ABSTRACT
Macrophages, in response to different environmental cues, undergo the classical polarization (M1 macrophages) as well as the alternative polarization (M2 macrophages) that involve the functions of stimulus-specific transcription factors.
Kruppel-like factor 4 (KLF4), a member of a subfamily of the zinc-finger class of DNA-binding transcription factors, plays as a critical regulator of macrophage polarization. KLF4 has been reported as a SUMOylated protein. In this study, we showed that SUMOylation of KLF4, is induced by IL-4 treatment in macrophages. IL4-induced KLF4 SUMOylation promotes RAW264.7 cells and bone marrow derived macrophages (BMDMs) to polarize into M2 subset. Thus, we identified an important post-translational modification (PTM), SUMOylation, plays a crucial role in regulating KLF4 activity during IL-4 induced macrophage M2 polarization. SUMOylation of KLF4 can be a potential therapeutic target in the resolution of inflammation.
Introduction
Macrophages play critical roles in organism development, homeostasis and immune responses to pathogens.Citation1,2 They exhibit remarkable plasticity in response to environmental signals. To provide a simple framework for discussion, macrophages are functionally classified into 2 major subsets, which are the classically activated macrophages (M1 macrophages) and alternatively activated macrophages (M2 macrophages).Citation3,4 M1 macrophages are considered to be proinflammatory while M2 macrophages represent the anti-inflammatory state.Citation4 Macrophage polarization is driven by cues in tissue microenvironment such as cytokines and growth factors. In response to stimulation with TLR (toll-like receptor) ligands and IFN-γ, macrophages undergo M1 polarization. However, upon stimulation with IL-4/IL-13, macrophages undergo M2 polarization.Citation5
Transcriptional regulation is central to the macrophage subset polarization.Citation6,7 Via activating stimulus-specific pathways, the phenotype of macrophages is shaped by environmental signals.Citation6,7 There are several key transcription factors that translate signals in the microenvironment into a polarized macrophage phenotype. For examples, the STATs (Signal Transducers and Activators of Transcription) family seems to be pivotal factors in macrophage M1/M2 polarization.Citation8-10 STAT1 is an essential mediator of M1 macrophage polarization while STAT6 is required to drive M2 macrophage activation during TH2 cell-mediated immune responses in the presence of IL-4 and/or IL-13.Citation11,12
Kruppel-like factors (KLFs) are a subfamily of the zinc-finger class of DNA-binding transcription regulators. Members of this gene family have been shown to play important roles in a diverse array of cellular processes, including macrophage polarization.Citation13 To date, KLF2, KLF4 and KLF6 have been shown to regulate macrophage function.Citation14-16 KLF4, one of the 4 well defined transcription factors in generating induced pluripotent stem (iPS) cells, from mouse somatic cells or human dermal fibroblasts,Citation17,18 functions to promote monocyte differentiation.Citation19,20 Several studies have investigated the biologic role of KLF4 in macrophage polarization.Citation21-24 For instance, macrophages deficient in KLF4 exhibit impaired expression of M2 markers in the presence of IL-4.Citation25 On the contrary, overexpression of KLF4 in RAW264.7 macrophages enhances IL-4 induced M2 gene expression.Citation26,27 Moreover, IL-4 stimulated macrophages show significant increases in KLF4 expression.Citation25 Mechanistically, IL-4 induces STAT6 phosphorylation to promote the KLF4 gene expression, which in turn cooperates with STAT6 to promote an M2 gene profile.Citation10 Kapoor et al demonstrated that STAT6 and KLF4 implement IL-4 induced M2 polarization via the dual catalytic activities of MCPIP.Citation28 However currently, regulation on KLF4 activity during macrophage M2 polarization is still not fully illustrated.
At transcriptional level, it has been reported that ATRA (all-trans retinoic acid) stimulation augments KLF4 mRNA level in vascular smooth muscle cells (VSMCs).Citation29 During monocyte/macrophage differentiation, AICDA mediated active demethylation of the KLF4 promoter is necessary for transcriptional regulation of KLF4 by PU.1.Citation30 In both differentiated and stem cells, the Von Hippel-Lindau gene product, pVHL, physically interacts with KLF4 and promotes its degradation.Citation31 Phosphorylation of KLF4 by ERK1/2 recruits the F-box proteins βTrCP1 or βTrCP2 (components of an ubiquitin E3 ligase) to the KLF4 N-terminal domain, subsequently induces KLF4 ubiquitination and degradation.Citation32 In addition, ATRA can regulate KLF4 activity by inducing HDAC phosphorylation, which triggers its dissociation from KLF4. Furthermore, dissociation KLF4 from HDAC increases its acetylation and binding activity to target genes in VSMCs,Citation33 therefore enhances its target gene expression.
As an important posttranslational modification, SUMOylation can alter the activity of its target proteins as well as their cellular localizations in various biologic processes.Citation34-38 KLF4 SUMOylation is capable of enhancing it transcriptional activity but having no effects on its stability.Citation39,40 Recently, Nie et al revealed KLF4 SUMOylation acts as a switch in transcriptional programs that control VSMC proliferation.Citation41 In the present study, we demonstrated that IL-4 stimulation induces KLF4 SUMOylation in RAW264.7 macrophages. SUMOylation of KLF4 plays critical roles in IL-4 stimulated macrophage genetic M2 program and polarization in RAW264.7 cells and mice bone marrow-derived macrophages (BMDMs). Thus, we identified SUMOylation as a regulatory mechanism on KLF4 activity during the macrophage M2 polarization process.
Materials and methods
Plasmids and antibodies
The plasmids pShuttle-CMV and pAdEasy-1 for generation of recombinant adenovirus were provided by Dr. Yibin Wang. HA-KLF4 and pShuttle-CMV-KLF4 were generated using standard cloning procedures (Vazyme Biotech Co.,Ltd). HA-KLF4-K278R and pShuttle-CMV-KLF4-K278R were generated using site-directed mutagenesis (Strategene). Antibodies against FLAG M2 and HA were purchased from Sigma, KLF4 and STAT6 from Santa Cruz, SUMO1 from Abcam, and ACTIN from Cell Signaling Technology.
Cell culture
BMDMs were differentiated with M-CSF as described previously. MEF, BMDM, 293T, and RAW264.7 cells were cultured in DMEM (Hyclone) supplemented with 10% fetal bovine serum (Invitrogen) and 1% antibiotics (penicillin/streptomycin) (Invitrogen).
Immunoprecipitation and immunoblotting
Transfected cells were lysed in radioimmune precipitation assay buffer (50 mMTris-HCl (pH 7.4), 400 mMNaCl, 1 mM EDTA, 1% Nonidet P-40, 0.1% SDS, 1% sodium deoxycholate, and a mixture of protease inhibitors) and cleared by centrifugation. Cleared cell lysates were incubated with 10 μl of anti-FLAG M2-agarose affinity gel (Sigma) or 10 μl of anti-HA-agarose affinity gel (Sigma) for 2 h. After extensive washing, beads were boiled at 100 °C for 10 min. Proteins were resolved by SDS-PAGE and transferred onto PVDF membranes (Millipore), followed by immunoblotting using corresponding antibodies according to the instructions of the manufacturer. Immunoblots were analyzed using the LAS-4000 system (Fujifilm).
Stable cell lines
FLAG-KLF4 or FLAG-KLF4-K278R lentiviral plasmids were transfected into HEK293T cells with lentivirus packaging vectors by calcium phosphate-DNA coprecipitation method. Viral supernatants were collected 48 h after transfection. RAW264.7 cells were infected by lentiviral supernatant in the presence of 8 μg/ml Polybrene for 12 h. Cells were sorted for GFP positive cells by flow cytometry 72 h later after the infection.
RNA isolation and qPCR
Total RNA was isolated from cells by using Tripure isolation reagent (Roche). For mRNA analysis, an aliquot containing 2 μg of total RNA was reverse-transcribed using the cDNA synthesis kit (Takara). Real-time PCR was performed using SYBR Green PCR master mix (Applied Biosystems) and detected by the ABI Prism 7500 sequence detection system (Applied Biosystems). The primers for real-time RT-PCR were as follows:
18S, sense: 5-AGTCCCTGCCCTTTGTACACA-3,
antisense: 5-CGATCCGAGGGCCTCACTA-3;
Arg-1, sense: 5-TTTTTCCAGCAGACCAGCTT-3and
antisense: 5-AGAGATTATCGGAGCGCCTT-3;
Mrc-1, sense: 5-CAGGTGTGGGCTCAGGTAGT-3 and
antisense: 5-TGGCATGTCCTGGAATGAT-3;
Mgl1, sense: 5-CAGGATCCAGACAGATACGGA-3 and
antisense: 5-GGAAGCCAAGACTTCACACTG-3.
Adenovirus
The Adenoviruses (Vector, FLAG-KLF4, FLAG-KLF4-K278R) were generated by AdEasy system and has been described previously, and for overexpression, BMDM cells were infected with the empty control virus (Vector) or the adenovirus carrying the human KLF4 gene (FLAG-KLF4) or the adenovirus carrying the human KLF4-K278R mutant gene (FLAG-KLF4-K278R).
ChIP-qPCR assay
ChIP-qPCR assay was performed as described previously.Citation42 Briefly, RAW264.7-Vector, RAW264.7-KLF4, and RAW264.7-KLF4 (K278R) (with FLAG tag) were stimulated with IL-4 (20 ng/ml) for 4 h before crosslinking with 1% formaldehyde, and sonicated. Solubilized chromatin was immunoprecipitated with anti-FLAG M2-agarose affinity gel (Sigma), washed, and then eluted. After crosslink reversal and proteinase K treatment, immunoprecipitated DNA was extracted with phenol-chloroform, ethanol precipitated. The DNA fragments were further analyzed by qPCR. The specific primers used to amplify Arg-1 promoter region was as followed: sense: 5-TCACGCGTGGTAGCCGACGAGAG-3; antisense: 5-CGCACGCGTAAAGTGGCACAACTCACGTA-3.
Flow cytometry
Cells were resuspended in 200 ml FACS buffer (PBS with 5% bovine calf serum) and placed on ice for 15 min to block. Cells were incubated with fluorescently labeled antibodies on ice in the dark for 20 min and then washed with PBS, centrifuged at 500 × g for 5 min, resuspended in 200 ml FACS buffer, and evaluated on a FACSCanto (Becton Dickinson) flow cytometer. Data were further analyzed using FlowJo software (Tree Star).
Statistical analysis
Statistical analyses were performed with a 2-tailed unpaired Student's t test. All data shown represent the results obtained from triplicated independent experiments with SEM (mean ± SD). The values of p < 0.05 were considered statistically significant.
Results and discussion
IL-4 treatment increases KLF4 SUMOylation in macrophages
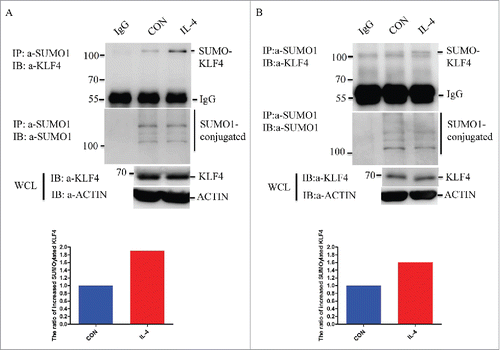
Since KFL4 played a key role in macrophage M2 polarization, to illustrate the regulatory role of SUMOylation on KLF4 activity during this process, 20 ng/ml IL-4 or 40 ng/ml IL-13 was applied to treat RAW264.7 cells for 24 h, and whole cell lysates were collected before subject to Western blot analysis. We observed that the SUMOylation of endogenous KL4 protein was increased post IL-4 () or IL-13 stimulation (Fig. S1). The lower panel in as well as in Fig. S1 showed the quantitative analysis of the ratio of SUMOylated/total KLF4 proteins in RWA264.7 cells before and after IL-4 or IL-13 stimulation. As a result, the ratio was increased up to 1.8-fold () or 1.75-fold (Fig. S1) in IL-4 or IL-13 treated RAW264.7 cells, when compared with the non-treated control cells. Moreover, in isolated primary BMDMs,we also found IL-4 treatment increased the ratio of SUMOylated/total KLF4 proteins up to 1.6-fold (). Liao and colleagues demonstrated upon treatment with IL-4 or IL-13, the expression of klf4 gene in mouse PMs (peritoneal macrophages) and BMDMs, was greatly induced both at the mRNA and protein levels.Citation25 And mechanistically, they found KLF4 cooperated with STAT6 to induce an M2 genetic program and inhibits M1 targets. Although we did not detect a significant induction of KLF4 protein upon IL-4 treatment in macrophages reported as Liao et al, we did find increased SUMOylation of KLF4 by IL-4 was STAT6 dependent, since know-down of STAT6 in RAW264.7 cells abrogated the enhanced SUMOylation of KLF4 upon IL-4 treatment (Fig. S2). In a word, the results of induced effect of IL-4 on KLF4 SUMOylation in RAW264.7 cells and BMDMs shown by strongly hints its involvement in macrophage M2 polarization.
Figure 1. IL-4 treatment increases KLF4 SUMOylation in macrophages. (A) IL-4 treatment increases the SUMOylation of endogenous KLF4 protein in RAW264.7 macrophages. RAW264.7 cells were treated with or without 20 ng/ml IL-4 for 24 h, the alteration of KLF4 SUMOlyation was detected by immunoprecipitation. SUMO1 conjugated proteins were pulled down by non-specific IgG or SUMO1 antibody from these cell lysates. Bound proteins were blotted with KLF4 or SUMO1 antibody. Cell lysates (Input) were immunoblotted (IB) with KLF4 or ACTIN. The quantitative result for the ratio of SUMOylated KLF4/total KLF4 in RAW264.7 cells is shown in the lower panel. (B) IL-4 treatment increases the SUMOylation of endogenous KLF4 protein in BMDMs. Primary BMDMs were isolated from mice and treated with or without 20 ng/ml IL-4 for 24 h, the alteration of KLF4 SUMOlyation was detected by immunoprecipitation. SUMO1 conjugated proteins were pulled down by non-specific IgG or SUMO1 antibody from these cell lysates. Bound proteins were blotted with KLF4 or SUMO1 antibody. Cell lysates (Input) were immunoblotted (IB) with KLF4 or ACTIN. The quantitative result for the ratio of SUMOylated KLF4/total KLF4 in BMDMs is shown in the lower panel.
SUMOylation of KLF4 promotes IL-4 stimulated macrophage genetic M2 program
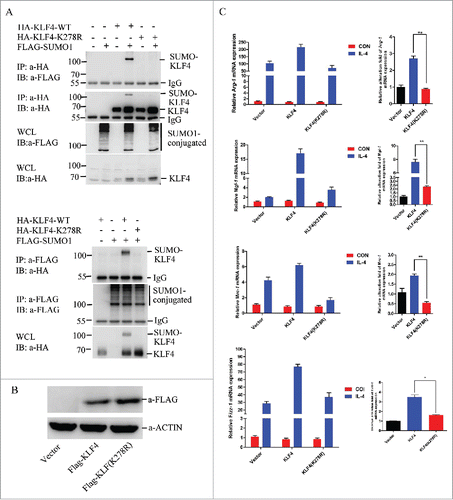
Du et al reported that mouse KLF4 protein is SUMOylated at lysine residue 275.Citation39 In this study, we identified human KLF4 is a SUMOylated protein at K278 (). When lysine 278 is mutated into arginine, the SUMOylation band of KLF4 totally disappeared (). Next we went on to address the key role of KLF4 SUMOylaiton in M2 polarization. Stable cell lines of RAW264.7 overexpressing human wild-type KLF4 or KLF4 (K278R) proteins were established, and their expressions were determined by Western blot assay using anti-Flag antibody (). To observe the M2 macrophage marker gene expression, stable transfectants stable transfectants RAW264.7-Vector, RAW264.7-KLF4, and RAW264.7-KLF4 (K278R) were stimulated either with or without IL-4 for 24 h, and cells were collected and total RNA was extracted from above mentioned stable cell lines before subject to qPCR analysis. illustrated that KLF4 overexpression significantly enhanced M2 gene expression (Arg-1, Mgl-1, Mrc-1, Fizz1) upon IL-4 stimuli. Interestingly, overexpression of KLF4 K278R mutant was still capable to increase M2 gene expression slightly in macrophages upon stimulation, but with less extended levels when compared with KLF4 wild type. The relatively reducing of M2 marker gene expression in macrophages, caused by SUMOylation abrogation on KLF4 through mutating its lysine 278 into arginine, was 3.0-fold for Arg-1, 3.3-fold for Mgl-1, 3.8-fold for Mrc-1, and 2.6-fold for Fizz 1, respectively upon IL-4 treatment. This result suggests the critical role of SUMOylation on KLF4 transcriptional activity regulation, during IL-4 stimulated M2 genetic program in RAW264.7 cells. Summarize of the results from and , we can conclude that IL-4 treatment induces the SUMOylation of KLF4, which in turn enhances its ability to induce its target gene transcription in M2 genetic program in macrophages.
Figure 2. SUMOylation of KLF4 increases the expression of M2 marker genes in RAW264.7 cells after IL-4 treatment. (A) Human KLF4 protein is SUMOylated at K278. In upper graph, 293T cells were transfected with HA-KLF4, HA-KLF4-K278R, or Flag-SUMO1 as indicated. HA-KLF4 proteins were pulled down by HA-beads from these cell lysates. Bound proteins were blotted with anti-Flag or anti-HA antibody. Cell lysates (Input) were immunoblotted (IB) with anti-Flag or anti-HA antibody. In lower graph, 293T cells were transfected with HA-KLF4, HA-KLF4-K278R, or Flag-SUMO1 as indicated. For IP, Flag-SUMO1 proteins were pulled down by Flag M2 beads from these cell lysates. Then bound proteins were blotted with anti-HA or anti-Flag antibody. Cell lysate (Input) was immunoblotted (IB) with anti-HA antibody. (B) Demonstration on establishment of KLF4 wild-type and KLF4 (K278R) stably overexpressed RAW264.7 cell lines. Cell lysates were immunoblotted with Flag or ACTIN. (C) KLF4, but not the K278R mutant, activates the expression of M2 marker gene expression after IL-4 treatment in RAW264.7 cells. mRNA levels of KLF4 target genes Arg-1, Mgl-1, Mrc-1 and Fizz1 were measured by realtime PCR in RAW264.7-Vector, RAW264.7-KLF4, and RAW264.7-KLF4 (K278R) cells with or without IL-4 treatment. The data are presented as means ± s.d. of 3 independent experiments. The relative alteration folds of the genes in 3 cell lines before and after IL-4 treatment were illustrated in the right graph. **P < 0.01, t-test.
SUMOylation of KLF4 enhances its binding ability to Arg-1 promoter
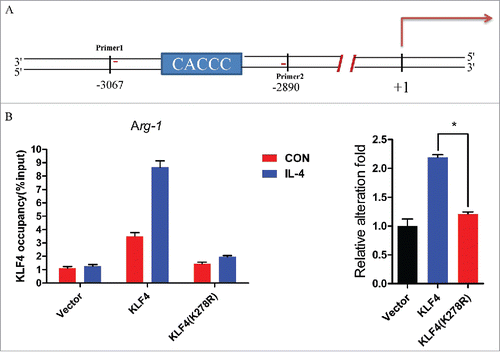
Arg-1 (arginase 1) is an important M2 marker that connects KLF4 to the biologic processes involved in M2 polarization.Citation43 Arg-1 encodes an L-arginine degrading enzyme limits NO production and polyamine synthesis.Citation44 Consensus KLF-binding sites (CACCC) have been identified on Arg-1 promoter and IL-4 can induce Arg-1 promoter activity through mediation of KLF4 binding.Citation25 has shown that the overexpression of wild-type KLF4 in RAW264.7 cells significantly induced the Arg-1 gene transcription upon IL-4 treatment. In contrast, KLF4 (K278R) has lost its ability to increase Arg-1 gene expression. To determine whether SUMOlyation influences the binding ability of KLF4 to Arg-1 gene promoter upon IL-4 treatment, ChIP assay was conducted in RAW264.7-Vector, RAW264.7-KLF4, and RAW264.7-KLF4 (K278R) stable cell lines, with or without IL-4 treatment of 4 h. The qPCR targeting region in ChIP assay is illustrated by a carton as . showed that in RAW264.7 cells, IL-4 treatment enhanced the binding ability of KLF4 to Arg-1 promoter about 2.2-fold, compared with in RAW264.7-vector cells. In contrast, when KLF4 cannot be SUMOylated, the ability of IL-4 promoted binding of KLF4 to Arg-1 gene promoter was almost abolished in RAW264.7-K278R cells. The result revealed in RAW264.7 cells, IL-4 promoted M2 marker gene expression mainly through enhancing its SUMOylated modification, not its expression.
Figure 3. SUMOylation of KLF4 enhances its binding to Arg-1 promoter after IL-4 treatment. (A) The primer for amplifying the region containing KLF4 binding site in Arg-1 promoter used in qChIP assay is illustrated in the carton. (B) The wild-type KLF4 and KLF4 (K278R) occupancy on endogenous Arg1 promoter were analyzed by qChIP in 3 RAW264.7 cell lines with or without IL-4 treatment. The data are presented as means ± s.d. of 3 independent experiments. The relative alteration folds of binding ability in 3 cell lines before and after IL-4 treatment were illustrated in the right graph. **P < 0.01, t-test.
SUMOylation of KLF4 promotes IL-4 stimulated BMDMs M2 polarization
M-CSF differentiated mouse bone marrow derived-macrophages (BMDMs) is a well-established cell system to study the macrophage polarization in vitro.Citation45 To further illustrate the promoting role of KLF4 SUMOylation in macrophage M2 polarization, BMDMs were purified from the mice, and by using Adenovirus system, we successfully overexpressed wild-type KLF4 or KLF (K278R) protein in BMDMs, verified by Western blot analysis ().
Figure 4. SUMOylation of KLF4 promotes IL-4 induced BMDM M2 polarization. (A) demonstration on establishment of KLF4 wild-type and KLF4 (K278R) overexpressed BMDMs. Cell lysates were immunoblotted with Flag or ACTIN. (B). KLF4, but not the K278R mutant, activates the expression of M2 marker gene expression after IL-4 treatment in BMDMs. mRNA levels of KLF4 target gene Arg-1, Mgl-1, Mrc-1 and Fizz1 were measured by realtime PCR in BMDM-Vector, BMDM-KLF4, and BMDM-KLF4 (K278R) cells with or without IL-4 treatment. The data are presented as means ± s.d. of 3 independent experiments. The relative alteration folds of the genes in 3 types of cells before and after IL-4 treatment were illustrated in the right graph. **P < 0.01, t-test. (C) SUMOylation of KLF4 promotes IL-4 induced BMDM M2 polarization. Representative FACS plots of percentages of CD206(+) cells in CD11b(+)F4/80(+) cells, in BMDM-Vector, BMDM-KLF4 and BMDM-KLF(K278R) cells, individually, with and without IL-4 treatment.
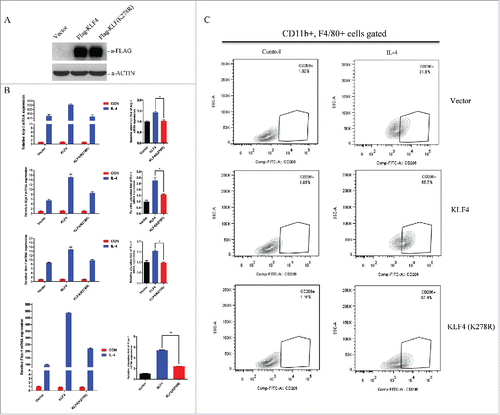
To determine the M2 macrophage marker gene expression, BMDM-Vector, BMDM-KLF4, and BMDM-KLF4 (K278R) stable transfectants either stimulated with or without M2 (IL-4) polarizing reagents for 24 h, were collected, total RNA was extracted and before subject to qPCR analysis. has demonstrated that KLF4 overexpression significantly increased M2 gene expression (Arg-1, Mgl-1, Mrc-1, Fizz1) upon IL-4 stimuli in BMDMs. Despite overexpression of KLF4 (K278R) increased the M2 gene expression in BMDMs upon IL-4 stimuli, its ability was significantly attenuated. We can see from the right graph of , when compared with in BMDM-KLF4, the M2 marker gene expressions were decreased to 1.5-fold for Arg-1, 1.8-fold for Mgl-1, 1.5-fold for Mrc-1, and 2.6-fold for Fizz 1, respectively, in BMDMs-KLF4 (K278R) stable transfectants, upon IL-4 stimulation. To further demonstrate that KLF4 SUMOylation promotes BMDMs M2 polarization, BMDM-Vector, BMDM-KLF4, and BMDM-KLF4 (K278R) cells, were collected at 24 h time point post IL-4 treatment. Cells were then fixed and stained with M2 macrophage surface marker CD206. Upon IL-4 stimuli, the percentage of CD206(+) cells in BMDM-Vector group increased to to 31.8% in comparison with non-treated group. We also observed that in BMDM-KLF4 group, due to the overexpression of KLF4 in macrophages, the same treatment of IL-4 on cells increased the percentage of M2 population to 65.7%. However, when KLF4 cannot be SUMOylated, its promoting role on M2 polarization was highly damped, the percentage of M2 population was shifted back to 37.4% of CD206(+) cells in BMDM-KLF4(K278R) cells (), which consistent with the qPCR results in . These results collectively indicate the crucial role of KLF4 SUMOylation on its functional regulation, during the process of macrophage M2 polarization induced by IL-4 treatment in vitro.
Herein, we characterized the crucial role of KLF4 SUMOylation in IL-4 induced macrophage M2 polarization, by using RAW264.7 as well as BMDM cell systems. The increase of KLF4 SUMOylation post IL-4 treatment is a novel and interesting findings (). But as illustrated by the working model in , how IL-4 induced KLF4 SUMOylation during macrophage M2 polarization still remains unclear. IL-4 treatment might increase KLF4 SUMOylation either by increasing its specific E3 ligase expression or activity, or by decreasing the expression or activity of its specific de-SUMOylating protease, which has not yet been detected. In addition, we also demonstrated that IL-4 induced KLF4 SUMOylation greatly promotes BMDMs polarization into M2 subset (). We identified how KLF4 induces macrophage M2 polarization upon IL-4 treatment, in addition to its induced expression during the process reported previously.
Figure 5. Model depicting the role of KLF4 SUMOylation in regulating macrophage M2 polarization induced by IL-4.
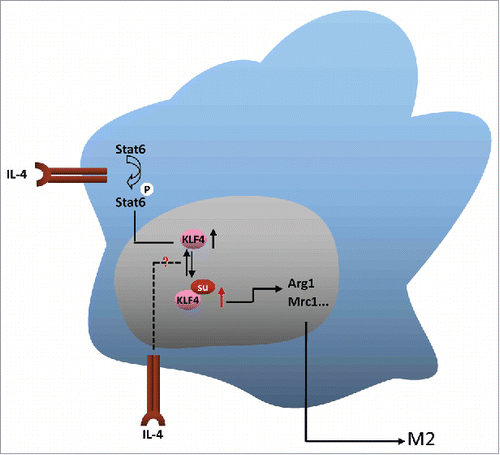
Our study has also revealed a new mechanism of how KLF4 activity is regulated during IL-4 induced macrophage M2 polarization. Through enhanced SUMOylation on KLF4, its transcriptional activity is increased by IL-4 (). Du's study found that KLF4 is both associated with SUMO1 via a SIM (SUMO interacting motif) and SUMOylated at a single site.Citation39 They provide direct evidence that SUMOylation of KLF4 doesn't influence its protein level and SIM in KLF4 functions as a transcriptional domain.Citation39 In 2015 and 2016, the studies of Zheng et al and Nie et al demonstrated the biologic role of SUMOylated KLF4 in VSMC (vascular smooth muscle cell).Citation40,41 Zheng et al found that KLF4 enhances, but SUMOylated KLF4 inhibits, transactivation activity of miR-200c during the process of VSMC proliferation.Citation40 Nie et al presented evidence that VSMC proliferation is regulated by KLF4 and Ubc9. They found that KLF4 activated p21 expression, and PDGF-BB blocked the KLF4-mediated induction of p21 by enhancing the SUMOylation of KLF4 and the subsequent recruitment of the co-repressors NCoR, HDAC2 and LSD1.Citation41 These 2 studies indicate that SUMOylation enables KLF4 to have dual transcriptional regulation roles in response to cellular and environmental cues. Although ChIP assay illustrated SUMOylation of KLF4 enhances its binding ability to its target gene promoter in macrophages () upon IL-4 treatment, how SUMO conjugation influences KLF4 activity remains still unknown. Maybe IL-4 induced SUMO1 conjugation to KLF4 make it more easily to be recruited by coactivators to its target gene promoters, or SUMOylation make KLF4 subsequent dissociation from its corepressors. It's still a question waiting to be answered in the future.
Given the importance of macrophages subsets in health and disease, they have been the subjects of intense investigation over the past few years.Citation46 In vivo experiments demonstrated that mice bearing myeloid-specific deletion of KLF4 exhibited delayed wound healing, developed diet-induced obesity, glucose intolerance, and insulin resistance.Citation25 Therefore, using KLF4 SUMOlyated site mutant allela knock-in mice as a research model, to study the biologic consequences of KLF4 SUMOylation in macrophage polarization and function in vivo, is interesting and promising. The enhanced SUMOylation of KLF4 during IL-4 treatment, promoted macrophages to M2 polarization, which can hopefully act as a potential therapeutic target in the treatment of inflammatory diseases.
Abbreviations
| BMDMs | = | bone marrow-derived macrophages |
| KLF4 | = | Kruppel-like factor 4 |
| STATS | = | Signal Transducers and Activators of Transcription |
| SUMO | = | small-ubiqutin like modifier |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1269045_Supplemental_Material.zip
Download Zip (246.1 KB)Acknowledgments
We thank Dr. Jiao Ma for manuscript reading and editing.
Funding
This work was supported in part by National Basic Research Program of China (973 Program) (2015CB910400), Natural Science Foundation of China (81572691).
References
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013; 496:445-55; PMID:23619691; http://dx.doi.org/10.1038/nature12034
- Epelman S, Lavine KJ, Randolph GJ. Origin and functions of tissue macrophages. Immunity 2014; 41:21-35; PMID:25035951; http://dx.doi.org/10.1016/j.immuni.2014.06.013
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 2014; 41:14-20; PMID:25035950; http://dx.doi.org/10.1016/j.immuni.2014.06.008
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity 2010; 32:593-604; PMID:20510870; http://dx.doi.org/10.1016/j.immuni.2010.05.007
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008; 8:958-69; PMID:19029990; http://dx.doi.org/10.1038/nri2448
- Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 2011; 11:750-61; PMID:22025054; http://dx.doi.org/10.1038/nri3088
- Tugal D, Liao X, Jain MK. Transcriptional control of macrophage polarization. Arterioscler Thromb Vasc Biol 2013; 33:1135-44; PMID:23640482; http://dx.doi.org/10.1161/ATVBAHA.113.301453
- Darnell JE, Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994; 264:1415-21; PMID:8197455; http://dx.doi.org/10.1126/science.8197455
- Levy DE, Darnell JE, Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 2002; 3:651-62; PMID:12209125; http://dx.doi.org/10.1038/nrm909
- Stark GR, Darnell JE, Jr. The JAK-STAT pathway at twenty. Immunity 2012; 36:503-14; PMID:22520844; http://dx.doi.org/10.1016/j.immuni.2012.03.013
- Varinou L, Ramsauer K, Karaghiosoff M, Kolbe T, Pfeffer K, Muller M, Decker T. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity 2003; 19:793-802; PMID:14670297; http://dx.doi.org/10.1016/S1074-7613(03)00322-4
- Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature 1996; 380:627-30; PMID:8602263; http://dx.doi.org/10.1038/380627a0
- Cao Z, Sun X, Icli B, Wara AK, Feinberg MW. Role of Kruppel-like factors in leukocyte development, function, and disease. Blood 2010; 116:4404-14; PMID:20616217; http://dx.doi.org/10.1182/blood-2010-05-285353
- Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem 2005; 280:38247-58; PMID:16169848; http://dx.doi.org/10.1074/jbc.M509378200
- Mahabeleshwar GH, Kawanami D, Sharma N, Takami Y, Zhou G, Shi H, Nayak L, Jeyaraj D, Grealy R, White M, et al. The myeloid transcription factor KLF2 regulates the host response to polymicrobial infection and endotoxic shock. Immunity 2011; 34:715-28; PMID:21565532; http://dx.doi.org/10.1016/j.immuni.2011.04.014
- Qi W, Holian J, Tan CY, Kelly DJ, Chen XM, Pollock CA. The roles of Kruppel-like factor 6 and peroxisome proliferator-activated receptor-gamma in the regulation of macrophage inflammatory protein-3alpha at early onset of diabetes. Int J Biochem Cell Biol 2011; 43:383-92; PMID:21109018; http://dx.doi.org/10.1016/j.biocel.2010.11.008
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663-76; PMID:16904174; http://dx.doi.org/10.1016/j.cell.2006.07.024
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131:861-72; PMID:18035408; http://dx.doi.org/10.1016/j.cell.2007.11.019
- Alder JK, Georgantas RW, 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, Civin CI. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol 2008; 180:5645-52; PMID:18390749; http://dx.doi.org/10.4049/jimmunol.180.8.5645
- Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J 2007; 26:4138-48; PMID:17762869; http://dx.doi.org/10.1038/sj.emboj.7601824
- Kaushik DK, Gupta M, Das S, Basu A. Kruppel-like factor 4, a novel transcription factor regulates microglial activation and subsequent neuroinflammation. J Neuroinflammation 2010; 7:68; PMID:20946687; http://dx.doi.org/10.1186/1742-2094-7-68
- Liu J, Liu Y, Zhang H, Chen G, Wang K, Xiao X. KLF4 promotes the expression, translocation, and releas eof HMGB1 in RAW264.7 macrophages in response to LPS. Shock 2008; 30:260-6; PMID:18197146.
- Liu Y, Wang J, Yi Y, Zhang H, Liu J, Liu M, Yuan C, Tang D, Benjamin IJ, Xiao X. Induction of KLF4 in response to heat stress. Cell Stress Chaperones 2006; 11:379-89; PMID:17278886; http://dx.doi.org/10.1379/CSC-210.1
- Zhou Q, Amar S. Identification of signaling pathways in macrophage exposed to Porphyromonas gingivalis or to its purified cell wall components. J Immunol 2007; 179:7777-90; PMID:18025224; http://dx.doi.org/10.4049/jimmunol.179.11.7777
- Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, et al. Kruppel-like factor 4 regulates macrophage polarization. J Clin Invest 2011; 121:2736-49; PMID:21670502; http://dx.doi.org/10.1172/JCI45444
- Liu J, Yang T, Liu Y, Zhang H, Wang K, Liu M, Chen G, Xiao X. Kruppel-like factor 4 inhibits the expression of interleukin-1 beta in lipopolysaccharide-induced RAW264.7 macrophages. FEBS Lett 2012; 586:834-40; PMID:22449968; http://dx.doi.org/10.1016/j.febslet.2012.02.003
- Liu J, Zhang H, Liu Y, Wang K, Feng Y, Liu M, Xiao X. KLF4 regulates the expression of interleukin-10 in RAW264.7 macrophages. Biochem Biophys Res Commun 2007; 362:575-81; PMID:17719562; http://dx.doi.org/10.1016/j.bbrc.2007.07.157
- Kapoor N, Niu J, Saad Y, Kumar S, Sirakova T, Becerra E, Li X, Kolattukudy PE. Transcription factors STAT6 and KLF4 implement macrophage polarization via the dual catalytic powers of MCPIP. J Immunol 2015; 194:6011-23; PMID:25934862; http://dx.doi.org/10.4049/jimmunol.1402797
- Shi JH, Zheng B, Chen S, Ma GY, Wen JK. Retinoic acid receptor alpha mediates all-trans-retinoic acid-induced Klf4 gene expression by regulating Klf4 promoter activity in vascular smooth muscle cells. J Biol Chem 2012; 287:10799-811; PMID:22337869; http://dx.doi.org/10.1074/jbc.M111.321836
- Karpurapu M, Ranjan R, Deng J, Chung S, Lee YG, Xiao L, Nirujogi TS, Jacobson JR, Park GY, Christman JW. Kruppel like factor 4 promoter undergoes active demethylation during monocyte/macrophage differentiation. PLoS One 2014; 9:e93362; PMID:24695324; http://dx.doi.org/10.1371/journal.pone.0093362
- Gamper AM, Qiao X, Kim J, Zhang L, DeSimone MC, Rathmell WK, Wan Y. Regulation of KLF4 turnover reveals an unexpected tissue-specific role of pVHL in tumorigenesis. Mol Cell 2012; 45:233-43; PMID:22284679; http://dx.doi.org/10.1016/j.molcel.2011.11.031
- Kim MO, Kim SH, Cho YY, Nadas J, Jeong CH, Yao K, Kim DJ, Yu DH, Keum YS, Lee KY, et al. ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4. Nat Struct Mol Biol 2012; 19:283-90; PMID:22307056; http://dx.doi.org/10.1038/nsmb.2217
- Meng F, Han M, Zheng B, Wang C, Zhang R, Zhang XH, Wen JK. All-trans retinoic acid increases KLF4 acetylation by inducing HDAC2 phosphorylation and its dissociation from KLF4 in vascular smooth muscle cells. Biochem Biophys Res Commun 2009; 387:13-8; PMID:19486889; http://dx.doi.org/10.1016/j.bbrc.2009.05.112
- Hay RT. SUMO: a history of modification. Mol Cell 2005; 18:1-12; PMID:15808504; http://dx.doi.org/10.1016/j.molcel.2005.03.012
- Yeh ET. SUMOylation and De-SUMOylation: wrestling with life's processes. J Biol Chem 2009; 284:8223-7; PMID:19008217; http://dx.doi.org/10.1074/jbc.R800050200
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 2010; 11:861-71; PMID:21102611; http://dx.doi.org/10.1038/nrm3011
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 2007; 8:947-56; PMID:18000527; http://dx.doi.org/10.1038/nrm2293
- Xiao N, Li H, Mei W, Cheng J. SUMOylation attenuates human beta-arrestin 2 inhibition of IL-1R/TRAF6 signaling. J Biol Chem 2015; 290:1927-35; PMID:25425640; http://dx.doi.org/10.1074/jbc.M114.608703
- Du JX, McConnell BB, Yang VW. A small ubiquitin-related modifier-interacting motif functions as the transcriptional activation domain of Kruppel-like factor 4. J Biol Chem 2010; 285:28298-308; PMID:20584900; http://dx.doi.org/10.1074/jbc.M110.101717
- Zheng B, Bernier M, Zhang XH, Suzuki T, Nie CQ, Li YH, Zhang Y, Song LL, Shi HJ, Liu Y, Zheng CY, Wen JK. miR-200c-SUMOylated KLF4 feedback loop acts as a switch in transcriptional programs that control VSMC proliferation. J Mol Cell Cardiol 2015; 82:201-12; PMID:25791170; http://dx.doi.org/10.1016/j.yjmcc.2015.03.011
- Nie CJ, Li YH, Zhang XH, Wang ZP, Jiang W, Zhang Y, Yin WN, Zhang Y, Shi HJ, Liu Y, et al. SUMOylation of KLF4 acts as a switch in transcriptional programs that control VSMC proliferation. Exp Cell Res 2016; 342:20-31; PMID:26945917; http://dx.doi.org/10.1016/j.yexcr.2016.03.001
- Cai R, Yu T, Huang C, Xia X, Liu X, Gu J, Xue S, Yeh ET, Cheng J. SUMO-specific protease 1 regulates mitochondrial biogenesis through PGC-1alpha. J Biol Chem 2012; 287:44464-70; PMID:23152500; http://dx.doi.org/10.1074/jbc.M112.422626
- Pauleau AL, Rutschman R, Lang R, Pernis A, Watowich SS, Murray PJ. Enhancer-mediated control of macrophage-specific arginase I expression. J Immunol 2004; 172:7565-73; PMID:15187136; http://dx.doi.org/10.4049/jimmunol.172.12.7565
- Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH, Scotton C, Locati M, Mantovani A, Sozzani S. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J Immunol 2005; 174:6561; author reply -2; PMID:15905489; http://dx.doi.org/10.4049/jimmunol.174.11.6561
- Rutherford MS, Schook LB. Differential immunocompetence of macrophages derived using macrophage or granulocyte-macrophage colony-stimulating factor. J Leukoc Biol 1992; 51:69-76; PMID: 1740646
- Biswas SK, Chittezhath M, Shalova IN, Lim JY. Macrophage polarization and plasticity in health and disease. Immunol Res 2012; 53:11-24; PMID:22418728; http://dx.doi.org/10.1007/s12026-012-8291-9
