Szyniarowski P, Munoz I, Macartney T, Hickson I, Rouse J, Alessi DR. USP45 deubiquitylase controls ERCC1-XPF endonuclease-mediated DNA damage responses. EMBO J 2015; 34:326-43; PMID:25538220; http://dx.doi.org/10.15252/embj.201489184
- Vissers JH, Nicassio F, van Lohuizen M, Di Fiore PP, Citterio E. The many faces of ubiquitinated histone H2A: insights from the DUBs. Cell Div 2008; 3:8; PMID:18430235; http://dx.doi.org/10.1186/1747-1028-3-8
- Wang Z, Zhang H, Liu J, Cheruiyot A, Lee JH,Ordog T, Lou Z, You Z, Zhang Z. USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response. Gen Dev 2016; 30:946-59; PMID:27083998; http://dx.doi.org/10.1101/gad.271841.115
- He J, Zhu Q, Wani G, Sharma N, Wani AA. Valosin-containing Protein (VCP)/p97 Segregase Mediates Proteolytic Processing of Cockayne Syndrome Group B (CSB) in Damaged Chromatin. J Biol Chem 2016; 291:7396-408; PMID:26826127; http://dx.doi.org/10.1074/jbc.M115.705350
- Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S, Tahara H, Neilan EG, Honma M, Nohmi T, et al. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet 2012; 44:593-7; PMID:22466612; http://dx.doi.org/10.1038/ng.2228
- Sarasin A. UVSSA and USP7: new players regulating transcription-coupled nucleotide excision repair in human cells. Genome Med 2012; 4:44; PMID:22621766; http://dx.doi.org/10.1186/gm343
- Fei J, Chen J. KIAA1530 protein is recruited by Cockayne syndrome complementation group protein A (CSA) to participate in transcription-coupled repair (TCR). J Biol Chem 2012; 287:35118-26; PMID:22902626; http://dx.doi.org/10.1074/jbc.M112.398131
- Higa M, Zhang X, Tanaka K, Saijo M. Stabilization of Ultraviolet (UV)-stimulated Scaffold Protein A by Interaction with Ubiquitin-specific Peptidase 7 Is Essential for Transcription-coupled Nucleotide Excision Repair. J Biol Chem 2016; 291:13771-9; PMID:27129218; http://dx.doi.org/10.1074/jbc.M116.724658
- Nakazawa Y, Sasaki K, Mitsutake N, Matsuse M, Shimada M, Nardo T, Takahashi Y, Ohyama K, Ito K, Mishima H, et al. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat Genet 2012; 44:586-92; PMID:22466610; http://dx.doi.org/10.1038/ng.2229
- van Cuijk L, Vermeulen W, Marteijn JA. Ubiquitin at work: the ubiquitous regulation of the damage recognition step of NER. Exp Cell Res 2014; 329:101-9; PMID:25062985; http://dx.doi.org/10.1016/j.yexcr.2014.07.018
- Wilson MD, Harreman M, Svejstrup JQ. Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim Biophys Acta 2013; 1829:151-7; PMID:22960598; http://dx.doi.org/10.1016/j.bbagrm.2012.08.002
- Harreman M, Taschner M, Sigurdsson S, Anindya R, Reid J, Somesh B, Kong SE, Banks CA, Conaway RC, Conaway JW, et al. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci U S A 2009; 106:20705-10; PMID:19920177; http://dx.doi.org/10.1073/pnas.0907052106
- Beaudenon SL, Huacani MR, Wang G, McDonnell DP, Huibregtse JM. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol 1999; 19:6972-9; PMID:10490634; http://dx.doi.org/10.1128/MCB.19.10.6972
- Anindya R, Aygun O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell 2007; 28:386-97; PMID:17996703; http://dx.doi.org/10.1016/j.molcel.2007.10.008
- Yasukawa T, Kamura T, Kitajima S, Conaway RC, Conaway JW, Aso T. Mammalian Elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J 2008; 27:3256-66; PMID:19037258; http://dx.doi.org/10.1038/emboj.2008.249
- Lafon A, Taranum S, Pietrocola F, Dingli F, Loew D, Brahma S, Bartholomew B, Papamichos-Chronakis M. INO80 Chromatin Remodeler Facilitates Release of RNA Polymerase II from Chromatin for Ubiquitin-Mediated Proteasomal Degradation. Mol Cell 2015; 60:784-96; PMID:26656161; http://dx.doi.org/10.1016/j.molcel.2015.10.028
- Ranes M, Boeing S, Wang Y, Wienholz F, Menoni H, Walker J, Encheva V, Chakravarty P, Mari PO, Stewart A, et al. A ubiquitylation site in Cockayne syndrome B required for repair of oxidative DNA damage, but not for transcription-coupled nucleotide excision repair. Nucleic Acids Res 2016; 44:5246-55; PMID:27060134; http://dx.doi.org/10.1093/nar/gkw216
