ABSTRACT
FEM1A, FEM1B, and FEM1C are evolutionarily-conserved VHL-box proteins, the substrate recognition subunits of CUL2-RING E3 ubiquitin ligase complexes. Here, we report that FEM1 proteins are ancient regulators of Stem-Loop Binding Protein (SLBP), a conserved protein that interacts with the stem loop structure located in the 3′ end of canonical histone mRNAs and functions in mRNA cleavage, translation and degradation. SLBP levels are highest during S-phase coinciding with histone synthesis. The ubiquitin ligase complex SCFcyclin F targets SLBP for degradation in G2 phase; however, the regulation of SLBP during other stages of the cell cycle is poorly understood. We provide evidence that FEM1A, FEM1B, and FEM1C interact with and mediate the degradation of SLBP. Cyclin F, FEM1A, FEM1B and FEM1C all interact with a region in SLBP's N-terminus using distinct degrons. An SLBP mutant that is unable to interact with all 4 ligases is expressed at higher levels than wild type SLBP and does not oscillate during the cell cycle. We demonstrate that orthologues of SLBP and FEM1 proteins interact in C. elegans and D. melanogaster, suggesting that the pathway is evolutionarily conserved. Furthermore, we show that FEM1 depletion in C. elegans results in the upregulation of SLBP ortholog CDL-1 in oocytes. Notably, cyclin F is absent in flies and worms, suggesting that FEM1 proteins play an important role in SLBP targeting in lower eukaryotes.
Introduction
Cullin-RING ligases (CRLs) are a superfamily of multisubunit ligases that mediate the degradation of a large number of substrates, thereby regulating numerous cellular processes.Citation1 CRLs share a core structure based upon an elongated Cullin protein that acts as a molecular scaffold. In mammals, there are 8 Cullin proteins (CUL1, CUL2, CUL3, CUL4A, CUL4B, CUL5, CUL7 and CUL9), each interacting with a distinct family of substrate-receptor proteins to recruit different targets for ubiquitination. In the case of CRL2 and CRL5 complexes, the N-terminus of CUL2 and CUL5 interact with the heterodimeric Elongin B/Elongin C complex, which functions as an adaptor that recruits a substrate receptor belonging either to the class of VHL-box proteins (of which there are ∼20 in humans) or the related SOCS-box proteins (of which there are ∼30).Citation1 VHL-box and SOCs-box proteins are classified based on their specificity for CUL2 or CUL5 complexes, respectively.Citation2 Substrate receptor proteins typically interact with their cognate substrates via short, specific motifs known as degrons (for degradation motif).Citation3
Mammalian FEM1A, FEM1B, FEM1C are highly conserved VHL-box proteins encoded by distinct genes that have orthologues in metazoans. In C. elegans, FEM1 mediates the degradation of transcriptional repressor TRA-1 to allow transcriptional de-repression of genes required for male sex differentiationCitation4 thus controlling male sex differentation.Citation5 In H. sapiens, FEM1B targets the TRA-1 homolog, Gli1, for degradation by the proteasome,Citation6 indicating that this interaction is conserved. In mouse, FEM1B has been shown to target Ankrd37 for proteasome-mediated degradation.Citation7 As yet, there are no reported substrates for FEM1A or FEM1C.
SLBP (stem-loop binding protein) is involved in the processing, translation and degradation of replication-dependent histone mRNAs (H1, H2A, H2B, H3, H4) (reviewed inCitation8) as well as H2AFX mRNA.Citation9,10 SLBP binds to the conserved 25–26 bp nucleotide sequence that includes the stem-loop structure found at the 3′ end of histone mRNAs. SLBP mediates the co-transcriptional processing of histone mRNAs together with cleavage factors (which include ZFP100, Symplekin, CPSF100, CPSF73, and the U7 small nuclear ribonucleoprotein)Citation11-13; histone mRNA translation in concert with translation factors (which include SLIP1, CBP80/20-dependent translation initiation factor (CTIF), and CBP80)Citation14-16; and histone mRNA decay with mRNA degradation proteins (UPF1, PNRC2 and SMG5).Citation15-18
SLBP protein levels correlate with histone mRNA levels and are highest during DNA replication (since histones are required to package newly replicated DNA).Citation19 Degradation of SLBP at the end of S-phase depends on phosphorylation of Thr61 by casein kinase II and Thr62 by CDK1-cyclin A.Citation19,20 SLBP degradation in G2 phase is mediated by SCFcyclinF. SLBP interacts with cyclin F using an atypical RxL motif (RxL97/99), and mutation of this motif (RxL97/99AA) prevents SLBP degradation in G2, which results in increased levels of histone variant H2AX.Citation10 SLBP is also regulated by proteasome-mediated degradation in G1 phase.Citation21 Notably, cyclin F is not expressed in G1, indicating that another E3 ligase must act in G1 phase. In addition to ubiquitin ligases targeting SLBP for degradation, CRL4WDR23 has been shown to monoubiquitinate SLBP, a process that is required for efficient histone mRNA 3′ end processing and for sufficient histone supply during DNA replication.Citation22,23
Here, we report that FEM1A, FEM1B, and FEM1C interact with and mediate the downregulation of SLBP in human cells. C. elegans FEM-1 mediates the downregulation of the SLBP ortholog CDL1 in oocytes, indicating that the pathway is conserved in lower eukaryotes. The characterization of this molecular interaction is described herein.
Results
SLBP interacts with FEM1A, FEM1B, and FEM1C in human cells
We previously demonstrated that SCFcyclin F mediates the degradation of SLBP during G2 phase of the cell cycle.Citation10 During this study, we noticed that SLBP underwent cyclin-F independent oscillations during the cell cycle, suggesting that alternative mechanism(s) regulate its levels. To determine the timing of oscillations more precisely, cells expressing a cyclin F-insensitive, G2-stable SLBP(RxL97/99AA) (known from here on as SLBP(Fdegron)) mutant were synchronized using double thymidine method. Whereas, as expected, cyclin-F insensitive SLBP was stabilized during G2 phase (at 9 and 12 hrs following double thymidine release), its levels began to disappear at 15 hours following release, as cells exited mitosis, as judged by decreasing levels of phosphohistone H3 levels (Ser10) (Supplementary Figure 1A). Thus, we hypothesized that SLBP(Fdegron) may be targeted for degradation by a distinct ubiquitin ligase(s) outside of G2 phase.
To identify the ubiquitin ligase(s) targeting SLBP for degradation, FLAG-STREP SLBP was transfected into 293T cells. Samples were subjected to tandem-affinity purification and analyzed by nanoflow LC-MS, as described.Citation10 In addition to cyclin F and known SLBP interactors (i.e. EIF4G1, UPF1, TUT7, CBP80 and SLIP1),Citation10 peptides belonging to FEM1A, FEM1C and CUL2 (but not FEM1B) were identified in the immunoprecipitates (data not shown). FEM1A, FEM1B, and FEM1C are VHL proteins that are highly related to each other, each containing multiple ankyrin repeats, as well as a C-terminal BC-box and CUL2 boxCitation24 (Fig. S1B,C).
To establish the significance of the interaction between FEM1 proteins and SLBP, we sought to identify the minimal binding regions in SLBP required for their interaction(s). SLBP's domain structure includes 2 nuclear localization sequences and a translational activation domain (TAD) in its N-terminus and an RNA-binding domain in the middle of the protein (). We transfected a series of SLBP deletion mutants into 293T cells and found that FEM1A, FEM1B, and FEM1C each interacted more strongly with the first 99 amino acids of SLBP (), suggesting that all 3 FEM1 proteins interact with these first 99 amino acids of SLBP and that a region in SLBP downstream of amino acid 99 inhibits SLBP's interaction with FEM1 proteins. To further define the binding region(s) on SLBP, we systematically mutated SLBP(1–99) and analyzed the binding between SLBP point mutants and individual FEM1 proteins. We found that FEM1A was unable to precipitate with SLBP(1–99) when amino acids MARY92–95 were mutated to AAAF (). Intriguingly, this mutant (which we will refer to as SLBP[1–99,Adegron]) retained its interaction with FEM1B, FEM1C, and CUL2, suggesting that the different FEM1 proteins utilize distinct binding motifs on SLBP. We found that mutating any group of 4 amino acids between aa 84 and 90 to alanine in SLBP[1–99,Adegron] abolished the binding with FEM1C () (creating SLBP[1–99,Adegron,Cdegron]. Finally, using alanine scanning we found that mutation of CSDW72–75AAAA or SV77/79AA of SLBP[1–99,Adegron,Cdegron] abrogated its interaction with FEM1B (), indicating that FEM1B needs amino acid 72–79 for its binding to SLBP. SLBP was no longer able to interact with CUL2 when its interactions with FEM1A, FEM1B, and FEM1C were abolished. Importantly, the region in SLBP that interacts with FEM1B (i.e., amino acids 72–79) overlaps with the translation activation motif in SLBP (which is located between amino acids 68 and 81) (). We concluded from these results that FEM1A, FEM1B, and FEM1C interact with SLBP, using distinct binding motifs within the N-terminus of SLBP (i.e., FEM1A binds to MARY92–95, FEM1C binds to MRTR84–87, and FEM1B binds to SV77/79) ().
Figure 1. Unique binding motifs in the N-terminus of SLBP interact with FEM1A, FEM1B, and FEM1C. (A) Diagram representing the domain structure of SLBP. The amino acid sequence and substrate receptor binding motifs representing the “degron hotspot” are shown. TAD, translational activation domain; NLS, nuclear localization sequence; RBD, RNA binding domain. (B) FEM1A, FEM1B, and FEM1C interact with amino acids 1–99 of SLBP. C-E Mapping the FEM1A, FEM1B and FEM1C binding regions in SLBP. HEK293T cells were transfected with either empty vector (EV) or FS-tagged SLBP constructs. MLN4924 was added to the cells for 4 hours before collection. Cell lysates were affinity precipitated with anti-STREP resin, and affinity precipitations were probed with the indicated antibodies. (F) The ligase-deficient SLBP(ABCdegron) mutant is unable to bind to CTIF. HEK293T cells were transfected with FLAG-tagged SLBP constructs. Cell lysates were supplemented with SUPERase-In™ RNase Inhibitor and immunoprecipitated with anti-FLAG resin. The immunoprecipitations were probed with the indicated antibodies.
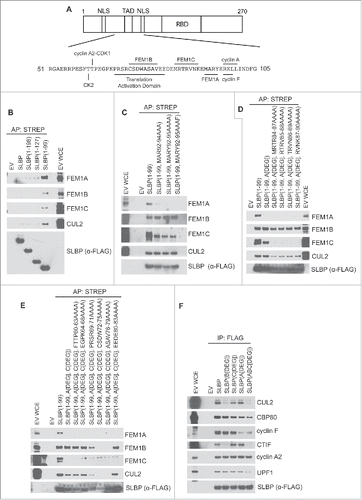
We generated these mutants in the context of the full-length SLBP, referring to the individual mutants as SLBP(Adegron), SLBP(Bdegron), SLBP(Cdegron), and the combination of mutants as FLAG-SLBP(ABCdegron). To examine whether these mutants retain their ability to interact with known SLBP interacting partners, we performed immunoprecipitation experiments (). We found that FLAG-SLBP(ABCdegron) (as well as individual FEM1-binding mutants of SLBP) retained their interactions with CBP80, UPF1, cyclin F (although at reduced levels likely due to the proximity of FEM1A's degron to cyclin F's degron), and cyclin A2. However, mutation of the FEM1B motif in SLBP (which overlaps with the translational activation domain in SLBP) abolished the interaction with CTIF, a cofactor required for histone mRNA translation. Therefore, we were unable to generate a functional (translation-competent) SLBP mutant unable to interact with FEM1 proteins.
FEM1A, FEM1B, and FEM1C independently mediate the downregulation of SLBP in human cells
To examine whether the FEM1 proteins mediate the downregulation of SLBP in human cells, we simultaneously silenced FEM1A, FEM1B, and FEM1C in HeLa cells by siRNA and synchronized cells using double thymidine method (). The extent of mRNA knockdown was quantified using quantitative PCR (). Cyclin F and phosphohistone H3 (Ser10) levels were used as synchronization markers for G2 and mitosis, respectively. The degradation of SLBP in G2 phase (which is mediated by cyclin F) was unaffected by FEM1A/FEM1B/FEM1C depletion (see the downregulation of SLBP at 12 hours after release). However, increased levels of SLBP following mitosis were observed between 16 and 20 hrs after release from double thymidine arrest, coinciding with loss of phosphohistone H3 (Ser10) levels (i.e., G1 phase).
Figure 2. SLBP levels increase after depletion of FEM1A, FEM1B, or FEM1C. (A) Depletion of FEM1A, FEM1B, and FEM1C results in increased levels of SLBP in G1 phase. HeLa cells were synchronized at G1/S by double-thymidine block before trypsinization and release into fresh media. Cells were transfected with FEM1A, FEM1B, and FEM1C siRNA 48 hours before the first timepoint. Right: OligodT primed cDNAs corresponding to the immunoblot samples were analyzed by qPCR for FEM1A, FEM1B, and FEM1C mRNA. The data are presented as mean ± SD of one representative experiment. (B-D) Depletion of individual FEM1 proteins results in increased levels of SLBP in G1 phase. HeLa cells were synchronized at G1/S by double-thymidine block before trypsinization and release into fresh media. Cells were transfected with respective siRNAs 48 hours before the first timepoint. Samples were collected at the listed time points, and immunoblotted as indicated. *Asterisks denote non-specific bands.
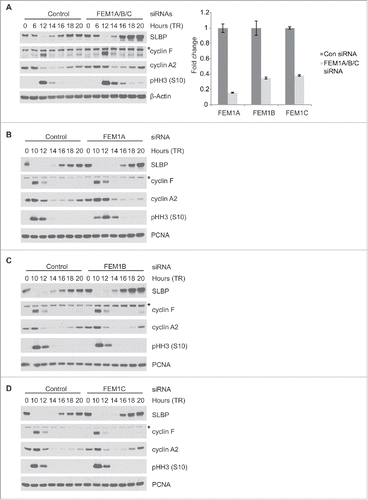
To determine the individual contribution of each FEM1 protein to downregulation of SLBP, we silenced each FEM1 protein independently. HeLa cells were transfected with siRNA oligos against FEM1A, FEM1B, or FEM1C and synchronized by double-thymidine block method (). Depletion of FEM1A, FEM1B, or FEM1C individually led to increased levels of SLBP at 18–20 hours after release from thymidine, although the effect was not as pronounced when all 3 FEM1 proteins were depleted. The depletion of FEM1A caused a slight delay in cell cycle progression, as observed by immunoblotting phosphohistone H3 (Ser10) (). Overall, these results indicate that individual FEM1 proteins are able to mediate the downregulation of SLBP.
The interaction between SLBP and FEM1A, FEM1B, and FEM1C is required for SLBP degradation
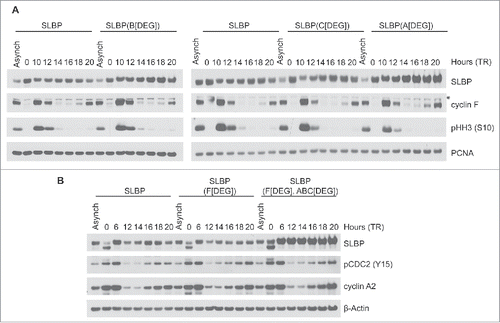
We next assessed whether the downregulation of SLBP depends on its interaction(s) with FEM1 proteins by analyzing the levels of SLBP mutants unable to bind to individual FEM1 proteins (see ). HeLa cells stably expressing FLAG-SLBP, FLAG-SLBP(Adegron), FLAG-SLBP(Bdegron), or FLAG-SLBP(Cdegron) were synchronized at G1/S with double-thymidine block (). We found that each of these mutants were expressed at higher levels than FLAG-SLBP at 14–20 hours after thymidine release, coinciding with G1 phase, indicating that each FEM1 protein contributes to the degradation of SLBP. To generate a mutant unable to bind FEM1A, FEM1B, FEM1C, or cyclin F, we mutated all 4 ligase degrons in concert [to create SLBP(ABCdegron/Fdegron)]. We stably expressed FLAG-SLBP, FLAG-SLBP(Fdegron), or FLAG-SLBP(ABCdegron/Fdegron) in HeLa cells and synchronized them with double-thymidine block. FLAG-SLBP(ABCdegron/Fdegron) was present at higher levels than FLAG-SLBP or FLAG-SLBP(Fdegron) and did not oscillate during the cell cycle (). The low levels of exogenous SLBP at the G1/S transition (i.e., time 0) likely reflects low rates of transcription from the retroviral promoter, as previously reported in.Citation10 Thus, the downregulation of SLBP depends on its interaction with FEM1A, FEM1B, and FEM1C, suggesting that FEM1 proteins target SLBP for proteasome-mediated degradation.
Figure 3. The interaction between SLBP and FEM1A, FEM1B, and FEM1C is required for SLBP degradation. (A) FLAG-SLBP(Bdegron), FLAG-SLBP(Cdegron), and FLAG-SLBP(Adegron) are each expressed at higher levels than FLAG-SLBP. HeLa cells infected with retroviruses expressing FLAG-tagged SLBP or FLAG-tagged SLBP mutants were synchronized at G1/(S)by double-thymidine block before trypsinization and release into fresh medium. Cells were collected at the indicated times, lysed, and immunoblotted. *Asterisk denote non-specific bands. (B) FLAG-SLBP(ABCdegron/Fdegron) is expressed at higher levels than FLAG-SLBP and FLAG-SLBP(Fdegron) and does not oscillate during the cell cycle. HeLa cells infected with retroviruses expressing FLAG-tagged SLBP, FLAG-tagged SLBP(Fdegron), or FLAG-SLBP(ABCdegron/Fdegron) were synchronized at the G1/(S)transition by double-thymidine block before trypsinization and release into fresh medium. Cells were collected at the indicated times, lysed, and immunoblotted.
The interaction between SLBP and FEM1 is conserved through evolution
Homologs of SLBP and FEM1 proteins are conserved in other metazoans. The D. melanogaster genome contains dFEM1A and dFEM1B genes, but no dFEM1C gene, whereas C. elegans only have a single FEM1 gene. We sought to understand whether the interaction between FEM1 and SLBP is conserved in these model systems. First, we explored the interaction between these proteins in D. melanogaster by co-expressing FLAG-dFEM1A or FLAG-dFEM1B with HA-dSLBP in HEK293T. HA-dSLBP was able to co-precipitate with both FLAG-dFEM1A and FLAG-dFEM1B (). Interestingly, in contrast to human cells, in which CUL2 interacted with FEM1A, FEM1B, and FEM1C, endogenous CUL2 only immunoprecipitated with dFEM1A, whereas CUL5 coprecipitated with both FLAG-dFEM1A and FLAG-dFEM1B. It therefore appears that dFEM1B forms a CUL5 complex, while FEM1A is able to form both CUL2 and CUL5 complexes.
Figure 4. The FEM1-SLBP degradation pathway is conserved in metazoans. (A) FLAG-dFEM1A and FLAG-dFEM1B interact with HA-dSLBP. HEK293T cells were co-transfected with empty vector (EV), FLAG-tagged dFEM1A, or FLAG-tagged dFEM1B, and HA-tagged dSLBP. MLN4924 was added to the cells for 4 hours before collection. Cell lysates were immunoprecipitated with anti-FLAG resin, and immunoprecipitations were probed with the indicated antibodies. (B) FLAG-FEM1 interacts with HA-CDL-1. HEK293T cells were co-transfected with empty vector (EV) or FLAG-tagged FEM1. Where indicated, MG-132 was added to the cells for 4 hours before collection. Cell lysates were immunoprecipitated with anti-FLAG resin, and immunoprecipitations were probed with the indicated antibodies. (C) RNAi depletion of FEM-1 in C. elegans results in increased CDL-1 levels in oocytes. Representative epifluorescence images of GFP::CDL-1 expressed in the oocytes of hermaphrodite adults treated with fem-1 RNAi or control RNAi. The scale bar represents 10 uM. D. Graph showing the increase in GFP::CDL-1 levels in C. elegans oocytes after fem-1 RNAi treatment. The levels were normalized to the control RNAi signal in the oocyte at -1 position, which was set to 100. The graph represents mean ± SEM.
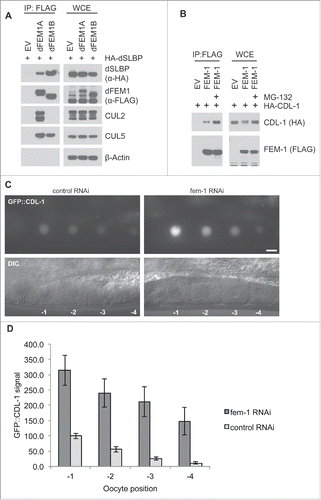
Next, we tested whether C. elegans FEM-1 interacts with the CDL-1, the nematode ortholog of SLBP. We co-expressed HA-tagged CDL-1 and FLAG-tagged FEM1 in 293T cells. Expression of FLAG-tagged FEM-1 resulted in the downregulation of HA-CDL-1, a state that was reversed by the addition of proteasome inhibitor MG-132. FLAG-tagged FEM-1 was able to coprecipitate CDL-1, demonstrating that the interaction is conserved in C. elegans (). Furthermore, MG-132 addition resulted in increased binding between HA-CDL-1 and FLAG-FEM-1, supporting our hypothesis that FEM-1 mediates the proteasome-mediated degradation of CDL-1.
To determine if C. elegans FEM-1 is able to negatively regulate CDL-1 levels, we used RNAi to deplete fem-1 and assessed the effect on a cdl-1 transgene expressed in the germline. An N-terminal GFP::CDL-1 translational fusion was expressed as a transgene under the control of the germline-specific pie-1 promoter. In the germline, GFP::CDL-1 accumulates in oocyte nuclei, with oocytes closest to the fertilization chamber containing the highest protein levels (). RNAi depletion of fem-1 significantly increased level of GFP::CDL-1 in oocytes, with a greater than 3-fold increase in levels (). This indicates that C. elegans FEM-1 can negatively regulate CDL-1 levels.
Discussion
It has been known for over a decade that SLBP levels are subject to intricate regulation by the ubiquitin proteasome system,Citation19,25 however the mechanisms involved have remained unknown. Recently, we demonstrated that SCFcyclin F recognizes an RXL motif in SLBP's N-terminus (RL97/99AA) and mediates the polyubiquitination and degradation of SLBP in the G2 phase of the cell division cycle. Expression of a G2-stable SLBP (unable to bind cyclin F) results in increased H2AX levels in G2, which ultimately renders cells susceptible to DNA damage-induced apoptosis.Citation10 In addition to polyubiquitination and degradation in G2 phase by SCFcyclin F, SLBP is monoubiquitinated on Lys156 (in the stem-loop binding region) by CRL4WDR23, which enables the efficient processing of canonical histone mRNA.Citation22,23 Notably, the monoubiquitination of WDR23 does not result in SLBP degradation. The present study demonstrates that the VHL-box proteins FEM1A, FEM1B, and FEM1C interact with and mediate the degradation of mammalian SLBP to keep SLBP levels in normal range.
Intriguingly, FEM1A, FEM1B, and FEM1C each utilize distinct (though proximal) degrons in SLBP's N-terminus. Each individual FEM1 protein is able to mediate the downregulation of SLBP. It is therefore possible that individual FEM1 ligases are responsible for the degradation of distinct pools of SLBP, perhaps in different subcellular locations. In fact, SLBP shuttles between the nucleus and cytoplasm using 2 bona-fide nuclear localization signals in the N-terminusCitation26 ().
We were unable to generate a functionally-active, non-degradable mutant of SLBP because mutation of the FEM1B binding site in SLBP also disrupts CTIF binding. CTIF and CBP80 are the principal proteins required for recruitment of the mRNA translational machinery.Citation15 Therefore, we were unable to assess the consequences of failure to degrade SLBP on histone metabolism in mammalian cells. Furthermore, we were unable to assess the impact of stabilization of SLBP on the development of C. elegans or D. melanogaster.
Our results shed light on how different organisms regulate SLBP levels. D. melanogaster SLBP is not degraded in G2 phase, however, it is downregulated in G1, as cells exit the cell cycle.Citation27 Furthermore, depletion of FEM-1 in C. elegans results in the increased levels of CDL-1, supporting our hypothesis that the FEM1 proteins also mediate SLBP degradation in other organisms. Cyclin F is not present in D. melanogaster or C. elegans,Citation10 suggesting that FEM1 ligases may be the sole E3 ligases targeting SLBP in lower eukaryotes.
Redundancy is present in human cells in that an individual E3 ligase may have multiple protein substrates and an individual substrate may be targeted by multiple E3 ligases. SCF ligases mediate the degradation of several substrates in G2 and M phases that are targeted by degradation by APC/CCdh1 in G1 phase. For example, the degradation of Cdc25A, Claspin, and USP37 is mediated by SCFβTrCP in S and G2 and by APC/CCdh1 in G1.Citation28-34 Similarly, RRM2 is degraded in G2 via SCFcyclin F and through APC/CCdh1 in G1.Citation35,36 SLBP is not stabilized when Cdh1 is depleted and does not bind either Cdh1 or its paralog Cdc20 (our unpublished results). In contrast, CRL2FEM1A/B/C complexes interact with and mediate the degradation of SLBP. Thus, CRL2FEM1A/B/C complexes join the APC/CCdh1 complex in cooperating with SCF complexes to tightly regulate the levels of specific substrates. Our results indicate that in human cells, CRL2FEM1A/B/C ligases cooperate with SCFcyclin F to regulate SLBP levels, thereby synchronizing histone metabolism with the cell cycle.
Methods
Cell culture procedures
HEK293T, U2OS, T98G, RPE-I, and HeLa cells were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and Pennicillin/Streptomycin/L-Glutamine. MLN4924 (Active Biochem) was used at 2 μM where indicated. T98G cells were synchronized in G0 by starving them for 72 hours in DMEM supplemented with 0.01% FBS. HeLa cells were synchronized at G1/S using the double-thymidine block method. HEK293T cells were transfected with DNA using polyethylenimine (Polysciences).
Antibodies
The following antibodies were used: SLBP (1:10,000, Bethyl A303–968A), cyclin F (1:1500, Santa Cruz Biotechnology sc-952), FLAG (1:7,000 (WB); Sigma-Aldrich F7425), pHH3 (S10) (1:1000, Cell Signaling 9701S), PCNA (1:5000, Zymed 13–3900), cyclin A2 (1:5000, produced in our laboratory), CTIF (1:500, Sigma-Aldrich HPA016865), CBP80 (1:1000, Bethyl A301–793A-T), UPF1 (1:10000, Bethyl A301–902A), β-Actin (1:7000, Sigma-Aldrich A5441), FEM1A (1:1500, Abcam ab92282), FEM1B (1:1500, Sigma-Aldrich HPA041920), FEM1C (1:1500, Genetex GTX121493), CUL5 (Bethyl A302–173A), HA.11 (16B12) (1:5000, Covance MMS-101P), CUL2 (1:10000, Bethyl A302–475A), pCDC2 (Y15) (1:1000, Cell Signaling 9111S), and p21(1:1000, BD Transduction Laboratories 610233).
Plasmids and siRNA
The following ON-TARGETplus siRNA oligos from Dharmacon were used: FEM1A custom siRNA oligo #1 AACGCCUCCAGAUCUUCCG, FEM1B siRNA oligo #6 GCCUAAUGAUUGCGGCAUA, FEM1C siRNA oligo #20 CAACACGACUUUAACCAAU. An ON-TARGETplus Non-targeting siRNA #1 (GE Healthcare cat. No. D-001810–01) served as a negative control. The siRNA duplexes were transfected into cells using RNAiMax (Invitrogen) according to the manufacturer's instructions.
SLBP, FEM1A, FEM1B, FEM1C, dSLBP, dFEM1A, dFEM1B, CDL-1, and C. elegans FEM-1 cDNAs were cloned into pcDNA3 mammalian expression vector or pBABE mammalian retroviral expression vector. Truncation mutants and phosphosite-mutants were generated by site-directed mutagenesis (QuikChange, Agilent Technologies).
Biochemical methods
Cell lysis was performed with lysis buffer (50 mM Tris, at pH 8.0, 150 mM NaCl, glycerol 10%, 1mM EDTA, 50 mM NaF, and NP-40 0.5%) supplemented with protease and phosphatase inhibitors. SUPERase-In™ RNase Inhibitor (Thermo Fisher Scientific) was used at 1U/μL where indicated. Lysates were immunoprecipitated with either Strep-Tactin (IBA) Superflow resin or anti-FLAG M2 affinity agarose gel (Sigma). Elution of the immunoprecipitate was performed with either D-Desthiobiotin (IBA) or FLAG peptide for the Strep-Tactin Superflow resin or anti-FLAG agarose, respectively. Immunoblotting has been described previously. The mass spectrometry identification of SLBP-interacting proteins has been described previously.Citation10
qRT-PCR
Total RNA was generated using RNeasy mini kits (Qiagen). cDNA was generated using Random Hexamers or OligodT EcoDry kits (Takara Clontech). qPCR was performed using Absolute SYBR green (Thermo Fisher Scientific) on a Roche Lightcycler 480. Analysis of the qPCR experiments was conducted via absolute relative quantification with in-experiment standard curves for each primer set to control for primer efficiency. The oligos used for qRT-PCR analysis were: FEM1A (F: 5′ -GCTACACATAGCAGCCCAGA-3′, R: 5′-TCTTCTTGAAGGCATTGGTG-3′), FEM1B (F: 5′-AAAGGTGGTACGCTTGCTCT-3′, R: 5′TCAATGACATACCCGTCGAA-3′) and FEM1C (F: 5′-GACAAAGCCCGAGTGACC-3′, R: 5′-CGTCCTACTGCTTTCCAACA-3′).
C. elegans analysis
The expression vector pPID3.01b/GFP::CDL-1 was created by cloning genomic cdl-1 coding sequence into the pPID3.01b germline expression vector37using the Gateway recombination system. RNAi was performed by inducing dsRNA production by growth of HT115 bacteria containing the RNAi constructs on 1 mM IPTG plus carbenicllin NGM agar plates.Citation38 Gravid animals were placed on the RNAi plates and their progeny were analyzed as adults. Animals were visualized on a Zeiss Axioskop microscope, and images were taken with a Hamamatsu ORCA-ER digital camera using Openlab 4.0.2 software (Agilent Technologies). Images were processed and analyzed with Adobe Photoshop software. Matched images (from 5–8 animals) were taken with the same exposure and were processed and analyzed identically with background signal subtracted.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1284715_Supplemental_Material.zip
Download Zip (727.3 KB)Funding
This work was funded by grants from the National Institute of Health (R01-GM057587 and R01-CA076584) to M.P., a grant from the National Institute of Health (R01GM074212) to E.T.K, a fellowship from the T32-CA009161 grant to J.F.D, and fellowships from the National Health and Medical Research Council of Australia and the Lymphoma Research Foundation to J.K.P. M.P. is an Investigator with the Howard Hughes Medical Institute.
References
- Lydeard JR, Schulman BA, Harper JW. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep 2013; 14:1050-61; PMID:24232186; http://dx.doi.org/10.1038/embor.2013.173
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev 2004; 18:3055-65; PMID:15601820; http://dx.doi.org/10.1101/gad.1252404
- Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 2013; 14:369-81; PMID:23657496; http://dx.doi.org/10.1038/nrm3582
- Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell 2007; 13:127-39; http://dx.doi.org/10.1016/j.devcel.2007.05.008
- Doniach T, Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol 1984; 106:223-35; PMID:6541600; http://dx.doi.org/10.1016/0012-1606(84)90077-0
- Gilder AS, Chen YB, Jackson RJ, 3rd, Jiang J, Maher V. Fem1b promotes ubiquitylation and suppresses transcriptional activity of Gli1. Biochem Biophys Res Commun 2013; 440:431-6; PMID:24076122; http://dx.doi.org/10.1016/j.bbrc.2013.09.090
- Shi YQ, Liao SY, Zhuang XJ, Han CS. Mouse Fem1b interacts with and induces ubiquitin-mediated degradation of Ankrd37. Gene 2011; 485:153-9; PMID:21723927; http://dx.doi.org/10.1016/j.gene.2011.06.025
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 2008; 9:843-54; PMID:18927579; http://dx.doi.org/10.1038/nrg2438
- Brooks L, 3rd, Lyons SM, Mahoney JM, Welch JD, Liu Z, Marzluff WF, Whitfield ML. A multiprotein occupancy map of the mRNP on the 3' end of histone mRNAs. RNA 2015; 21:1943-65; PMID:26377992; http://dx.doi.org/10.1261/rna.053389.115
- Dankert JF, Rona G, Clijsters L, Geter P, Skaar JR, Bermudez-Hernandez K, Sassani E, Fenyö D, Ueberheide B, Schneider R. Cyclin F-Mediated Degradation of SLBP Limits H2A.X Accumulation and Apoptosis upon Genotoxic Stress in G2. Mol Cell 2016; 64:507-19; PMID:27773672; http://dx.doi.org/10.1016/j.molcel.2016.09.010
- Dominski Z, Marzluff WF. Formation of the 3' end of histone mRNA. Gene 1999; 239:1-14; PMID:10571029; http://dx.doi.org/10.1016/S0378-1119(99)00367-4
- Wagner EJ, Marzluff WF. ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S phase. Mol Cell Biol 2006; 26:6702-12; PMID:16914750; http://dx.doi.org/10.1128/MCB.00391-06
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3'-end maturation of histone mRNAs. Genes Dev 2005; 19:2583-92; PMID:16230528; http://dx.doi.org/10.1101/gad.1371105
- Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol Cell Biol 2008; 28:1182-94; PMID:18025107; http://dx.doi.org/10.1128/MCB.01500-07
- Choe J, Ahn SH, Kim YK. The mRNP remodeling mediated by UPF1 promotes rapid degradation of replication-dependent histone mRNA. Nucleic Acids Res 2014; 42:9334-49; PMID:25016523; http://dx.doi.org/10.1093/nar/gku610
- Choe J, Kim KM, Park S, Lee YK, Song OK, Kim MK, Lee BG, Song HK, Kim YK. Rapid degradation of replication-dependent histone mRNAs largely occurs on mRNAs bound by nuclear cap-binding proteins 80 and 20. Nucleic Acids Res 2013; 41:1307-18; PMID:23234701; http://dx.doi.org/10.1093/nar/gks1196
- Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol 2005; 25:6879-88; PMID:16055702; http://dx.doi.org/10.1128/MCB.25.16.6879-6888.2005
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'. Genes Dev 2008; 22:50-65; PMID:18172165; http://dx.doi.org/10.1101/gad.1622708
- Zheng L, Dominski Z, Yang XC, Elms P, Raska CS, Borchers CH, Marzluff WF. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol Cell Biol 2003; 23:1590-601; PMID:12588979; http://dx.doi.org/10.1128/MCB.23.5.1590-1601.2003
- Koseoglu MM, Graves LM, Marzluff WF. Phosphorylation of threonine 61 by cyclin a/Cdk1 triggers degradation of stem-loop binding protein at the end of S phase. Mol Cell Biol 2008; 28:4469-79; PMID:18490441; http://dx.doi.org/10.1128/MCB.01416-07
- Djakbarova U, Marzluff WF, Koseoglu MM. Translation regulation and proteasome mediated degradation cooperate to keep stem-loop binding protein low in G1-phase. J Cell Biochem 2014; 115:523-30; PMID:24122909; http://dx.doi.org/10.1002/jcb.24686
- Brodersen MM, Lampert F, Barnes CA, Soste M, Piwko W, Peter M. CRL4(WDR23)-Mediated SLBP ubiquitylation ensures histone supply during DNA Replication. Mol Cell 2016; 62:627-35; PMID:27203182; http://dx.doi.org/10.1016/j.molcel.2016.04.017
- Djakbarova U, Marzluff WF, Koseoglu MM. DDB1 and CUL4 associated factor 11 (DCAF11) mediates degradation of Stem-loop binding protein at the end of S phase. Cell Cycle 2016; 15:1986-96; PMID:27254819; http://dx.doi.org/10.1080/15384101.2016.1191708
- Okumura F, Matsuzaki M, Nakatsukasa V, Kamura T. The role of Elongin BC-containing Ubiquitin Ligases. Front Oncol 2012; 2:10; PMID:22649776; http://dx.doi.org/10.3389/fonc.2012.00010
- Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 2008; 322:923-9; PMID:18988848; http://dx.doi.org/10.1126/science.1160462
- Erkmann JA, Wagner EJ, Dong J, Zhang Y, Kutay U, Marzluff WF. Nuclear import of the stem-loop binding protein and localization during the cell cycle. Mol Biol Cell 2005; 16:2960-71; PMID:15829567; http://dx.doi.org/10.1091/mbc.E04-11-1023
- Lanzotti DJ, Kupsco JM, Yang XC, Dominski Z, Marzluff WF, Duronio RJ. Drosophila stem-loop binding protein intracellular localization is mediated by phosphorylation and is required for cell cycle-regulated histone mRNA expression. Mol Biol Cell 2004; 15:1112-23; PMID:14999087; http://dx.doi.org/10.1091/mbc.E03-09-0649
- Donzelli M, Squatrito M, Ganoth D, Hershko A, Pagano M, Draetta GF. Dual mode of degradation of Cdc25 A phosphatase. EMBO J 2002; 21:4875-84; PMID:12234927; http://dx.doi.org/10.1093/emboj/cdf491
- Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. Degradation of Cdc25A by ßTrcp during S phase and in response to DNA damage. Nature 2003; 426:87-91; PMID:14603323; http://dx.doi.org/10.1038/nature02082
- Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell 2006; 23:319-29; PMID:16885022; http://dx.doi.org/10.1016/j.molcel.2006.06.013
- Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell 2008; 134:256-67; PMID:18662541; http://dx.doi.org/10.1016/j.cell.2008.05.043
- Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, Kirkpatrick DS, Jackson PK, Fang G, Dixit VM. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell 2011; 42:511-23; PMID:21596315; http://dx.doi.org/10.1016/j.molcel.2011.03.027
- Burrows AC, Prokop J, Summers MK. Summers, Skp1-Cul1-F-box ubiquitin ligase (SCF(betaTrCP))-mediated destruction of the ubiquitin-specific protease USP37 during G2-phase promotes mitotic entry. J Biol Chem 2012; 287:39021-9; PMID:23027877; http://dx.doi.org/10.1074/jbc.M112.390328
- Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature 2003; 426:87-91; PMID:14603323; http://dx.doi.org/10.1038/nature02082
- Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci U S A 2003; 100:3925-9; PMID:12655059; http://dx.doi.org/10.1073/pnas.0330774100
- D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 2012; 149:1023-34; PMID:22632967; http://dx.doi.org/10.1016/j.cell.2012.03.043
- Pellettieri J, Reinke V, Kim SK, Seydoux G. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev Cell 2003; 5:451-462; PMID:12967564; http://dx.doi.org/10.1016/S1534-5807(03)00231-4
- Sulston J, Hodgkin J, in The Nematode Caenorhabditis elegans, Wood WB, Ed. (Cold Spring Harbor Laboratory, 1988), pp. 587-606.
