ABSTRACT
Accumulating evidence suggests that long noncoding RNAs (lncRNAs) play an important role in oncogenesis and tumor progression. However, our knowledge of lncRNAs in thyroid cancer is still limited. To explore the crucial lncRNAs involved in oncogenesis of papillary thyroid cancer (PTC), we acquired data of differentially expressed lncRNAs between PTC tissues and paired adjacent noncancerous thyroid tissues through lncRNA microarray. In the microarray data, we observed that a newly identified lncRNA, HIT000218960, was significantly upregulated in PTC tissues and associated with a well-known oncogene, high mobility group AT-hook 2 (HMGA2) gene. Both in normal thyroid tissues and PTC tissues, the expression of HIT000218960 was significantly positively correlated with that of HMGA2 mRNA. Knockdown of HIT000218960 in PTC cells resulted in downregulation of HMGA2. In addition, functional assays indicated that inhibition of HIT000218960 in PTC cells suppressed cell proliferation, colony formation, migration and invasion in vitro. Increased HIT000218960 expression in PTC tissues was obviously correlated with lymph node metastasis and multifocality, as well as TNM stage. Those findings suggest that HIT000218960 might acts as a tumor promoter through regulating the expression of HMGA2.
Introduction
Thyroid cancer is the most common endocrine malignancies and represents approximately 1–5% of newly diagnosed cancers. Contrary to many other cancers, the incidence of thyroid cancer is increasing rapidly in the last 3 decades.Citation1 Papillary thyroid carcinoma (PTC) is the most common type, accounting for about 80–90% of all thyroid cancers in the United States.Citation2 Although the majority of PTCs are curable, there is approximately 30% cases with lymph node metastases which prognoses unfavorable outcomes.3The biological behavior of PTC varies widely. However, the ideal genetic marker for PTC prognosis has not yet been identified. Therefore, it is essential to understand the molecular mechanisms that underlie thyroid cancer, as a primary step to find valuable biomarkers.
Long noncoding RNAs (lncRNAs) are a class of transcripts that are 200 nucleotides in length or larger and have limited protein coding potential.Citation4 Many studies have demonstrated that lncRNAs associate with a surprisingly wide array of celluer functions, such as proliferation, apoptosis, or cell migration.Citation5 Although the precise mechanisms of lncRNAs remain poorly understood, there is accumulating evidence to suggest that the aberrant expression of lncRNAs in certain diseases may have excellent diagnostic and prognostic values.Citation6-10 At present, a few of lncRNAs were reported to play an important role in oncogenesis and progress of thyoid cancer. For instance, papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3), which is exclusively expressed in thyroid tissue and downregulated in PTC tissue, was found to impact PTC carcinogenesis through the S100A4 pathway.Citation11,12 And lncRNA NONHSAT037832 was reported to have a high diagnostic value for differentiating between PTC and noncancerous disease.Citation13 However, relative to other malignant diseases, our knowledge of lncRNAs in thyroid cancer is still limited. It necessary to identify more lncRNAs associated thyroid cancer and research into their functional mechanisms.
In the present study, we investigated the different expression profiles of lncRNAs between PTC tissue and paired adjacent noncancerous thyroid tissue using microarray. Quantitative real time polymerase chain reaction (qRT-PCR) was applied for validation of the differentially expressed lncRNAs. In addition, we identified lncRNA-HIT000218960 associated with PTC, and focus on the role of lncRNA-HIT000218960 in the oncogenesis and progress of PTC.
Results
LncRNAs and mRNAs expression profiles in PTC
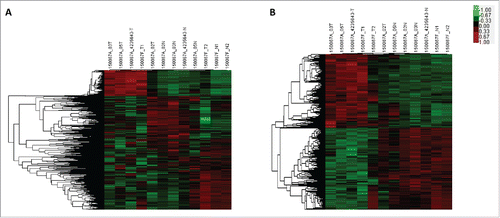
The microarray analysis dectected a total of 1593 lncRNAs and 1952 mRNAs which were differentially expressed (fold change ≥ 2.0 and P < 0.05) in PTC tissues compared with paired adjacent noncancerous thyroid tissues. The top 20 differentially expressed lncRNAs and mRNAs were listed in Table S1 and Table S2. Hierarchical clustering analysis was used to arrange specimens into groups according to their expression levels ().
Figure 1. Microarray analysis of lncRNA and mRNA expression in thyroid tissues. Hierarchical clustering analysis of differentially expressed lncRNA (A) and mRNA (B) between PTC tissues and noncancerous thyroid tissues (Fold change ≥ 2, p < 0.05). Red indicates high relative expression, while green indicates low relative expression. In the heat map, columns represent samples and rows represent each lncRNA or mRNA.
Validation of the microarray data
To validate microarray analysis findings, we selected the top 3 up- and downregulated lncRNAs and another random 4 differentially expressed lncRNAs (TCONS_00028337, TCONS_00020457, ENST00000539653.1, and uc.324) and measured their expression in 55 pairs of PTC and corresponding noncancerous thyroid tissues using qRT-PCR. These qRT-PCR results are consistent with the microarray data, in that all 10 lncRNAs were differentially expressed with the same trend (up- or downregulated) and reached statistical significance (P < 0.05) (Fig. S1). In addition, the microarray includes probes of 10 lncRNAs which have been reported to be associated with PTC in previous study. Although some of the reported lncRNAs were not significantly differentially expressed, perhaps because of the small sample size, their microarray results showed the same trend of up- or downregulation as previous studies (Table S3).
Gene ontology (GO) and kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
To explore potential biological associations, we ran GO and KEGG pathway analysis with the differentially expressed lncRNAs and mRNAs. GO analysis indicated several categories of biological process, cellular component, and molecular function were enriched. The resulted GO terms with 20 minimum P values were summarized in Fig. S2. Among them, single-organism process, glycosaminoglycan binding, and intrinsic to membrane were the most closely associated with PTC. KEGG Pathway analysis showed that 35 pathways were significantly enriched among the differentially expressed transcripts (Fig. S3). Many of there pathways were associated with cancer, such as “P53 signaling pathway,” “Pathways in cancer,” “PI3K-Akt signaling pathway,” “TGF-β signaling pathway” and “Transcriptional misregulation in cancer.”
Identification of lncRNA-HIT000218960 potentially associated with PTC oncogenesis
In order to select lncRNAs with fundamental biological functions for further investigation, we explore into the detailed information of the first 5 up- or downregulated lncRNAs in microarray (). Among that, we noticed an unannotated lncRNA-HIT000218960, the second most upregulated in microarray data (fold change = 44.14, p = 0.0011), putatively targets a well-known oncogene, high mobility group AT-hook 2 (HMGA2) gene. It has been reported that HMGA2 is overexpressed in PTC and can be used as a biomarker for distinguishing benign from malignant thyroid tumors.Citation14,15,22,25,27 The KEEG pathway analysis in this study showed that HMGA2 was enriched in the pathway of “transcriptional misregulation in cancer” .The correlation coefficient between HIT000218960 and HMGA2 was 0.999083. In addition, qRT-PCR results indicated that HIT000218960 was significantly upregulated in PTC tissues compared with noncancerous thyroid tissues, which was consistent with microarray data ().Therefore, we speculated that HIT000218960 may play an important role in PTC and deserve further research.
Table 1. Target gene prediction of top 5 differently expressed lncRNAs in microarray.
Figure 2. Expression of HIT000218960 and HMGA2 mRNA in papillary thyroid cancer (PTC) specimens and cells. (A, B) Relative expression of HIT000218960 and HMGA2 mRNA in PTC tissues compared with paired noncancerous thyroid tissues (T: PTC tissues, N: noncancerous thyroid tissues, n = 55). (C) Results from qRT-PCR showing HIT000218960 and HMGA2 mRNA expression levels in PTC cell lines (TPC1 and BCPAP) compared with normal human thyroid follicular epithelial cell line (Nthy-ori3–1). (D, E) Correlation between expression of HIT000218960 and HMGA2 in tissue specimens. (F, G) Relative expressions of HIT000218960 and HMGA2 mRNA in TPC1 and BCPAP cell lines infected with LV-shHIT000218960 or LV-NC. (H) Expression of HMGA2 protein in PTC cell lines infected with LV-shHIT000218960 or LV-NC. *P < 0.05, ** P < 0.01.
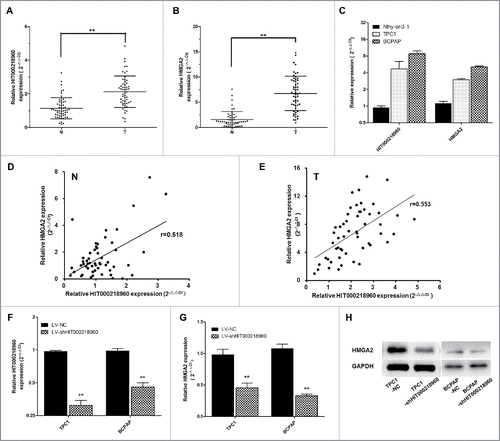
To validate the correlation between HIT000218960 and HMGA2, we first examined the expression of HIT000218960 and HMGA2 mRNA in 2 PTC cell lines (TPC1 and BCPAP) and a normal human thyroid follicular epithelial cell line (Nthy-ori3–1) by qRT-PCR. Results showed that HIT000218960 and HMGA2 expression were obviously overexpressed in PTC cell lines (). Then we examined the expression of HIT000218960 and HMGA2 mRNA in 55 pairs of PTC and adjacent noncancerous thyroid tissues. We found that HMGA2 mRNA expression was significantly upregulated in PTC specimens in comparison with noncancerous thyroid tissues, with the same trend of HIT000218960 (). Both in PTC tissues and noncancerous thyroid tissues, a significant positive correlation was observed between HIT000218960 and HMGA2 mRNAs (Spearmen's correlation, in noncancerous thyroid tissues, r = 0.518, p = 0.000, ; in PTC tissues, r = 0.553, p = 0.000, ), supporting the role of HIT000218960 in the expression of HMGA2.
Furthermore, we established HIT000218960 knockdown PTC cell lines using TPC1 and BCPAP cells and lentivirus vectors. Decreased expression of HIT000218960 in cells infected with LV-sh HIT000218960 was confirmed by qRT-PCR (). Then we examined HMGA2 levels in TPC1-shHIT0002189 and BCPAP-shHIT0002189 cells compared with TPC1-NC and BCPAP-NC cells. The results showed that both mRNA () and protein expression () levels of HMGA2 were significantly downregulated in the shHIT0002189 group, which verified that HMGA2 was a potential target gene of HIT000218960 and suggested the potential role of HIT000218960 in PTC.
HIT000218960 promotes PTC cell proliferation, colony formation, migration and invasion in vitro
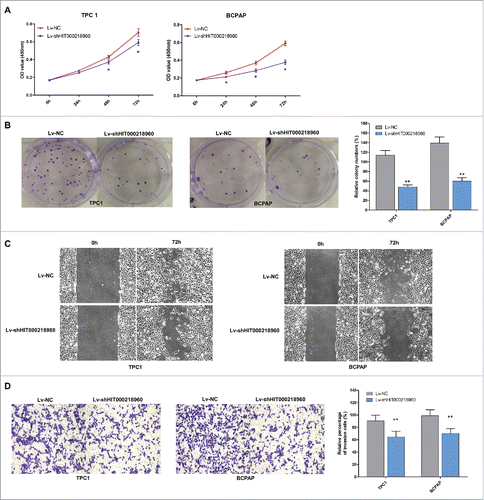
To evaluate the biological effects of HIT000218960 in PTC cells, we compared the proliferation, colony formation, migration and invasion ability of TPC1-shHIT0002189 and BCPAP-shHIT0002189 cells with TPC1-NC and BCPAP-NC cells. CCK-8 assays revealed that cell proliferation was suppressed in cells infected with LV-shHIT000218960 compared with negative controls (). The colony numbers of cells infected with LV-shHIT000218960 were notably lower than those infected with LV-NC (). Furthermore, knockdown of HIT000218960 resulted in attenuated migration and invasion of PTC cells measured by wound healing () and transwell assays ().
Figure 3. Tumor promoting effects of HIT000218960 in PTC cell lines. (A) The proliferation curve of TPC1 and BCPAP cells after infected with LV-shHIT000218960 or LV-NC by CCK8 assay. (B) Colony formation assay after knockdown of HIT000218960 expression. (C) Wound healing assays demonstrated that inhibition of HIT000218960 attenuated motility of PTC cells. (D) The effects of HIT000218960 knockdown on invasive ability of PTC cells were assessed by transwell method. Quantification was performed by counting the stained cells that invaded to the lower chamber under a light microscopy. *P < 0.05, ** P < 0.01.
Upregulation of HIT000218960 correlates with lymph node metastasis, multifocality and TNM stage in PTC patients
To investigate the roles of HIT000218960 in PTC, we examined the correlation between clinical characteristics and HIT000218960 expression in 55 PTC patients. We used the mean expression level of HIT000218960 in cancerous tissues to dichotomise the PTC cases. Univariate analysis showed that high HIT000218960 expression group displayed a higher incidence of increased lymph node metastasis (p = 0.009), multifocality (p = 0.005) and advanced TNM stage (p = 0.022). However, no significant differences were observed with regard to age, gender, or T stage ().
Table 2. Clinical characteristics of 55 PTC patients according to HIT000218960 expression levels.
Discussion
Papillary thyroid cancer (PTC) is the most common type of thyroid malignancies and has attracted increasing attention for its increased incidence over the last 3 decades. Knowledge about molecular biology of PTC has significant increased but the exact molecular mechanisms underlying PTC initiation and progress remain unclear. Long noncoding RNAs (lncRNAs) have been confirmed to play critical roles in numerous biological processes including oncogenesis. However, our knowledge of lncRNAs in thyroid cancer is still limited.
In this study, we performed microarray assay to explore the aberrant expression profile of lncRNAs and mRNAs in PTC tissue compared with adjacent noncancerous thyroid tissue. The results that a large number of lncRNAs were significantly differently expressed suggested the important role of lncRNAs in PTC. Applying bioinformatics approach, we noticed that lncRNA HIT000218960 was significantly upregulated in PTC and predicted to regulate a member of the high mobility group AT-hook (HMGA) gene family, HMGA2, which had been reported to be overexpressed in several cancers including thyroid cancer and function as an oncogene.Citation16-18
HIT000218960 is a lncRNA located on chromosome 11 and had never been reported before. In the present study, we first validated that the expression of HIT000218960 was significantly increased in PTC tissues compared with normal thyroid tissues. In addition, inhibition of HIT000218960 was demonstrated to suppress proliferation, colony formation, migration and invasion of PTC cells in vitro. High HIT000218960 expression in PTC tissues was correlated with increased lymph node metastasis, multifocality and advanced TNM stage. These results imply that HIT000218960 acts as a promoter of PTC progress.
In this study, we also showed for the first time that the expression of HMGA2 might be regulated by a lncRNA-HIT000218960. The expression of HIT000218960 in PTC tissues was positively correlated with HMGA2 mRNA expression. Knockdown of HIT000218960 resulted in downregulation of HMGA2. HMGA proteins are knowed as architectural transcription factors influencing a diverse array of normal biological processes.Citation19 Overexpression of HMGA2 was observed in numerous human malignancies including thyroid cancer, suggesting causal role of HMGA2 for oncogenesis and tumor progression.Citation16 With regard to the roles in PTC, HMGA2 protein has been demonstrated to play a key role in the neoplastic transformation of thyroid cells.Citation18,20 HMGA2 knockdown cells showed an epithelial morphology and suppressed proliferative ability,Citation16 suggesting the similar biological function of HMGA2 with that of HIT000218960 showed in our study. The reported studies on the association between HMGA2 and clinicopathological features mostly focused on the diagnostic utility of HMGA2 to distinguish between benign and malignant thyroid tumors.Citation21-26 In other malignancies, such as nasopharyngeal carcinoma, high HMGA2 expression is correlated with N stage, 2-year metastasis status and poor survival prognosis.Citation27,28 The correlation between HIT000218960 expression and clinicopathological features observed in our study was consistent with that of HMGA2. Taken together, these findings indicate that HIT000218960 plays its tumor promoting function via upregulation of HMGA2.
Although HIT000218960 has been suggested to have a role in upregulating the expression of HMGA2 and promoting progression of PTC, its mechanisms of modulating HMGA2 expression remains undetermined. Previous researches highlighted the post-transcriptional repression of HMGA2 by several miRNAs, such as let-7Citation29, miR-33aCitation30 and miR-204Citation31. Gong et al found that lncRNA H19 promoted HMGA2-mediated epithelial-mesenchymal transition through antagonizing let-7Citation32. Study of Deng et al also showed that a lncRNA, CCAT1 functions as a molecular sponge for let-7 and leads to the de-repression of its endogenous targets HMGA2Citation33. Does HALNR also function as an endogenous competitor with miRNAs targeting HMGA2? The mechanisms underlying the relationship between HALNR and HMGA2 must be explored in future studies.
In summary, this study demonstrated that HIT000218960 is significantly upregulated in PTC and the increased HIT000218960 expression levels was associated with metastatic phenotype such as lymph node metastasis and multifocality. Further more, our results showed the expression of HIT000218960 was significantly associated with HMGA2, which indicates that HIT000218960 may promote tumor progress of PTC putatively through upregulating HMGA2.
Materials and methods
Patient samples
Sixty-one PTC tissue and paired adjacent noncancerous thyroid tissue samples were obtained from the Department of General Surgery, Peking University Peoples' Hospital. The samples were immediately snap-frozen in liquid nitrogen after surgical resection, and then transferred to the freezer at −80°C until RNA extraction. The diagnosis of PTC was confirmed pathologically. Six pairs of samples were used for microarray analysis, while the other 55 pairs were used for qRT-PCR validation. The clinicopathologic characteristics of the 61 patients were shown in Table S4. None of the patients had received preoperative radiotherapy or chemotherapy. All patients provided written informed consent before samples were collected. The study was approved by the local research Ethics Committee of Peking University.
Cell lines and cell culture
Human PTC cell lines TPC1 and BCPAP were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). They were all tested and authenticated for genotypes by DNA fingerprinting. These cell lines were passaged for less than 6 months after resuscitation, and no reauthorization was done. TPC1 was cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS) (all from Gibco), and BCPAP was cultured in RPMI1640 medium supplemented with 10% FBS and 1% non-essential amino acid at 37°C with 5% CO2.
RNA extraction and purification
Total RNA containing small RNA was extracted from tissue samples by using the Trizol reagent (Invitrogen) and purified with mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to manufacter's protocol. The purity and concentration of RNA were determined from OD260/280 readings using spectrophotometer (NanoDrop ND-1000). RNA integrity was determined by 1% formaldehyde denaturing gel electrophoresis.
LncRNA and mRNA microarray expression profiling
The microarray profiling was conducted in the laboratory of Capitalbio Technology Corporation in Beijing, China. The sample labeling, microarray hybridization and washing were performed based on the manufacturer's standard protocols. Briefly, each purified RNA sample was transcribed to double strand cDNA, then synthesized into cRNA and labeled with Cy3-dCTP. The labled cRNAs were hybridized onto the human lncRNA + mRNA Array V4.0 (4 × 180 K, Agilent), including the global profiling of 40,916 human lncRNAs and 34,235 mRNAs. Then the arrays were washed and scanned with the Agilent Scanner G2505C (Agilent Technologies). The lncRNA+mRNA array data were analyzed for data summarization, normalization and quality control by using the GeneSpring software V13.0 (Agilent). To select the differentially expressed genes, we used thresholdvalues of ≥ 2 and ≤−2-fold change and a Benjamini-Hochberg corrected p vlaue of 0.05.
Functional group analysis
Gene ontology (GO) analysis was applied to analyze the main function of the differentially expressed mRNAs according to the Gene Ontology, which is the key functional classification of NCBI.Citation34,35 Similarly, pathway analysis was used to find out the significant pathway of the differentially expressed mRNAs based on the latest KEGG (Kyoto Encyclopedia of Genes and Genomes) database.Citation36-38 Fisher's exact test and χ2 test were used to select the significant function and pathway, the threshold of significance was defined by P-value and the false discovery rate (FDR).Citation39
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
The reverse transcription was performed using a reverse transcription kit (Takara, Japan). The expression of lncRNAs were measured by qRT-PCR with SYBR Green PCR Kit (Takara) on CFX96 Real-Time PCT Detection System (Bio-Rad, Hercules, CA). Primer sequences are shown in Table S5. Human β-Actin RNA was amplified as an internal control respectively for lncRNA. The lncRNA levels were calculated according to 2−△△Ct.
LncRNA target prediction
To identify the target genes of differentially expressed lncRNAs via cis- or trans- regulatory effects. The Cis-acting lncRNA prediction were performed by their tight correlation (Pearson's correlation coefficient minimum of 0.99) to a group of expressed protein-coding genes. The lncRNA resided at genomic loci where a protein-coding gene and an lncRNA gene were within 10 kb of each other along the genome.Citation40 The trans-prediction was conducted using blat tools (Standalone BLAT v. 35 × 1 fast sequence search command line tool, download from: http://hgdownload.cse.ucsc.edu/admin/exe/) to compare the full sequence of the lncRNA with the 3′ UTR of its co-expression mRNAs, with the default parameter setting.
Generation of HIT000218960 knockdown cell lines
The short hairpin RNA (shRNA) sequences against HIT0002189 were designed, synthesized and inserted into lentiviral vector by GenePharma Corporation (Suzhou, China). Empty vectors were used as negative control. TPC1 and BCPAP cells were seeded onto plate wells, and infected with either LV-shHIT0002189 or LV-NC plus 5 μg/ml polybrene (GenePharma Co., Suzhou, China) the next day. Then the cells were selected with 5 μg/ml puromycin to create stable knockdown cell lines. The infected TPC1 and BCPAP cells were designated as TPC1-shHIT0002189, BCPAP-shHIT0002189, TPC1-NC and BCPAP-NC in accordance with the lentiviral construct used in each. The expression of HIT0002189 in the infected cells was confirmed by qRT-PCR.
Western blot analysis
The total protein from the cells was extracted using cell lysis buffer with proteinase inhibitors (Roche Diagnostics, Basel, Switzerland). Lysates were denatured with sodium dodecyl sulfate (SDS) buffer at 100°C for 10 min, separated in 10% or 15% polyacrylamide gels and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Hertfordshire, UK). The membranes were blocked with 5% non-fat milk powder in Tris-buffered saline containing 0.1% Tween 20 (TBST) and probed with HMGA2 antibody (ab184616; Abcam plc., Cambridge, UK) overnight at 4°C. The following day, the membranes were treated with secondary antibodies. The band signals were visualized through enhanced chemiluminescence (ECL; Pierce, Rockford, IL, USA) after exposure to a ChemiDoc™ XRSC system (Bio-Rad).
Cell proliferation assay
Cells were seeded onto 96-well plates and incubated at 37°C after transfection. After incubation for 1 to 5 days, each well was added 10 μl of CCK8 solution and the plates were continued to be incubated for another 4 h at 37°C. The absorbance was measured at 450nm using a microplate reader (Bio-Rad). The CCK8 assay was repeated 3 times with 6 replicates.
Colony formation assay
The colony formation was performed as follows: after transfection, cells were seeded onto each well of 6-well plate and then incubated for another 6 d. The wells were then washed with PBS, fixed with 1% paraformaldehyde, and stained with 0.1% crystal violet. The colony assay was repeated 3 times in duplicate.
Wound healing assay
Cells were seed in 6-well plates and incubated until 100% confluent. The confluent monolayer of cells was scratched with a plastic apparatus to create a cell-free clear zone, about 1mm in width. Subsequently, the cells were incubated in medium without FBS and the wound distance was measured regularly.
Matrigel invasion assay
The Matrigel invasion chamber was used to assess cell invasion ability (24-well plates, 8-μm pore size, Corning). In brief, cells were seeded in the upper chamber with the media containing 0.1% bovine serum albumin, while the media containing 30% FBS was placed in the lower well. After incubation for 24 h at 37°C, the non-invading cells were removed with cotton swabs. Invasive cells at the bottom of the membrane were fixed with 1% formalin, stained with 0.1% crystal violet and counted under microscopic observation. This invasion detection was repeated 3 times in duplicate.
Statistical analysis
All results are expressed as mean ± SD. Difference between groups were assessed using Student's t-test and Fisher's exact test. P < 0.05 was considered to be statistically significant. All data, unless otherwise explained specifically, were analyzed using the SPSS 20.0 software.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary files
Download Zip (3.7 MB)Funding
This study was supported by grants from the National Natural Science Foundation of China (81572379, 81372291) and the National High-tech R&D Program (863 Program) (2015AA020110).
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: A Cancer Journal for Clinicians 2015; 65:5-29.
- Davies L, Welch H. Increasing Incidence of Thyroid Cancer in the United States, 1973–2002. JAMA 2006; 18:2164-7; http://dx.doi.org/10.1001/jama.295.18.2164
- Shoup M, Stojadinovic A, Nissan A, Ghossein RA, Freedman S, Brennan MF, Shah JP, Shaha AR. Prognostic indicators of outcomes in patients with distant metastases from differentiated thyroid carcinoma. J Am Coll Surg 2003; 197:191-7; PMID:12892796; http://dx.doi.org/10.1016/S1072-7515(03)00332-6
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. CELL 2009; 136:629-41; PMID:19239885; http://dx.doi.org/10.1016/j.cell.2009.02.006
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10:155-9; PMID:19188922; http://dx.doi.org/10.1038/nrg2521
- Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology 2012; 56:2231-41; PMID:22706893; http://dx.doi.org/10.1002/hep.25895
- Tanos V, Ariel I, Prus D, De-Groot N, Hochberg A. H19 and IGF2 gene expression in human normal, hyperplastic, and malignant endometrium. Int J Gynecol Cancer 2004; 14:521-5; PMID:15228427; http://dx.doi.org/10.1111/j.1048-891x.2004.014314.x
- Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. ONCOGENE 2007; 26:851-8; PMID:16878148; http://dx.doi.org/10.1038/sj.onc.1209846
- Kamel MM, Matboli M, Sallam M, Montasser IF, Saad AS, El-Tawdi AH. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl Res 2016; 168:134-45; PMID:26551349; http://dx.doi.org/10.1016/j.trsl.2015.10.002
- Yang Z, Guo X, Li G, Shi Y, Li L. Long noncoding RNAs as potential biomarkers in gastric cancer: Opportunities and challenges. Cancer Lett 2016; 371:62-70; PMID:26577810; http://dx.doi.org/10.1016/j.canlet.2015.11.011
- Jendrzejewski J, Thomas A, Liyanarachchi S, Eiterman A, Tomsic J, He H, Radomska HS, Li W, Nagy R, Sworczak K, et al. PTCSC3 Is Involved in Papillary Thyroid Carcinoma Development by ModulatingS100A4 Gene Expression. J Clin Endocrinol Metab 2015; 100:E1370-7; PMID:26274343; http://dx.doi.org/10.1210/jc.2015-2247
- Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R, de la Chapelle A. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci 2012; 109:8646-51; http://dx.doi.org/10.1073/pnas.1205654109
- Lan X, Sun W, Zhang P, He L, Dong W, Wang Z, Liu S, Zhang H. Downregulation of long noncoding RNA NONHSAT037832 in papillary thyroid carcinoma and its clinical significance. Tumor Biol 2016; 37:6117-23; http://dx.doi.org/10.1007/s13277-015-4461-4
- Prasad NB, Kowalski J, Tsai H, Talbot K, Somervell H, Kouniavsky G, Wang Y, Dackiw APB, Westra WH, Clark DP, et al. Three-Gene Molecular Diagnostic Model for Thyroid Cancer. Thyroid 2012; 22:275-84; PMID:22280184; http://dx.doi.org/10.1089/thy.2011.0169
- Chiappetta G, Ferraro A, Vuttariello E, Monaco M, Galdiero F, De Simone V, Califano D, Pallante P, Botti G, Pezzullo L, et al. HMGA2 mRNA expression correlates with the malignant phenotype in human thyroid neoplasias. Eur J Cancer 2008; 44:1015-21; PMID:18375116; http://dx.doi.org/10.1016/j.ejca.2008.02.039
- Pallante P, Sepe R, Puca F, Fusco A. High mobility group A proteins as tumor markers. Fron Med 2015; 2:15; PMID:25729750; http://dx.doi.org/10.3389/fmed.2015.00015
- Ayoubi TA, Jansen E, Meulemans SM, Van de Ven WJ. Regulation of HMGIC expression: an architectural transcription factor involved in growth control and development. Oncogene 1999; 18:5076-87; PMID:10490844; http://dx.doi.org/10.1038/sj.onc.1202881
- Berlingieri MT, Manfioletti G, Santoro M, Bandiera A, Visconti R, Giancotti V, Fusco A. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol Cell Biol 1995; 15:1545-53; PMID:7862147; http://dx.doi.org/10.1128/MCB.15.3.1545
- Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review). Int J Oncol 2008; 32:289-305; PMID:18202751
- Vallone D, Battista S, Pierantoni GM, Fedele M, Casalino L. Neoplastic transformation of rat thyroid cells requires the junB and fra-1 gene induction which is dependent on the HMGI-C gene product. Embo J 1997; 16(17):5310–5321; PMID:9311991
- Belge G, Meyer A, Klemke M, Burchardt K, Stern C, Wosniok W, Loeschke S, Bullerdiek J. Upregulation ofHMGA2 in thyroid carcinomas: A novel molecular marker to distinguish between benign and malignant follicular neoplasias. Genes, Chromosomes and Cancer 2008; 47:56-63; http://dx.doi.org/10.1002/gcc.20505
- Chiappetta G, Bandiera A, Berlingieri MT, Visconti R, Manfioletti G, Battista S, Martinez-Tello FJ, Santoro M, Giancotti V, Fusco A. The expression of the high mobility group HMGI (Y) proteins correlates with the malignant phenotype of human thyroid neoplasias. Oncogene 1995; 10:1307-14; PMID:7731681
- Jin L, Lloyd RV, Henry MR, Erickson LA, Sebo TJ, Rumilla KM, Zhang J. The diagnostic utility of combination of HMGA2 and IMP3 qRT-PCR testing in thyroid neoplasms. Appl Immunohistochem Mol Morphol 2015; 23:36-43; PMID:25356939; http://dx.doi.org/10.1097/PAI.0000000000000031
- Jin L, Lloyd RV, Nassar A, Lappinga PJ, Sebo TJ, Swartz K, Seys AR, Erickson-Johnson MR, Roth CW, Evers BR, et al. HMGA2 expression analysis in cytological and paraffin-embedded tissue specimens of thyroid tumors by relative quantitative RT-PCR. Diagn Mol Pathol 2011; 20:71-80; PMID:21532495; http://dx.doi.org/10.1097/PDM.0b013e3181ed784d
- Barros-Filho MC, Marchi FA, Pinto CA, Rogatto SR, Kowalski LP. High Diagnostic Accuracy Based onCLDN10 ,HMGA2 , andLAMB3 Transcripts in Papillary Thyroid Carcinoma. The J Clin Endocrinol Metab 2015; 100:E890-9; PMID:25867809; http://dx.doi.org/10.1210/jc.2014-4053
- Nagar S, Ahmed S, Peeples C, Urban N, Boura J, Thibodeau B, Akervall J, Wilson G, Long G, Czako P. Evaluation of genetic biomarkers for distinguishing benign from malignant thyroid neoplasms. Am J Surg 2014; 207:596-601; PMID:24713092; http://dx.doi.org/10.1016/j.amjsurg.2013.06.012
- Xia YY, Yin L, Tian H, Guo WJ, Jiang N, Jiang XS, Wu J, Chen M, Wu JZ, He X. HMGA2 is associated with epithelial-mesenchymal transition and can predict poor prognosis in nasopharyngeal carcinoma. Onco Targets Ther 2015; 8:169-76; PMID:25653540; http://dx.doi.org/10.2147/OTT.S74397
- Xia YY, Yin L, Jiang N, Guo WJ, Tian H, Jiang XS, Wu J, Chen M, Wu JZ, He X. Downregulating HMGA2 attenuates epithelial-mesenchymal transition-induced invasion and migration in nasopharyngeal cancer cells. Biochem Biophys Res Commun 2015; 463:357-63; PMID:26025649; http://dx.doi.org/10.1016/j.bbrc.2015.05.068
- Damanakis AI, Eckhardt S, Wunderlich A, Roth S, Wissniowski TT, Bartsch DK, Di Fazio P. MicroRNAs let7 expression in thyroid cancer: correlation with their deputed targets HMGA2 and SLC5A5. J Cancer Res Clin 2016; 142:1213-20; http://dx.doi.org/10.1007/s00432-016-2138-z
- Rice SJ, Lai SC, Wood LW, Helsley KR, Runkle EA, Winslow MM, Mu D. MicroRNA-33a Mediates the Regulation of High Mobility Group AT-Hook 2 Gene (HMGA2) by Thyroid Transcription Factor 1 (TTF-1/NKX2-1). J BIOL CHEM 2013; 288:16348-60; PMID:23625920; http://dx.doi.org/10.1074/jbc.M113.474643
- Wu ZY, Wang SM, Chen ZH, Huv SX, Huang K, Huang BJ, Du JL, Huang CM, Peng L, Jian ZX, et al. MiR-204 regulates HMGA2 expression and inhibits cell proliferation in human thyroid cancer. Cancer Biomark 2015; 15:535-42; PMID:26406941; http://dx.doi.org/10.3233/CBM-150492
- Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7s suppression on its target HMGA2-mediated EMT. Tumour Biol 2014; 35:9163-9; PMID:24920070; http://dx.doi.org/10.1007/s13277-014-2185-5
- Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res 2015; 34:18; PMID:25884472; http://dx.doi.org/10.1186/s13046-015-0136-7
- The Gene Ontology (GO) project in 2006. NUCLEIC ACIDS RES 2006; 34:D322-6.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-9.
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res 2004; 32:D277-80; PMID:14681412; http://dx.doi.org/10.1093/nar/gkh063
- Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC BIOINFORMATICS 2006; 7:30; PMID:16423281; http://dx.doi.org/10.1186/1471-2105-7-30
- Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res 2007; 17:1537-45; PMID:17785539; http://dx.doi.org/10.1101/gr.6202607
- Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol 2007; 25:663-8; PMID:17486083; http://dx.doi.org/10.1038/nbt1305
- Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 2010; 16:1478-87; PMID:20587619; http://dx.doi.org/10.1261/rna.1951310
