ABSTRACT
miRNAs are increasingly being implicated as key regulators of cell proliferation, apoptosis, and differentiation. miRNA-34c appears to play a crucial role in cancer pathogenesis wherein it exerts its effect as a tumor suppressor. However, the role of miR-34c in myoblast proliferation remains poorly understood. Here, we found that overexpression miR-34c inhibited myoblasts proliferation by reducing the protein and mRNA expression of cell cycle genes. In contrast, blocking the function of miR-34c promoted myoblasts proliferation and increased the protein and mRNA expression of cell cycle genes. Moreover, miR-34c directly targeted YY1 and inhibited its expression. Similar to overexpression miR-34c, knockdown of YY1 by siRNA suppressed myoblasts proliferation. Our study provides novel evidence for a role of miR-34c in inhibiting myoblasts proliferation by repressing YY1. Thus, miR-34c has the potential to be used to enhance skeletal muscle development and regeneration.
Introduction
Myoblasts are derived from multipotent precursor cells and play a critical role in myogenesis.Citation1,2 It is a highly orchestrated process during which myoblasts proliferate, subsequently withdraw from the cell cycle, enter terminal differentiation and fuse into large multinucleated myotubes.Citation3-5 Increasing evidence suggest that miRNAs have been implicated in myoblasts proliferation and differentiation.Citation6 miR-1, miR-206, miR-29, miR-26a, and miR-148a are upregulated during myoblast differentiation, where they inhibit myoblast proliferation and promote this process.Citation7-13 However, many miRNAs that regulate myoblast proliferation and differentiation are yet undiscovered.
miR-34c is known as a tumor suppressor.Citation14 It can inhibit the development of various cancers including osteosarcoma, lung cancer, uveal melanoma, prostate cancer, laryngeal carcinoma, breast cancer, and gastric cancer.Citation15-21 Previous studies show that sperm-borne miR-34c is important for the first cleavage division by Bcl-2.Citation22 It also modulates male germ cell development.Citation23-25 Furthermore, miR-34c inhibits osteoblast proliferation and differentiation and regulates bone development through the Notch pathway by targeting SATB2.Citation26,27 However, the functions of miR-34c in myoblasts proliferation and differentiation are still unknown.
YY1 is a ubiquitous transcription factor and plays an important role in many biological processes. YY1 is an essential factor for embryo development, and homozygosity of a mutated YY1 allele results in embryonic lethality in mice.Citation28 During mouse spermatogenesis, loss of YY1 impacts the heterochromatic state and meiotic double-strand breaks.Citation29 Skeletal-muscle-specific YY1 knockout resulted in significant aberrations in mitochondrial morphology with severely defective oxidative function that were associated with exercise intolerance.Citation30 During skeletal muscle development, YY1 is stimulated by NF-κB and acts as a repressor in myogenesis.Citation31 It binds to the enhancer or promoter of myofibrillar genes and recruits PcG member Ezh2 as well as the histone deacetylase protein HDAC1 to inhibit the expression of these genes.Citation31-33 Recent studies show that, miR-29 and miR-1 promote myogenic differentiation by targeting YY1, meanwhile YY1 can downregulate the expression of these miRNAs.Citation11,34
Given the role of other miRNAs in myogenesis and the lack of information regarding the role of miR-34c in this process, we hypothesized that this miRNA in myoblast proliferation and differentiation. In this study, we found that miR-34c is upregulated during myogenesis. We demonstrated that miR-34c could inhibit myoblast proliferation through targeting YY1. Our study provides novel evidence for the role of miR-34c in muscle development.
Results
miR-34c exhibited low expression in proliferating myoblasts and were upregulated during skeletal muscle regeneration
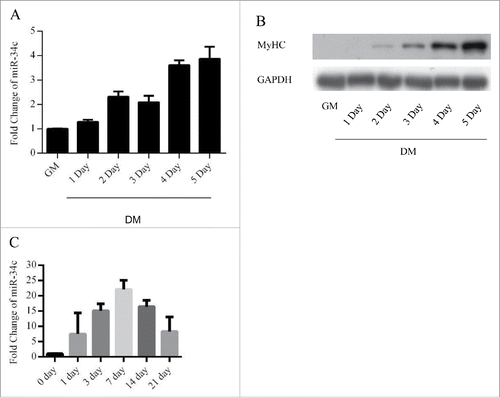
To investigate the roles of miR-34c in myoblasts, we assessed the expression of miR-34c during C2C12 myogenesis by using qPCR. The results indicated that miR-34c was maintained at a low level in proliferating C2C12 myoblasts and was upregulated during differentiation (). We next analyzed the expression of miR-34c during skeletal muscle regeneration. We injected cardiotoxin (CTX) into the mouse tibialis anterior muscle to induced skeletal muscle damage and regeneration, the expression of miR-34c was sharply increased following 1–21 d post injection of CTX ().
Figure 1. miR-34c exhibited low expression in proliferating myoblasts and were upregulated during skeletal muscle regeneration. (A) miR-34 expression was determined by qPCR from C2C12 myoblasts cultured in growth medium and differentiation medium for 1–5 d. (B) Immunoblotting was performed to detect MyHC protein expression as an indication of the differentiation status at the indicated times. (C) miR-34c was upregulated on days 1–21 post-CTX injury based on q-PCR analysis. miR-34c expression in skeletal muscle before CTX injection was set to 1.0. The results are expressed as the mean ± SD of 3 replicates.
miR-34c inhibited C2C12 myoblasts proliferation
To observe the function of miR-34c in myoblast proliferation, we transfected myoblasts cultured in growth medium (GM) with miR-34c mimics or negative control (NC) mimics (). Flow cytometry results revealed that the proportion of cells arrested in G0/G1 phase was significantly higher in cells transfected with miR-34c mimics than in those transfected with NC mimics (). Furthermore, EdU staining also demonstrated that the EdU+ cells of miR-34c transfected cells were significantly reduced compared with that of the control cells (). In addition, with overexpression of miR-34c, the mRNA and protein expression levels of cell cycle genes, specifically CCND1, CDK6, CCNE1, and CDK2, were reduced ( and ).
Figure 2. miR-34c inhibited C2C12 myoblasts proliferation. (A) After transfection 24 h, miR-34c expression was determined by qPCR in C2C12 myoblasts transfected with miR-34c mimics or negative control (NC) mimics. (B) C2C12 myoblasts were collected for cell cycle analysis. Flow cytometry was used to identify the percentage of cells in G0/G1, S, and G2 phases. (C) C2C12 cells were stained with EdU. The scale bar represents 200 μm. The percentage of EdU+ C2C12 cells was quantified (right). (D) The mRNA expression of cell cycle genes was detected by qPCR. (E) The protein expression of cell cycle genes was detected by western blotting. (F) After transfection 36 h, miR-34c expression was determined by qPCR in C2C12 myoblasts transfected with miR-34c inhibitor or NC inhibitor. (G) C2C12 myoblasts were collected for cell cycle analysis. Flow cytometry was used to determine the percentage of cells in G0/G1, S, and G2 phases. (H) C2C12 cells were stained with EdU. The scale bar represents 200 μm. The percentage of EdU+ C2C12 cells was quantified (right). (I) The mRNA expression of cell cycle genes was detected by qPCR. (J) The protein expression of cell cycle genes was detected by western blotting. All of the results are expressed as the mean ± SD *P < 0.05; **P < 0.01.
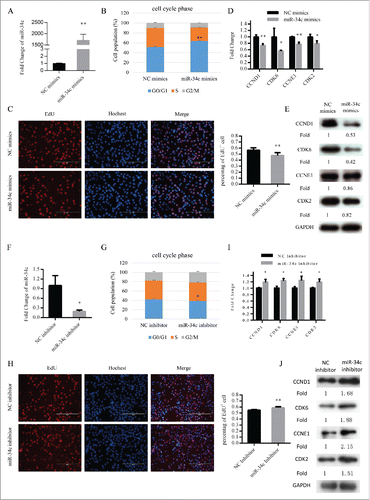
Moreover, we decreased the expression of miR-34c in C2C12 cells by transfecting them with miR-34c inhibitor (). Subsequently, the proportion of cells in the G0/G1 phase was reduced (), EdU+ cells were increased (), and the mRNA and protein levels of cell cycle genes were upregulated ( and ). Together, the data demonstrated that miR-34c can repress C2C12 myoblast proliferation and induce G0/G1 arrest.
miR-34c inhibited primary myoblasts proliferation
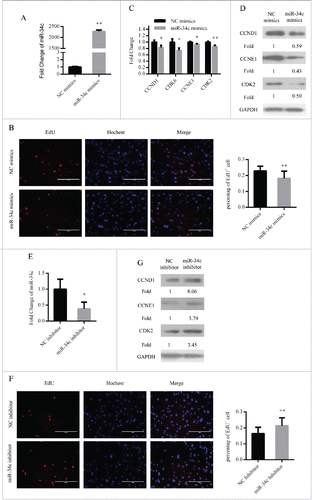
In addition, we assessed the function of miR-34c in primary myoblasts. We overexpressed miR-34c in primary myoblasts by transfecting miR-34c mimics (). As similar with C2C12 myoblasts, the number of EdU+ cells was decreased () and the mRNA and protein expression levels of cell cycle genes were reduced ( and ). Conversely, we downregulated the expression of miR-34c in primary myoblasts by transfecting miR-34c inhibitor (). With this treatment, the proportion of EdU+ cells was increased () and the protein expression levels of cell cycle genes were enhanced (). Therefore, these results demonstrated that miR-34c can repress primary myoblast proliferation.
Figure 3. miR-34c inhibited primary myoblasts proliferation. (A) After transfection 24 h, miR-34c expression was determined by qPCR in primary myoblasts transfected with miR-34c mimics or negative control (NC) mimics. (B) Primary myoblasts cells were stained with EdU. The scale bar represents 200 μm. The percentage of EdU+ primary myoblasts cells was quantified (right). (C) The mRNA expression of cell cycle genes was detected by qPCR. (D) The protein expression of cell cycle genes was detected by western blotting. (E) After transfection 36 h, miR-34c expression was determined by qPCR in primary myoblasts transfected with miR-34c inhibitor or NC inhibitor. (F) Primary myoblasts cells were stained with EdU. The scale bar represents 200 μm. The percentage of EdU+ C2C12 cells was quantified (right). (G) The protein expression of cell cycle genes was detected by western blotting. All of the results are expressed as the mean ± SD *P < 0.05; **P < 0.01.
miR-34c promoted C2C12 myoblasts differentiation
Cell cycle arrest in G0/G1 phase is a critical step during myoblasts differentiation, therefore, we also tested the effect of miR-34c on myoblasts differentiation. C2C12 myoblasts were transfected with the miR-34c mimics or NC mimics. Twenty-four hours after transfection, the GM was replaced with differentiation medium (DM) to induce myoblast differentiation (). The mRNA and protein expression of MyoG were significantly upregulated at DM 1 day ( and ). Moreover, overexpression of miR-34c increased the number of myotubes () and the protein expression levels of MyHC at DM 3 day ().
Figure 4. miR-34c promoted C2C12 myoblasts differentiation. (A–E) Cells were transfected with miR-34c mimics in growth medium and samples were collected at DM1 and DM3. (A) The mRNA expression of miR-34c was detected by qPCR. (B) The mRNA expression of MyoG was detected by qPCR at DM1. (C) The protein expression of MyoG was detected by western blotting at DM1. (D) Cells were fixed and immunostained for MyHC at DM3, and nuclei were stained blue with DAPI (left); the scale bar represents 400 μm, and the analysis of the number of fused myotubes per field is shown (right). (E) The protein expression of MyHC was detected by western blotting at DM3. (F–J) Cells were transfected with miR-34c mimics in GM and samples were collected at DM1 and DM3. (F) The mRNA expression of miR-34c was detected by qPCR. (G) The mRNA expression of MyoG was detected by qPCR at DM1. (H) The protein expression of MyoG was detected by western blotting at DM1. (I) Cells were fixed and immunostained for MyHC at DM3, and nuclei were stained blue with DAPI (left); the scale bar represents 400 μm. The analysis of the number of fused myotubes per field is shown (right). (J) The protein expression of MyHC was detected by western blotting at DM3. All results are expressed as the mean ± SD *P < 0.05; **P < 0.01.
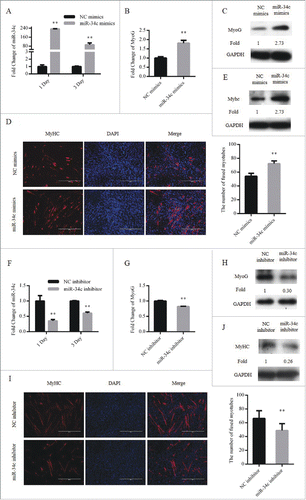
We also tested the influence of miR-34c loss-of-function during myoblast differentiation by transfecting cells with miR-34c inhibitor or NC inhibitor (). Through decreasing the expression of miR-34c, the mRNA and protein expression of MyoG was downregulated at DM 1 day (and ), and the number of myotubes was reduced (). In addition, the protein expression levels of MyHC at DM 3 day was downregulated (). Taken together, we suggest that miR-34c can promote C2C12 myoblasts differentiation.
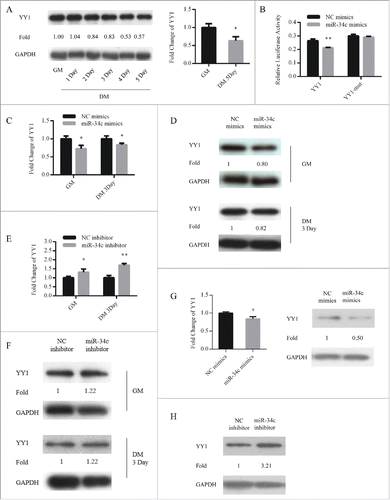
YY1 was a direct target of miR-34c
To account for these mechanisms through which miR-34c affected myoblasts proliferation and differentiation, YY1 was predicted to be a potential target by TargetScan (http://www.targetcan.org/mmu_71/). YY1 maintains transcriptional suppression of myofibrillar genes in undifferentiated myoblasts.Citation31 Contrary to miR-34c, YY1 was upregulated in proliferating myoblasts and downregulated during differentiation (). To determine whether miR-34c can directly target YY1, we constructed 2 dual-luciferase reporters that included the wild-type or mutant 3′ UTR of YY1. When the dual-luciferase reporters were co-transfected with miR-34c mimics or NC mimics into 293T cells, miR-34c significantly reduced the luciferase activity of the wild-type YY1 reporter compared with that in the negative control, and the mutant reporter no longer responded to miR-34c (). Next, we upregulated miR-34c in C2C12 cells cultured in GM or DM and assessed the effect on the mRNA and protein levels of YY1. qPCR and immunoblotting analysis revealed that mRNA and protein levels of YY1 were reduced by miR-34c ( and ). Conversely, inhibition of miR-34c upregulated the mRNA and protein levels of YY1 (). In addition, overexpression of miR-34c in primary myoblasts reduced the mRNA and protein level of YY1 (), inhibition of miR-34c in primary myoblasts increased the protein level of YY1 (). These results demonstrated that miR-34c directly targeted the YY1.
Figure 5. YY1 is a direct target of miR-34c. (A) The protein (left) and mRNA (right) expression of YY1 in proliferating and differentiated C2C12 myoblasts. (B) 293T cells were transfected with YY1 3′-UTR wild-type or mutant luciferase reporters and co-transfected with miR-34c mimics or negative control (NC) mimics. The relative luciferase activity was evaluated 24 h later. After C2C12 cells were transfected with miR-34c or NC mimics, (C) the levels of YY1 mRNA were assessed by qPCR, (D) and the levels of the YY1 protein were determined by western blotting. After C2C12 cells were transfected with miR-34C or NC inhibitor, (E) the levels of YY1 mRNA were assessed by qPCR, (F) and the levels of YY1 protein were visualized by western blotting. (G) After primary myoblasts cells were transfected with miR-34c or NC mimics, the levels of the YY1 mRNA were determined by qPCR (left) and the levels of the YY1 proteins were visualized by western blotting (right). (H) After primary myoblasts cells were transfected with miR-34c or NC inhibitor, the levels of the YY1 proteins were visualized by western blotting. All of the results are expressed as the mean ± SD *P < 0.05; **P < 0.01.
miR-34c regulated myoblasts proliferation by YY1
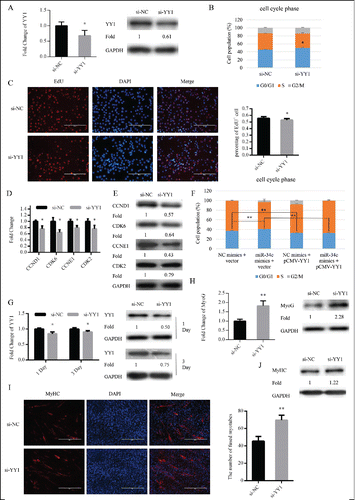
We transfected C2C12 myoblasts cultured in GM with si-YY1 or si-NC to knockdown the expression of YY1 (). Twenty-four hours after transfection, si-YY1 significantly enhanced cell cycle arrest in the G0/G1 phase () and reduced the proportion of EdU+ myoblasts (). Meanwhile, with knockdown of YY1, mRNA and protein expression levels of cell cycle genes were also reduced ( and ). Interestingly, the cell cycle arrest in the G0/G1 phase via overexpression miR-34c was rescued by YY1 overexpression in C2C12 cells (). After transfection, the GM was replaced with DM to induce myoblast differentiation (). Similar to the effect of miR-34c overexpression, the mRNA and protein expression of MyoG was significantly upregulated at DM 1 day (), the number of myotubes () and protein expression of MyHC was increased at DM 3 day (). Thus, miR-34c regulated myoblasts proliferation and the consequence differentiation process by targeting YY1.
Figure 6. miR-34c regulates myoblasts proliferation through YY1. (A–E) C2C12 cells were transfected with si-YY1 or si-NC (negative control) in growth medium (GM) and collected 24 h after transfection. (A) miR-34c mRNA expression was determined by qPCR (left) and protein expression was determined by western blotting (right). (B) Flow cytometry was used to determine the percentage of cells in G0/G1, S, and G2 phases. (C) C2C12 cells were stained with EdU. The scale bar represents 200 μm. The percentage of EdU+ C2C12 cells was quantified (right). (D) The mRNA expression of cell cycle genes was detected by qPCR. (E) The protein expression of cell cycle genes was determined by western blotting. (F) C2C12 cells were transfected with a YY1-expressing plasmid (pCMV-YY1) or a control vector and co-transfected with miR-34c mimics or NC mimics for 24 h. Flow cytometry was used to determine the percentage of cells in G0/G1, S, and G2 phases. (G–J) Cells were transfected with si-YY1 or si-NC in GM and collected at DM1 and DM3. (G) The mRNA expression of YY1 was detected by qPCR (left) and the protein expression of YY1 was detected by western blotting at the indicated times (right). (H) The mRNA expression of MyoG was detected by qPCR at DM1 (left). The protein expression of MyoG was assessed by western blotting at DM1 (right). (I) Cells were fixed and immunostained for MyHC at DM3, and nuclei were stained blue with DAPI (left), the scale bar represents 400 μm, and the number of fused myotubes per field was analyzed (right). (J) The protein expression of MyHC was determined by western blotting at DM3. All results are expressed as the mean ± SD *P < 0.05; **P < 0.01.
Discussion
miRNAs, especially the muscle-specific miRNAs called myomiRs, miR-206, miR-1, and miR-133a, are recently discovered as important regulators in myoblasts. These miRNAs are expressed at low levels in proliferating myoblasts and are upregulated during differentiation.Citation7 In this study, we identified that the expression pattern of miR-34c is similar to that of myomiRs in myoblasts, which indicates that it plays a role in myoblasts proliferation and differentiation.
Recent research indicates that miR-34c, acting as an important tumor suppressor, can inhibit proliferation of many kinds of cancer cell proliferation.Citation35-39 Previous studies have demonstrated that the miR-34 family can directly target cell cycle genes such as CCND1, CCNE2, CDK4, and CDK6.Citation40-44 These target genes are important for cell proliferation, overexpression of miR-34c induced cell cycle arrest in G0/G1 stage and reduced the number of EdU+ cells. This might at least partially provide a fundamental role for cell cycle arrest mediated by miR-34c in myoblasts.
As one of its indicated target genes, YY1 was reported to regulate gastric carcinogenesis associated with miR-34 family.Citation20 YY1, as a transcription factor, plays a critical role in cell proliferation in a dose-dependent manner. Knocking out YY1 induces cytokinesis failure and cell cycle arrest.Citation45 In differentiated cells, YY1 interaction with Rb favors progression into the S phase.Citation46 These results indicate that miR-34c can inhibit cell proliferation by targeting YY1 in both pathological and physiological conditions. Our present study identified YY1 as a target of miR-34c in myoblasts. A low level of miR-34c induced the production of sufficient quantities of YY1 to promote cell multiplication and proliferation; in addition, the upregulation of miR-34c downregulated YY1 to permit myoblast differentiation during the differentiation phase.
The proliferation and differentiation of myoblasts are important for skeletal muscle regeneration.Citation47,48 Cell cycle arrest is a critical step for myoblast differentiation.Citation3 Our results showed that miR-34c plays a positive regulator role in inducing myoblasts differentiation. Myoblasts proliferate and differentiate rapidly following 3–7 d post injection of cardiotoxin (CTX). During this period, the expression of miR-34c was sharply increased (). These results imply that miR-34c plays an important role in muscle regeneration, which is a key topic for further study.
In conclusion, our study provides direct evidence that miR-34c inhibits C2C12 myoblasts and primary myoblasts proliferation. Overexpression of miR-34c leading to G0/G1 arrest accelerated differentiation. We also found that miR-34c inhibit myoblast proliferation, in part by repressing YY1 expression. miR-34c might therefore be a potential candidate for improving skeletal muscle development and regeneration.
Materials and methods
Plasmids
The following method was used to create the pcDNA-3.1-YY1 expression vector. Through enzymatic digestion using EcoR V and Xho I restriction sites, the YY1 coding sequence was obtained from a YY1 expression plasmid provided by Prof. Wang Huating (The Chinese University of Hong Kong, Hong Kong) and cloned into the pcDNA-3.1 vector.
To construct the YY1 3′ UTR luciferase reporter plasmid, the YY1 3′ UTR fragment was amplified from mouse genomic DNA using the following primers:
forward primer 5′-GATCCTCGAGGGTTTTGTTTGCTATCTT-3′;
reverse primer 5′-GATCCTCGAGGGTTTTGTTTGCTATCTT-3′.
The PCR product was cloned into the psiCHECK-2 dual-luciferase reporter vector (Promega) using Xho I and Not I restriction sites. The mutant fragment of the YY1 3′ UTR was created by mutating the seed region of the predicted mmu-miR-34c site (ACTGCCA to GTCATTG) by overlap PCR.
Mouse muscle regeneration model
Muscle degeneration and regeneration in mice were induced by injecting CTX (Sigma, C9759). Eight-week-old C57BL/6 mice were injected in their tibialis anterior muscles with 100 μl of 10 μM CTX. Tibialis anterior muscles from each mouse were collected at 0, 1, 3, 7, 14, and 21 d post-CTX injection.
Cell culture
C2C12 myoblasts, 293T cells, and RD cells were cultured in growth medium consisting of Dulbecco modified Eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Invitrogen). To induce myogenic differentiation, culture medium was switched to DMEM supplemented with 2% horse serum (Gibco) and 1% penicillin/streptomycin.
Primary myoblast isolation and culture
Primary myoblasts were isolated from hind limb muscles of 4–8-week-old mice as described previously.Citation49 Briefly, the muscles were finely minced and digested in 0.2% collagenase I (Sigma, C6885) and 2.5 U/ml dispase II (Roche, 04942078001) for 1 h, and the cell slurry was passed through a 40 μm cell filter, and then pre-plated for 1 h. Unattached cells were centrifuged at 350 × g and cultured on collagen-coated dishes in F10 medium (Sigma) supplemented with 20% FBS and 2.5 ng/ml fibroblast growth factor (bFGF, Invitrogen, 13256–029). Primary myoblasts were further purified by pre-plating and then were cultured in F10/DMEM medium (1:1) supplemented with 20% FBS, 2.5 ng/ml bFGF, and 1% penicillin/streptomycin. For myogenic differentiation, culture medium was shifted to DMEM supplemented with 2% horse serum and 1% penicillin/streptomycin.
q-PCR
Total RNA was isolated from cells with TRIzol reagent (Invitrogen) and treated with DNase I (Qiagen) to remove residual DNA. miRNA was isolated from cells using the mirVana miRNA kit (Ambion) following the manufacturer's protocol. Reverse transcription was performed with M-MLV reverse transcriptase (Promega). As the reverse transcription primer, a specific stem-loop primer was used to initiate cDNA synthesis of miRNA.Citation50 The qPCR primers used are as follows:
RT-miR-34c: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCAATCAG-3′;
RT-U6: 5′-AACGCTTCACGAATTTGCGT-3′.
LightCycler 480 SYBR Green I Master (Roche) was used for qPCR with a LightCycler 480 II (Roche) system. U6 and GAPDH were used to normalize miRNA and other genes expression, respectively. The qPCR primers used are as follows:
U6-F, 5′-CTCGCTTCGGCAGCACA-3′;
U6-R, 5′-AACGCTTCACGAATTTGCGT-3′;
miR-34c-F, 5′-GCTGCTGTAGGCAGTGTAGTTAG-3′;
miR-34c-R, 5′-CTCAACTGGTGTCGTGGAGTC-3′;
GAPDH-F, 5′-GTGCCGCCTGGAGAAACCT-3′;
GAPDH-R, 5′-AAGTCGCAGGAGACAAC-3′;
MyoG-F, 5′-GAATGCAACTCCCACAGC-3′;
MyoG-R, 5′-TCCACGATGGACGTAAGG-3′;
YY1-F, 5′-GAGGGATACCTGGCATTG-3′;
YY1-R, 5′-TTCTTGGAGCATCATCTTCT-3′;
CDK2-F, 5′-GGCATTCCTCTTCCCCTCA-3′;
CDK2-R, 5′-CATTGATAAGCAGGTTCTGGG-3′;
CDK6-F, 5′-ATGCCGCTCTCCACCATC-3′;
CDK6-R, 5′-GTCCGTCCGTGACACTGTG-3′;
CCND1-F, 5′-GCCCTCCGTATCTTACTTCAAG-3′;
CCND1-R, 5′-ACCTCCTCTTCGCACTTCTG-3′;
CCNE1-F, 5′-CCCCACCCCTAACAAAGAAG-3′;
CCNE1-R, 5′-TCCTCCAAACCTCTTCTCTATTG-3′.
RNA oligonucleotides and transfection
The miR-34c mimics, NC mimics, miR-34c inhibitor, NC inhibitor, and siRNA were purchased from GenePharma (GenePharma). Cells transfection was performed with the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's recommendations.
Immunoblotting and Immunofluorescence
Immunoblotting was performed using standard procedures and antibodies to MyHC (Sigma, M4276, 1:3000), MyoG (Abcam, ab1835, 1:1000), CDK6 (Cell Signaling Technology, 3136S, 1:2000), CCND1 (Cell Signaling Technology, 2978S, 1:1000), CDK2 (Cell Signaling Technology, 2546s, 1:1000), CCNE1 (Cell Signaling Technology, 4129S, 1:1000), YY1 (Abcam, ab12132, 1:1000), and GAPDH (Cell Signaling Technology, 2118L, 1:10000). For immunostaining, cells treated in 6-well plates were fixed in 4% formaldehyde for 20 min and then washed 3 times for 5 min each with PBS. The cells were then permeabilized with 0.1% triton X-100 for 15 min. The cells were incubated in blocking solution (Beyotime) for 1 h at room temperature. After blocking, the cells were incubated with anti-MYHC (Sigma, M4276, 1:500) overnight at 4 ºC. The next day, secondary Alexa Fluor 594 goat anti-mouse IgG (H+L) antibody (Invitrogen) was incubated with the samples for 2 h at room temperature. The cell nuclei were visualized with DAPI staining (Roche). MyHC+ myoblasts with more than 2 nuclei were defined as fused myotubes.
EdU assays
The EdU assay was performed using the EdU assay kit (Ribobio, C10310–2) according to the manufacturer's instructions. Briefly, 24 or 36 h after transfection, cells were exposed to 50 mM EdU for 3 h. Next, cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% triton X-100. Subsequently, cells were incubated in Apollo reaction solution for 1 h and stained with Hoechst 33342 for 30 min. The cells were further analyzed by calculating the ratio of EdU+ cellsto the total number of cells.
Cell cycle flow cytometry
After transfection 24 or 36 h, cells were fixed in 70% (v/v) ethanol overnight at -20 ºC. Following incubation in 50 μg/ml propidium iodide (Sigma) containing 100 μg/ml RNase A (Qiagen) and 0.2% (v/v) TritonX-100 (Sigma) for 30 min at 4 ºC, the cells were analyzed in a FACSCalibur flow cytometer (BD Biosciences) and ModFit software.
Dual luciferase reporter assay
293T cells were co-transfected with 100 ng of wide-type or mutant 3′ UTR luciferase reporter and 40 nM of miR-34c mimics or NC mimics using Lipofectamine 2000 reagent (Invitrogen) in 24-well plates. After transfection for 24 h, the activities of firefly and Renilla luciferase were measured using a dual-luciferase reporter assay system (Promega, E2920) following the manufacturer's instructions. The Renilla luciferase signal was normalized to the firefly luciferase signal.
Statistical analysis
All results were represented as mean ± SD. A 2-tailed Student's t-test was used for P-value calculations and P < 0.05 was considered significant (*P < 0.05; **P < 0.01).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Prof. Huating Wang for providing the YY1 expression plasmid.
Funding
This work was supported by the National Basic Research Program of China under Grant 2015CB943103; and National Transgenic Breeding Project of China under Grant 2016ZX08010004–002.
References
- Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development 2012; 139:641-56; PMID:22274696; https://doi.org/10.1242/dev.068353
- Buckingham M. Skeletal muscle formation in vertebrates. Curr Opin Genet Dev 2001; 11:440-8; PMID:11448631; https://doi.org/10.1016/S0959-437X(00)00215-X
- Walsh K, Perlman H. Cell cycle exit upon myogenic differentiation. Curr Opin Genet Dev 1997; 7:597-602; ; https://doi.org/10.1016/S0959-437X(97)80005-6
- Kitzmann M, Fernandez A. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell Mol Life Sci 2001; 58:571-9; PMID:11361092; https://doi.org/10.1007/PL00000882
- Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet 2000; 57:16-25; PMID:10733231; https://doi.org/10.1034/j.1399-0004.2000.570103.x
- O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD. Essential role for Dicer during skeletal muscle development. Dev Biol 2007; 311:359-68; PMID:17936265; https://doi.org/10.1016/j.ydbio.2007.08.032
- Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 2006; 174:677-87; PMID:16923828; https://doi.org/10.1083/jcb.200603008
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006; 38:228-33; PMID:16380711; https://doi.org/10.1038/ng1725
- Wei W, He HB, Zhang WY, Zhang HX, Bai JB, Liu HZ, Cao JH, Chang KC, Li XY, Zhao SH. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis 2013; 4:e668; PMID:23764849; https://doi.org/10.1038/cddis.2013.184
- Winbanks CE, Wang B, Beyer C, Koh P, White L, Kantharidis P, Gregorevic P. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem 2011; 286:13805-14; PMID:21324893; https://doi.org/10.1074/jbc.M110.192625
- Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 2008; 14:369-81; PMID:18977326; https://doi.org/10.1016/j.ccr.2008.10.006
- Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev 2012; 26:2180-91; PMID:23028144; https://doi.org/10.1101/gad.198085.112
- Zhang J, Ying ZZ, Tang ZL, Long LQ, Li K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J Biol Chem 2012; 287:21093-101; PMID:22547064; https://doi.org/10.1074/jbc.M111.330381
- Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res 2007; 67:8433-8; PMID:17823410; https://doi.org/10.1158/0008-5472.CAN-07-1585
- van der Deen M, Taipaleenmaki H, Zhang Y, Teplyuk NM, Gupta A, Cinghu S, Shogren K, Maran A, Yaszemski MJ, Ling L, et al. MicroRNA-34c inversely couples the biological functions of the runt-related transcription factor RUNX2 and the tumor suppressor p53 in osteosarcoma. J Biol Chem 2013; 288:21307-19; PMID:23720736; https://doi.org/10.1074/jbc.M112.445890
- Garofalo M, Jeon YJ, Nuovo GJ, Middleton J, Secchiero P, Joshi P, Alder H, Nazaryan N, Di Leva G, Romano G, et al. MiR-34a/c-dependent PDGFR-alpha/beta downregulation inhibits tumorigenesis and enhances TRAIL-Induced apoptosis in lung cancer. PLoS One 2013; 8:e67581; PMID:23805317; https://doi.org/10.1371/journal.pone.0067581
- Dong F, Lou D. MicroRNA-34b/c suppresses uveal melanoma cell proliferation and migration through multiple targets. Mol Vis 2012; 18:537-46; PMID:22419847.
- Hagman Z, Larne O, Edsjo A, Bjartell A, Ehrnstrom RA, Ulmert D, Lilja H, Ceder Y. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer 2010; 127:2768-76; PMID:21351256; https://doi.org/10.1002/ijc.25269
- Cai KM, Bao XL, Kong XH, Jinag W, Mao MR, Chu JS, Huang YJ, Zhao XJ. Hsa-miR-34c suppresses growth and invasion of human laryngeal carcinoma cells via targeting c-Met. Int J Mol Med 2010; 25:565-71; PMID:20198305; https://doi.org/10.3892/ijmm_00000378
- Wang AM, Huang TT, Hsu KW, Huang KH, Fang WL, Yang MH, Lo SS, Chi CW, Lin JJ, Yeh TS. Yin Yang 1 is a target of microRNA-34 family and contributes to gastric carcinogenesis. Oncotarget 2014; 5:5002-16; PMID:24970812; https://doi.org/10.18632/oncotarget.2073
- Achari C, Winslow S, Ceder Y, Larsson C. Expression of miR-34c induces G2/M cell cycle arrest in breast cancer cells. BMC Cancer 2014; 14:538; PMID:25064703; https://doi.org/10.1186/1471-2407-14-538
- Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF, Yeung WS. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A 2012; 109:490-4; PMID:22203953; https://doi.org/10.1073/pnas.1110368109
- Zhang S, Yu M, Liu C, Wang L, Hu Y, Bai Y, Hua J. MIR-34c regulates mouse embryonic stem cells differentiation into male germ-like cells through RARg. Cell Biochem Funct 2012; 30:623-32; PMID:23097316; https://doi.org/10.1002/cbf.2922
- Liang X, Zhou D, Wei C, Luo H, Liu J, Fu R, Cui S. MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. PLoS One 2012; 7:e33861; PMID:22479460; https://doi.org/10.1371/journal.pone.0033861
- Bouhallier F, Allioli N, Lavial F, Chalmel F, Perrard MH, Durand P, Samarut J, Pain B, Rouault JP. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 2010; 16:720-31; PMID:20150330; https://doi.org/10.1261/rna.1963810
- Wei J, Shi Y, Zheng L, Zhou B, Inose H, Wang J, Guo XE, Grosschedl R, Karsenty G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J Cell Biol 2012; 197:509-21; PMID:22564414; https://doi.org/10.1083/jcb.201201057
- Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet 2012; 21:2991-3000; PMID:22498974; https://doi.org/10.1093/hmg/dds129
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol 1999; 19:7237-44; PMID:10490658; https://doi.org/10.1128/MCB.19.10.7237
- Wu S, Hu YC, Liu H, Shi Y. Loss of YY1 impacts the heterochromatic state and meiotic double-strand breaks during mouse spermatogenesis. Mol Cell Biol 2009; 29:6245-56; PMID:19786570; https://doi.org/10.1128/MCB.00679-09
- Blattler SM, Verdeguer F, Liesa M, Cunningham JT, Vogel RO, Chim H, Liu H, Romanino K, Shirihai OS, Vazquez F, et al. Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor Yin Yang 1 in skeletal muscle. Mol Cell Biol 2012; 32:3333-46; PMID:22711985; https://doi.org/10.1128/MCB.00337-12
- Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J, Carathers M, Davuluri R, Guttridge DC. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol 2007; 27:4374-87; PMID:17438126; https://doi.org/10.1128/MCB.02020-06
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 2004; 18:2627-38; PMID:15520282; https://doi.org/10.1101/gad.1241904
- Vincent CK, Gualberto A, Patel CV, Walsh K. Different regulatory sequences control creatine kinase-M gene expression in directly injected skeletal and cardiac muscle. Mol Cell Biol 1993; 13:1264-72; PMID:8423791; https://doi.org/10.1128/MCB.13.2.1264
- Lu L, Zhou L, Chen EZ, Sun K, Jiang P, Wang L, Su X, Sun H, Wang H. A Novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. PLoS One 2012; 7:e27596; PMID:22319554; https://doi.org/10.1371/journal.pone.0027596
- Yang S, Wu B, Sun H, Ji F, Sun T, Zhao Y, Zhou D. Interrupted E2F1-miR-34c-SCF negative feedback loop by hyper-methylation promotes colorectal cancer cell proliferation. Biosci Rep 2016; 36:e00293; ; https://doi.org/10.1042/BSR20150290
- Choe N, Kwon JS, Kim YS, Eom GH, Ahn YK, Baik YH, Park HY, Kook H. The microRNA miR-34c inhibits vascular smooth muscle cell proliferation and neointimal hyperplasia by targeting stem cell factor. Cell Signal 2015; 27:1056-65; PMID:25683915; https://doi.org/10.1016/j.cellsig.2014.12.022
- Song J, Wang Q, Luo Y, Yuan P, Tang C, Hui Y, Wang Z. miR-34c-3p inhibits cell proliferation, migration and invasion of hepatocellular carcinoma by targeting MARCKS. Int J Clin Exp Pathol 2015; 8:12728-37; PMID:26722462.
- Zhou YL, Xu YJ, Qiao CW. MiR-34c-3p suppresses the proliferation and invasion of non-small cell lung cancer (NSCLC) by inhibiting PAC1/MAPK pathway. Int J Clin Exp Pathol 2015; 8:6312-22; PMID:26261507.
- Liu F, Wang X, Li J, Gu K, Lv L, Zhang S, Che D, Cao J, Jin S, Yu Y. miR-34c-3p functions as a tumour suppressor by inhibiting eIF4E expression in non-small cell lung cancer. Cell Prolif 2015; 48:582-92; PMID:26250586; https://doi.org/10.1111/cpr.12201
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447:1130-4; PMID:17554337; https://doi.org/10.1038/nature05939
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol 2007; 17:1298-307; PMID:17656095; https://doi.org/10.1016/j.cub.2007.06.068
- Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 2008; 582:1564-8; PMID:18406353; https://doi.org/10.1016/j.febslet.2008.03.057
- Fujita Y, Kojima K, Hamada N, Ohhashi R, Akao Y, Nozawa Y, Deguchi T, Ito M. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun 2008; 377:114-9; PMID:18834855; https://doi.org/10.1016/j.bbrc.2008.09.086
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res 2008; 68:4123-32; PMID:18519671; https://doi.org/10.1158/0008-5472.CAN-08-0325
- Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol 2006; 26:3565-81; PMID:16611997; https://doi.org/10.1128/MCB.26.9.3565-3581.2006
- Petkova V, Romanowski MJ, Sulijoadikusumo I, Rohne D, Kang P, Shenk T, Usheva A. Interaction between YY1 and the retinoblastoma protein - Regulation of cell cycle progression in differentiated cells. J Biol Chem 2001; 276:7932-6; PMID:11118439; https://doi.org/10.1074/jbc.M007411200
- Wagers AJ, Conboy IM. Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell 2005; 122:659-67; PMID:16143100; https://doi.org/10.1016/j.cell.2005.08.021
- Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev 2004; 84:209-38; PMID:14715915; https://doi.org/10.1152/physrev.00019.2003
- Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol 1994; 125:1275-87; PMID:8207057; https://doi.org/10.1083/jcb.125.6.1275
- Tang F, Hajkova P, Barton SC, Lao K, Surani MA. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res 2006; 34:e9; PMID:16434699; https://doi.org/10.1093/nar/gnj009
